Abstract
Ribonucleotide reductases (RNRs) catalyze the conversion of nucleoside 5′–diphosphates to the corresponding deoxynucleotides supplying the dNTPs required for DNA replication and DNA repair. Class I RNRs require two subunits, α and β, for activity. Humans possess two β subunits: one involved in S phase DNA replication (β) and a second in mitochondrial DNA replication (β′ or p53R2) and potentially DNA repair. Gemcitabine (F2C) is used clinically as an anticancer agent and its phosphorylated metabolites target many enzymes involved in nucleotide metabolism, including RNR. The present investigation with α (specific activity of 400 nmol/min/mg) and β′ (0.6 Y•/β′2 and a specific activity of 420 nmol/min/mg) establishes F2CDP is a substoichiometric inactivator of RNR. Incubation of this α/β′ with [1′-3H] F2CDP or [5-3H] F2CDP and re-isolation of the protein by Sephadex G50 chromatography resulted in recovery 0.5 eq. of covalently bound sugar and 0.03 eq. of tightly associated cytosine to α2. SDS PAGE analysis (loaded without boiling) of the inactivated RNR, showed that 60% of α migrates as a 90 kDa protein and 40% as a 120 kDa protein. Incubation of [1′-3H] F2CDP with active site mutants C444S/A, C218S/A, E431Q/D-α and the C-terminal tail C787S/A and C790S /A mutants, reveals that no sugar label is bound to the active site mutants of α and that in the case of C218S-α, α migrates as a 90 kDa protein. Analysis of the inactivated wt-α/β′ RNR by size exclusion chromatography indicates a quaternary structure of α6β′6. A mechanism of inactivation common with hα/β is presented.
Gemcitabine (2', 2'-dideoxy-difluorocytidine, F2C) is a drug that is used clinically in the treatment of non-small cell lung carcinomas and advanced pancreatic cancer (2–6). The mechanism of its cytotoxicity is multifactoral where its metabolites, the mono-, di- and triphosphates (F2CMP, F2CDP and F2CTP) inhibit a variety of steps in nucleic acid metabolism (4, 7). The essential step in apoptosis of the cells is F2CTP inhibition of DNA polymerase by its incorporation into the growing polymer chain, resulting in chain termination (3). Potentiation of the effects of F2CTP results from the inhibition of ribonucleotide reductase(s) (RNR), the enzymes that make deoxynucleoside 5′-diphosphates (dNDPs) from nucleoside diphosphates (8). RNRs are stoichiometrically inhibited by F2CDP (9–12). This inhibition leads to a reduction in dNDP pools and consequently a reduction in dNTP pools. Reduced concentrations of dNTPs reduce the competition for F2CTP to become incorporated into DNA by DNA polymerase.
Human RNRs belong to Class Ia RNRs are composed of α and β subunits existing in complex quaternary structures αn(β2)m (n = 2, 4, 6 and m = 1, 3) (13, 14). α houses the active site for nucleotide reduction and the binding sites for ATP/dNTP allosteric effectors that control the specificity and rate of nucleotide reduction. α contains five essential cysteines for catalysis: C429 the site of the thiyl radical that initiates catalysis, C218 and C444 that provide the reducing equivalents (eq.) during dNDP formation and C787 and C790 at the C-terminus that re-reduces the active site disulfide subsequent to dCDP production. β houses the essential diferric-tyrosyl radical (Y•) cofactor (15, 16). Recently, we have shown that hRNR, involved in DNA replication, is inactivated by 0.5 eq. of F2CDP per α (11). Our studies unexpectedly revealed that the inhibition resulted from tight association of the two subunits and formation of an RNR quaternary structure determined by size exclusion chromatography (SEC) to be α6β6. Recently a second RNR small subunit was discovered in humans (17, 18). It was designated p53R2 as its production is induced by p53 (17). Throughout the paper we will designate p53R2 as β'. In the present paper we report on the mechanism of inactivation of hRNR, αn(β'2)m, by F2CDP and compare the results with the replicative RNR, αn(β2)m.
The biology and biochemistry of β' are actively being investigated. The biological function(s) of β' is (are) complex and appears to be different in quiescent cells and cells which have been subjected to DNA damage. Recent studies of Pontarin et al (19) suggest that β' is localized to the cytosol and is not shuttled to the nucleus as previously suggested (17, 20, 21). They proposed that β' plays a primary role in supplying the dNTPs required for mitochondrial DNA replication. This role is supported by identification of children with mutations in β' that experience severe mitochondrial DNA depletion (22).
While p53 mediates transcriptional induction of β' and as a consequence led to the proposal of its involvement in DNA repair, the induction process is not sufficiently rapid to supply dNTPs given the rate of damage repair (19, 23). Recent observations, however, have shown that β' interacts with and is phosphorylated by ATM (ataxia telangiectasia mutated), a DNA damage-inducible kinase, and is also found in the MRE11 (double strand break repair proteins) complex, involved in DNA double strand break repair. β' has also been shown to interact with ERK1/2 (extracellular signal-regulated kinase) and to inhibit MEK (a kinase that phosphorlates ERK)-ERK signaling and prevent cancer invasion, demonstrating an important role in genomic stability (23, 24).
β' is composed of 351 amino acids and is 80% homologous to human β (hβ). Both proteins contain an essential diferric-Y• cofactor and are active in nucleotide reduction in the presence of α. hβ' has been reported to have activity 1.4 fold lower than β and a Y• content of 0.8/β'2, 67% of that of β2 (25, 26). However, the activity of hβ reported (25) is substantially lower (14 fold) than the activity recently reported from our lab (11) for reasons unknown. β' has also been proposed to have catalase activity in contrast to hβ, and to have a 2.5 fold less susceptibility to inactivation by hydroxyurea (27).
As noted above, F2CDP is a potent inhibitor of RNRs and its demonstrated efficacy against a range of solid tumors warrants a detailed investigation of the mechanism of inhibition. Studies have been carried out on the class Ia RNRs from E. coli and human αn(β2)m, and the class II RNR from Lactobacillus leichmannii. The L. leichmannii RNR is a monomeric enzyme that uses adenosylcobalamin as a cofactor. These studies have provided us with the following generalizations about the mechanism of inactivation. Using [5-3H] and [1'-3H] F2CDP (F2CTP) the stoichiometry of inhibition is 1 F2CDP/α2 for the class Ia RNRs (11) and 1 F2CTP/α for the class II RNR (28–30). Despite the stoichiometry of inactivation, as with all mechanism-based inhibitors of RNR, at least two pathways are responsible for inhibition (1, 31). Covalent modification by a sugar moiety of F2CDP is responsible for inactivation in one of these pathways. Destruction of the cofactor (Y• in the class Ia and adenosylcobalamin in the class II RNRs) (28) is responsible for the remaining inactivation. Subsequent to complete inactivation, stoichiometric amounts of cytosine are released, as are two fluoride ions. With the E. coli and L. leichmannii RNRs a new nucleotide radical (1) has been recently identified (10, 12, 28–30). This information has been used to formulate mechanisms of inactivation by the two pathways. Finally, with the oligomeric class Ia RNRs we have recently made the unexpected observation that the basis for their complete inactivation by 0.5 F2CDP/α is that the α and β subunits form a tight complex proposed to be α2β2 for E. coli RNR and α6β6 for hRNR based on SEC analysis (11).
The prevalence of gemcitabine as a clinically useful drug and literature evidence suggesting that controlling the levels of β' may increase potency of co-administered genotoxic therapeutics mandates a better understanding of the enzymology of β' to elucidate its function (32). The present communication reports that F2CDP is an inactivator of β', although the binding of F2CDP to the α/β' complex appears to be weaker than that observed with the corresponding α/β complex. Studies with [5-3H] and [1'-3H] F2CDP reveal that >90% inactivation occurs with 0.5 eq. inhibitor/α, that 0.5 sugars are attached covalently to α, and that one cytosine is released per inactivation. SDS PAGE analysis of the inactivated RNR reveals that α has been covalently modified. Forty percent of α migrates as a 120 kDa protein relative to unmodified α which migrates as a 90 kDa protein. Site directed mutagenesis studies of all of the residues involved in catalysis in α and studies of the effect of each mutation on the inactivation of RNR composed of hβ or β' suggest that C218, one of the cysteines required to deliver reducing eq. to make dNDP and a cysteine (C790 or C787) from the C-terminus of α, involved in rereduction of the disulfide in the active site subsequent to dNDP formation, are likely responsible for covalent modification of α and the unusual migratory properties of α by SDS PAGE analysis. SEC suggests that subsequent to inactivation, RNR migrates as a α6β'6 complex. β' thus can also be a target of gemcitabine and play a role in its cytotoxicity.
MATERIALS and METHODS
Competent E. coli BL21(DE3)–RIL cells were purchased from Stratagene. Complete EDTA-free protease inhibitor tablets and calf alkaline phosphatase (20 U/µL) were purchased from Roche Biochemicals. [3H]-CDP (17 Ci /µmol) is from ViTrax radiochemicals (CA). p53R2 (26) containing the gene for β′ was a gift from Lars Thelander (Department of Medical Biosciences, Medical Biochemistry, Umeå University, SE-901 87 Umeå, Sweden). phRRM1 containing the gene for α and phRRM2 containing gene for β′ was from Dr. Yun Yen (City of Hope National Medical Center, Duarte, CA) (25). Protein concentrations were determined using extinction coefficients (ε280 nm) per monomer [62,000 M−1 cm −1 for β′, 119,160 M−1 cm −1 for α]. E. coli thioredoxin (TR, specific activity of 40 U/mg) and thioredoxin reductase (TRR, specific activity of 1320 U/mg) were isolated as previously described (33, 34).
Expression and purification of human β′
The expression vector pET3a-β′, encoding for untagged β′ under the control of an IPTG inducible promoter, was transformed into BL21 Codon Plus (DE3)-RIL cells (Stratagene), plated on LB agar plates with 100 µg/mL ampicillin (Amp) and 34 µg/mL chloramphenicol (CM). A single colony was added to a culture (20 mL LB in 150 mL flask) and grown at 37°C to saturation overnight. The culture was then diluted into 2 L of LB in a 6 L flask containing 100 µg/mL Amp, 34 µg/mL CM and grown at 37°C to OD600nm 0.7. 1,10-Phenanthroline (100 µM) was then added to the media and the cells were grown for an additional 15 min (35). Isopropyl-β-D-thiogalactopyranoside (IPTG, 400 µM) was then added and the cells were grown at 37°C for an additional 5 h. The cells were harvested and frozen in liquid N2, typically yielding 2 g/L.
For the isolation of the apo β′, 16 g cells were resuspended (4 vol/g) in 50 mM Tris-HCl, pH 7.6, 1mM phenylmethanesulphonylfluoride (PMSF), 1mM EDTA at 4 °C. The suspension was passed through the French press at 14,000 psi. The cell lysate was centrifuged at 27,000 × g for 30 min at 4 °C. A solution of 10% (w/v) of streptomycin sulfate was added to the supernatant over 10 min while stirring at 4 °C, to a final concentration of 2.5%. After an additional 10 min of stirring, the pellet was removed by centrifugation (27, 000 ×g for 30 min, 4 °C). Solid ammonium sulfate (0.243 g/mL, 40% saturation) was added to the supernatant over 10 min at 4 °C. After an additional 30 min of stirring, the precipitate was recovered by centrifugation (27,000 × g for 30 min, 4 °C).
The pellet was dissolved in 4 mL of extraction buffer and loaded directly onto a Phenyl Sepharose 6 Fast flow column (20 mL, 2.5 × 10 cm) that was pre-equilibrated with 200 mL, 25 mM Tris pH 7.6, 10% glycerol, 30% w/v (NH4)2SO4 (buffer A). The column was washed with buffer A (100 mL) and then 300 mL, 25 mM Tris (pH 7.6), 10% glycerol, 5% w/v (NH4)2SO4 (buffer B). Finally, β′ was eluted with 50 mM Tris pH 7.6, 5% glycerol, 1 mM EDTA, 1mM PMSF (buffer C). Fractions (2 mL) containing β′ (based on Bradford assay) were collected.
The pooled protein fractions were then loaded onto a Q-Sepharose column (20 mL, 2.5 × 10 cm), which was pre-equilibrated with 100 mL buffer C at 4 °C. The column was washed with 200 mL buffer C and 150 mM KCl. The protein was eluted with a 70 mL × 70 mL linear gradient of 150 – 400 mM KCl in buffer C. Fractions (2 mL) containing β′ (based on Bradford assay and 10% SDS-PAGE analysis) were collected. The apo β′ eluted around 220 mM KCl. A typical yield of 0.4 mg β′/g cells was obtained.
Construction of the mutant Y138F-β′ gene and expression and isolation of Y138F-β′
The gene for the mutant Y138F-β′ was generated by site-directed mutagenesis using the Quick Change Kit (Stratagene). The gene for β′ was amplified by PCR using PfuUltra II polymerase (Stratagene) with primers for Y138F mutant (Table 1 of the Supporting Information). The sequence of the mutant plasmid was confirmed by sequencing at the MIT Biopolymers Laboratory. Y138F-β′ was expressed and purified as described for β′ with a similar yield.
Table 1.
Covalent labeling of human αn(β′2)m with [1'-3H]-F2CDP and [5-3H]-F2CDP analyzed by SEC.
| Protein | [x-3H]-F2CDP where x = |
Sephadex G-50 |
[3H]/α2b |
|---|---|---|---|
| human | 1' | native | 0.5 (0.83) |
| β'a/α | 1' | denaturing | 0.5 (0.83) |
| 5 | native | 0.03 (0.05) |
0.6 Y•/β′2.
Normalized 0.6 Y• to 1 Y•/β′2.
Conversion of apo β′2 to holo β′2
β'2 (60 µM) in 500 µL of 50 mM Hepes, 100 mM KCl, 10% glycerol, pH 7.6 and 2.5 mM ascorbate acid was deoxygenated by six cycles of evacuation (for 3 × 10 s) followed by argon flushing (2 min) on a Schlenk line. The deoxygenated β'2 solution was brought into the glove box (M. Braun, Stratham, NH) and 6 eq. of Fe (II) (deoxygenated ferrous ammonium sulfate in 50 mM Tris, 100 mM KCl, pH 7.6) per β'2 was added. The resulting mixture was incubated at 4 °C for 30 min. The protein was then removed from the glove box and 100 µL of ice cold O2 saturated 50 mM Tris, 100 mM KCl, pH 7.6 was added. O2 (g) was also blown over the surface of the protein solution for 1 min. Excess iron was removed by Sephadex G-25 chromatography (40 mL, 2.5 × 30 cm) and the protein fractions pooled to give a concentration of 3.5 mg/mL. An activity assay was carried out immediately and 250 µL of the protein solution was placed in an EPR tube and frozen in liquid N2 for measurement of the Y•. Typically 0.6 Y•/β'2 were observed with a specific activity of 420 nmol/min/mg.
Construction of active site and C-terminal tail mutant genes of α and expression and purification of the mutant proteins
Mutants of α [C218S(A), C429S(A), C444S(A), C787S(A), C790S(A), E431Q(D)] were generated by site-directed mutagenesis using the Quik Change Kit (Stratagene). For each mutant, the gene was amplified by PCR using PfuUltra II polymerase (Stratagene) with primers (Table S1 of the Supporting Information). The sequence of each mutant plasmid was confirmed by the MIT Biopolymers Laboratory. The mutants were expressed and purified as described below for wt α with similar yields.
Purification of α and α mutants by Ni NTA and dATP affinity chromatography
Cells (15 g) were suspended (5 vol/g) in 50 mM NaH2PO4, pH 7.0, 0.1% Triton X-100 and 10 mM β-mercaptoethanol (β-ME) with the complete protease inhibitor (Roche). The suspension was passed through the French press at 14,000 psi. The cell lysate was centrifuged at 20,000 × g for 30 min. Streptomycin sulfate (10%) was added to the supernatant over 10 min to a final concentration of 1% (w/v) and stirred for an additional 10 min. After the pellet was removed by centrifugation, the supernatant was incubated with Ni-NTA agarose resin (1 mL/g of cells, Qiagen) at 4°C for 1 h and then loaded into a column (2.5 × 10 cm). The column was washed with 30 column volumes of 50 mM NaH2PO4, 800 mM NaCl, 50 mM imidazole, pH 7.0, 0.1% Triton X-100 and 10 mM β-ME. The protein was eluted with 50 mM NaH2PO4, 300 mM NaCl, 125 mM imidazole, pH 7.0. The fractions containing protein were identified using the Bradford assay, pooled, and concentrated to < 10 mL, and then the imidazole was removed by Sephadex G-25 chromatography (200 mL, 2.5 × 50 cm) using 50 mM Tris, pH 7.6, 5% glycerol, 1 mM DTT as eluent. The pooled protein fractions were added to dATP affinity resin (30 mL, equilibrated with 50 mM Tris, pH 7.6, 5% glycerol, 1 mM DTT) (36) and gently mixed for 2 h at 4 °C in a 50 mL falcon tube. The resin was then loaded into a column (2.5 × 10 cm) and washed with 300 mL 50 mM Tris, pH 7.6, 5% glycerol, 300 mM KCl, 1mM DTT. The protein was eluted with 50 mM Tris, pH 7.6, 5% glycerol, 100 mM KCl, 5mM DTT, 10 mM ATP. The fractions containing protein were identified using the Bradford assay, pooled, and concentrated to < 0.5 mL. The ATP was removed by Sephadex G-25 chromatography (40 mL, 1.5 × 30 cm). α was stored in 50 mM Tris, 100 mM KCl, 15 mM MgCl2, 5 mM DTT, pH 7.6, 5% glycerol with a typical yield of 1 mg/g of cells after the Ni affinity column, and 0.2 mg/g of cell pellet after dATP affinity chromatography. High quality α purified by dATP chromatography was used in SEC studies only. All other experiments were carried out using α (wt or mutants) purified by Ni affinity chromatography only.
Activity assay
The reaction mixture contained the following in a final volume of 350 µL: 50 mM Hepes (pH 7.6), 15 mM MgCl2, 1 mM EDTA (assay buffer), 0.3 µM (or 3 µM) α, 3 µM (or 0.3 µM) β', 3 mM ATP, 1mM [3H]-CDP (specific activity 5115 cpm /nmol), 100 µM E. coli TR, 1.0 µM TRR, 2 mM NADPH. The assay mixture was pre-incubated at 37 °C for 2 min and the reaction was initiated by the addition of [3H]-CDP. Aliquots (30 µL) were removed over a 15 min time period and quenched in a boiling water bath for 2 min. dC production was analyzed, subsequent dephosphorylation with alkaline phosphatase as previously described (37) and analyzed by the method of Steeper and Steuart (38).
Time dependent inactivation assays
The inactivation mixture contained in a final volume of 100 µL: 6 µM α, 6 µM β', 3 mM ATP, 5 mM DTT, and assay buffer. The reaction was initiated by addition of 0.5 or 5 eq. of F2CDP (3 µM, 30 µM) and incubated at 37°C. Three types of assays were carried out. In assay 1, aliquots (10 µL) from the reaction mixture were removed at several time points from 30 s to 23 min and diluted 5 fold into 50 µL of assay buffer. dCDP production was measured as described above. A control experiment under identical conditions with the omission of F2CDP was always carried out to assess the stability of β'. In assays 2 and 3, the activity of α or β' was measured, respectively. Aliquots (2.5 µL) of the inactivation mixture were diluted into 50 µL of assay buffer containing 3 µM (10 fold excess) of the second subunit. dCDP production was measured as described above.
Quantitation of covalent labeling of human αn(β′2)m with [1'-3H]-F2CDP and [5-3H]-F2CDP
A typical reaction mixture contained in a final volume of 200 µL: 8.5 µM α and β', with 5 mM DTT, 3 mM ATP in assay buffer. The reaction was initiated with 10.6 µM of [1'-3H]-F2CDP (5889 cpm /nmol) or 10.6 µM of [5-3H]-F2CDP (6643 cpm/nmol). After 8 min at 37°C, an aliquot of 195 µL was either directly loaded onto a Sephadex G-50 column (1 cm × 20 cm, 20 mL) pre-equilibrated in assay buffer, or mixed with guanidine-HCl to a final concentration of 6 M, incubated for 5 min, and then loaded onto a Sephadex G-50 column in assay buffer containing 2 M guanidine-HCl. Fractions (1 mL) were collected and assayed for protein by Bradford assay and 500 µL of each fraction was analyzed by scintillation counting.
Quantitation of cytosine released during the inactivation of human αn(β′2)m by [5-3H]-F2CDP
The reaction mixture contained in 500 µL 1.2 µM α, β' and 1.2 µM [5-3H]-F2CDP (6643 cpm/nmol, 2 eq./α2). The reaction was incubated for 15 min at 37 °C. The mixture was then filtered through an YM-30 Centricon device (Millipore) at 4 °C. F2C (60 nmol) and cytosine (120 nmol) were added as carriers before filtration. The flow through was treated with 30 U of alkaline phosphatase (Roche) for 3 h at 37 °C and filtered through a second YM-30 Centricon device. The flow through was analyzed using a Waters 2480 HPLC with an Altech Adsorbosphere Nucleotide Nucleoside C-18 column (250 mm × 4.6 mm) at a flow rate of 1 mL/min. The elution buffer contained: Buffer I, 10 mM NH4OAc, pH 6.8; Buffer II: 100% methanol. A 10 min isocratic elution with Buffer I was followed by a linear gradient to 40% buffer II over 30 min. A linear gradient was then run to 100% buffer II over 5 min. Fractions (1 mL) were collected and 200 µL of each were analyzed by scintillation counting. The recovery of [5-3H]-cytosine and [5-3H]-F2C was calculated based on the UV spectrum (cytosine, λ267 nm, ε = 6100 M−1 cm−1, F2C, λ268 nm, ε = 9360 M−1 cm−1) and normalized for carrier added. The radioactivity recovered with [5-3H]-cytosine and [5-3H]-F2C was analyzed by scintillation counting.
SDS-PAGE of the inactivation mixture without boiling
The inactivation mixture contained in a final volume of 35 µL: 6 µM α or C218S-α, 6 µM β', 3 mM ATP, with or without 5 mM DTT, and assay buffer. The reaction was initiated by addition of 5 eq. of F2CDP (30 µM) and incubated at 37°C for 5 min. The inactivation mixture (8 µL) was mixed with 8 µL 2x loading buffer ± β-ME. [Note loading buffer contains SDS.] The samples were either heated at 90 °C for 2 min or not heated before loading on a 10% SDS-PAGE gel. The proteins were visualized with Coomassie blue staining. The band intensities were quantified using BioRad Quantity One software.
Incubation of [1'-3H] F2CDP with C218S(A)-, C429S(A)-, C444S(A)-, C787S(A)-, C790S(A)-, E431Q(D)-α and β' (β) and analysis for covalent labeling by Sephadex G50 chromatography
A typical reaction mixture contained in a final volume of 200 µL: 8.5 µM α mutant and β' (β), 5 mM DTT, and 3 mM ATP in assay buffer. The reaction was initiated with 10.6 µM of [1'-3H]-F2CDP (5889 cpm /nmol). After 8 min at 37°C, an aliquot of 195 µL was loaded onto a Sephadex G-50 column (1 cm × 20 cm, 20 mL) that was pre-equilibrated with the assay buffer. Fractions (1 mL) were collected and assayed for protein by the Bradford assay and 500 µL of each fraction was analyzed by scintillation counting.
SEC to examine the quaternary structure of αn(β′2)m subsequent to inactivation by F2CDP
SEC was performed using a Superdex 200 column (10 × 300 mm, GE Healthcare) attached to a Waters 2480 HPLC. Gel filtration molecular weight standards (GE healthcare) were ovalbumin, 43 kDa; conalbumin, 75 kDa; aldolase, 158 kDa; catalase 232 KDa; ferritin, 440 kDa; thyroglobulin, 669 kDa; and blue dextran, 2000 kDa. The elution buffer was 50 mM Hepes (pH 7.6), 15 mM MgCl2, 1 mM EDTA and 0.5 mM ATP. Molecular weight standards were run at the beginning of each experiment. The reaction mixture (400 µL) contained 15 µM α and β' (or Y138F-β'), 5 mM DTT, 3 mM ATP, 37.5 µM F2CDP or [1'-3H]-F2CDP (2.5 eq.) in assay buffer. α was purified using Ni affinity- and dATP affinity-chromatography and had a specific activity of 232 nmol/min/mg (measured with β2). After 10 min incubation, 390 µL was injected onto the column using a 500 µL loop. The elution rate was 0.5 mL/min and 0.5 mL fractions were collected and 300 µL aliquots of each fraction were analyzed by scintillation counting.
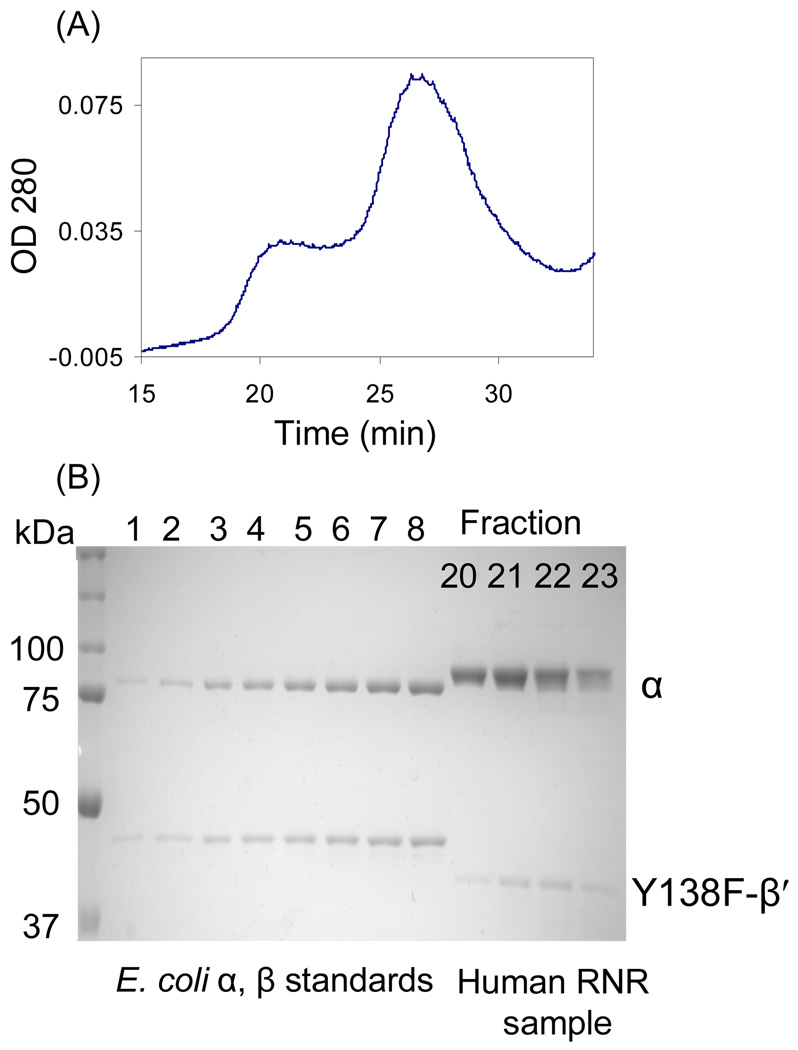
Quantitative analysis of the subunits of αn(β′2)m by SDS PAGE
The fractions collected from the SEC analysis above were analyzed directly (8 µL from each fraction) by 10% SDS- PAGE and compared with concentrations of α and β from E. coli RNR (0.05 µM to 0.4 µM) as standards. To analyze control experiments with Y138F-β′/α and with α/β′/ATP, each fraction of the first SEC peak was concentrated before loading on the gel. The proteins were visualized with Coomassie blue staining. The band intensities were quantified using BioRad Quantity One software. The concentrations of α and β' in the complex were determined from standard curves made with E. coli subunits as described above.
RESULTS
Purification and reconstitution of the β′2
Growth of β′2 was carried out in the presence of 1,10-phenanthroline resulting in apo protein (35). This procedure, subsequent to reconstitution of cluster, gave higher recovery of Y• and activity than standard growth conditions with addition of ferrous ammonium sulfate and ascorbate to the crude lysate prior to purification (39). β′2 was purified by addition of Phenyl Sepharose 6 Fast Flow and Q-Sepharose Fast Flow column chromatographies to the procedure of Thelander (26). These steps were required to obtain homogeneous protein by SDS-PAGE. A variety of methods to assemble the essential diferric-tyrosyl radical (Y•) cluster of β′2 were examined including use of tagged and non-tagged versions of β′2 and varying the ratio of Fe2+/β′. The detailed in vitro reconstitution procedure described in Methods gave 0.63 Y• per β′2 and an activity of 420 nmol/min/mg. The procedure reported by the Thelander group gave 1 Y• per β′2 (non-tagged), but the activity was 95 nmol/min/mg (26). A second purification procedure reported by Yen and coworkers using a tagged-β′2 gave 0.8 Y•/ β'2 and a specific activity of 50 nmol/min/mg (25). The low activity and high Y• content reported by these groups are at odds with activity being directly proportional to Y• (40, 41). The basis for the discrepancy is not understood. In our hands we have been unable, after many attempts with identical constructs to those used by Thelander and Yen, to obtain higher levels of Y•/β′2. However, with our previous purification of hβ, our group did obtain the same amount of radical (1 Y•/β2) as these groups, but with 7–14 fold higher activity. Our ability to only obtain 0.6 Y•/β′2 makes the quantitation of products from the F2CDP inactivation studies more complex as there is always 40% of β′2 and α that have not reacted. In the quantitations reported below we have placed numbers normalized to 1 Y•/β′2 in parentheses.
Time dependent inactivation studies of α, β′2 by F2CDP/ATP
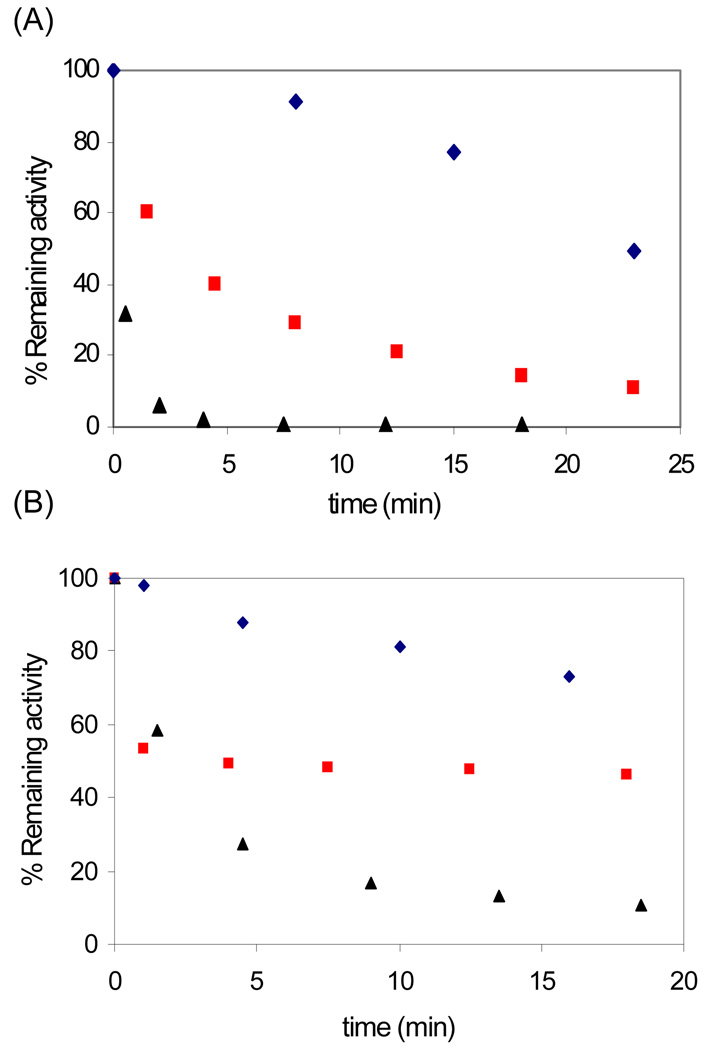
Our previous studies with F2CDP on human αn(β2)m and E. coli α2β2 showed that F2CDP is a sub-stoichiometric mechanism-based inhibitor resulting in covalent binding of 1 eq. of the sugar moiety from [1′-3H]-F2CDP per α2. To determine if β'2 behaves in a similar fashion, inactivation studies were carried out with α, β′ (6 µM each) and 0.5 or 5 eq. F2CDP/α in the presence of reductant (either DTT or TR/TRR/NADPH). Three different assays were carried out. In the first assay (Figure 1A) the enzyme was diluted 5 fold and assayed for dCDP production. Both concentrations of F2CDP resulted in > 90% inactivation with the reaction containing a 5 fold excess of inhibitor reaching completion in four minutes (▲). Even under these conditions, it is unlikely that the enzyme is saturated with F2CDP. As with hβ, substoichiometric amounts of inhibitor result in >90% inactivation, although the inactivation is slow
( ). The control with β'2 in the absence of inhibitor reveals that Y• is lost, precluding longer time points (
). The control with β'2 in the absence of inhibitor reveals that Y• is lost, precluding longer time points ( ). The half life of Y• loss is similar to our observations with hβ (11).
). The half life of Y• loss is similar to our observations with hβ (11).
Figure 1.
Time dependent inactivation assay of human β'/α by F2CDP 30 µM (▲), 3 µM ( ), 0 µM (
), 0 µM ( ). A. Aliquots were removed from the inactivation mixture at various times and diluted 5 fold for determination of RNR activity. B. Aliquots were removed and diluted 20 fold for determination of α (▲) or β' (
). A. Aliquots were removed from the inactivation mixture at various times and diluted 5 fold for determination of RNR activity. B. Aliquots were removed and diluted 20 fold for determination of α (▲) or β' ( ) activity in the presence of a 10 fold excess of β' or α, respectively. The activity of β' (
) activity in the presence of a 10 fold excess of β' or α, respectively. The activity of β' ( ) was adjusted using results from the control experiment (
) was adjusted using results from the control experiment ( ) in the absence of inhibitor.
) in the absence of inhibitor.
Because of the weak interactions between the two subunits in the class I RNRs, assays of the individual subunits, α (β'), are typically carried out with a ten fold excess of the second subunit, β' (α) present in the assay mixture. The result of a typical experiment under these conditions (0.5 eq. F2CDP) is shown in Figure 1B. Under these conditions α is 95% inactivated (▲), while β' retained 40% of its activity ( ) after correction for β' stability (
) after correction for β' stability ( ). These results are similar to those for the E. coli RNR and the hRNR with β. The results suggest that excess α can facilitate subunit dissociation and liberate the remaining active β'. The implications of these results will be addressed subsequently.
). These results are similar to those for the E. coli RNR and the hRNR with β. The results suggest that excess α can facilitate subunit dissociation and liberate the remaining active β'. The implications of these results will be addressed subsequently.
Quantitation of labeled RNR and small molecules generated by inactivation with [1'-3H] and [5-3H]-F2CDP
With [1'-3H]-F2CDP, our previous studies with hβ2 containing 1.2 Y•/β2, identified 0.9 eq./α2 of covalently bound sugar. A similar experiment using α and β' at 8.5 µM and 2.5 eq. [1'-3H] F2CDP was carried out for 8 min at 37°C. The reaction mixture was analyzed by Sephadex G50 column chromatography in the presence or absence of denaturant (guanidine-HCl). The results are summarized (Table 1) and reveal 0.5 eq. sugar/α2 (normalization for Y• gives 0.83 eq./α2). When the experiment was repeated with the [5-3H]-F2CDP under identical conditions in the absence of denaturant, 0.03 eq./α2 (0.05) was detected with protein (Table 1).
To test if 0.5 eq. sugar/inactivated enzyme is accompanied by 0.5 eq. of cytosine release, two eq. of [5-3H] F2CDPs (1.2 µM) were incubated with α2β'2 (0.6 µM) for 15 min at 37 °C. Subsequent to inactivation the nucleotides were recovered by ultrafiltration and analyzed by HPLC. The analysis revealed that 0.65 eq. (1.08) of cytosine was released and 1.35 eq. of F2C was recovered (Figure S1 of the Supporting Information). These results are in agreement with our previous studies on inactivation of human αn(β2)m and E. coli RNR and reveal that the extent of reaction correlates with the amount of Y• in β or β'.
Analysis of the inactivation mixture of wt and C218S-α by SDS PAGE without boiling
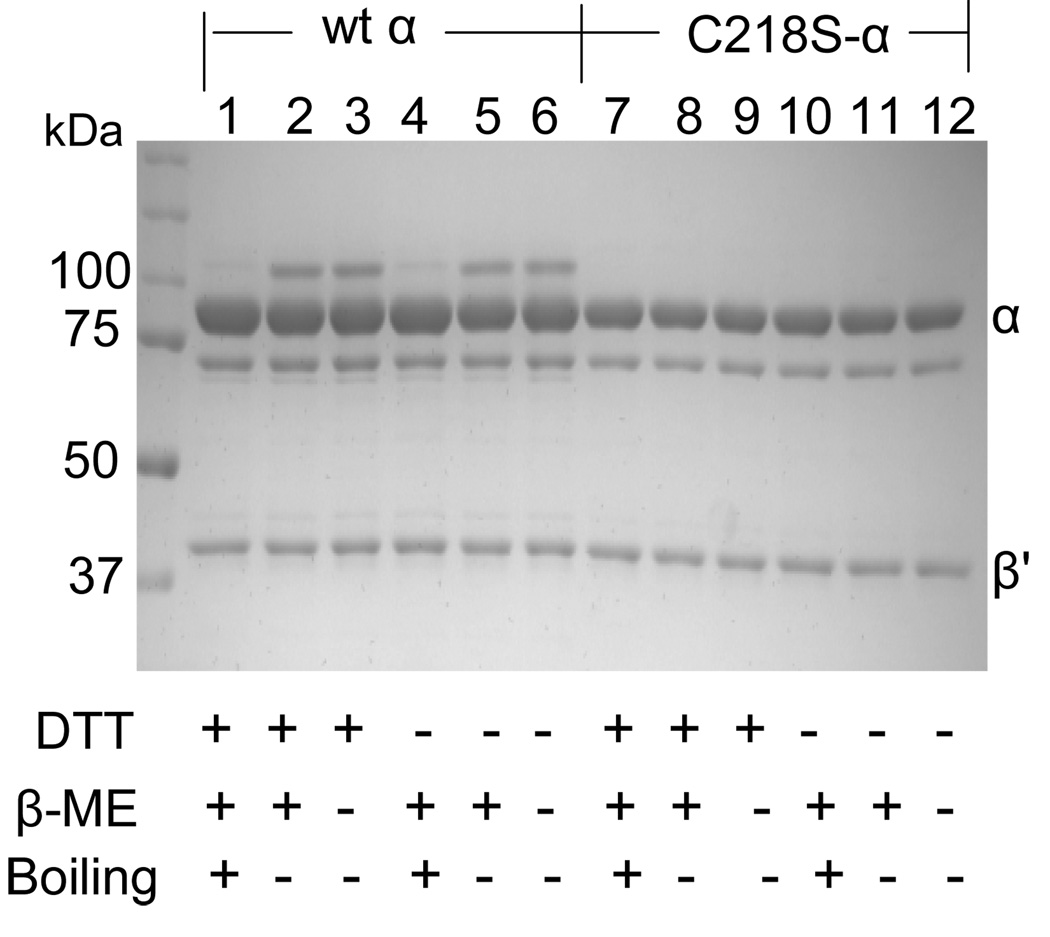
Our recent studies with E. coli RNR and L. leichmannii RNR revealed that when the enzyme inactivated with the inhibitor was analyzed by an SDS-PAGE gel without boiling, 30 to 50% of α migrated as a larger species (~110 kDa for E. coli and 110 kDa for L. leichmannii) (12, 29, 30). The α with the aberrant migratory properties is likely associated with the covalently bound sugar moiety from F2CDP. To determine if the substoichiometric labeling of hRNR was accompanied by production of a modified α, the reaction mixture subsequent to inactivation was analyzed directly without boiling by SDS PAGE (Figure 2). A similar experiment was carried out with β (Figure S2 of the Supporting Information). Twenty four percent (40%) and 33% of the α migrated as a larger species (~120 kDa) with β′ and hβ, respectively. This number in each case corresponds roughly to the amount of covalently bound sugar. With all RNRs examined to date, even the monomeric adenosylcobalamin RNR, production of an α with altered migratory properties is associated with covalent modification by the sugar moiety of F2CD(T)P.
Figure 2.
SDS-PAGE analysis of the α/β' (6 µM) inactivated by F2CDP (30 µM) and ATP (3 mM) at 37 °C for 5 min. Each sample was mixed with 2x loading buffer ± β-ME or boiling for 2 min before loading as indicated. A band at 120 kDa is observed in Lanes 2, 3 (inactivation in the presence of DTT, without boiling) and Lanes 5, 6 (inactivation in the absence of DTT, without boiling).
The same experiment was repeated with a mutant of one of the two active site cysteines involved directly in nucleotide reduction (C218S-α) based on observations reported in the next section. The results are also shown in Figure 2 (Lane 8–13) for β' and Figure S2 for β. With this mutant no altered conformation of α is observed suggesting that this residue may play an important role in its production. A proposal for these observations is presented subsequently.
Identification of potential sites of covalent modification of human RNR inactivated by [1'-3H] F2CDP using site-directed mutants of residues in α involved in catalysis
Our recent studies with L. leichmannii RTPR have established that 10% of the label associated with RNR subsequent to inactivation by [1′-3H]-F2CTP is due to alkylation of the sugar of the nucleotide by both cysteines at the C-terminus of the protein (29, 30). All efforts to identify the major labeled species failed, due to instability and rearrangement, even when NaBH4 was used to stabilize the alkyated protein. Furthermore, studies with the C225S mutant of E. coli RNR, suggested that this residue plays a key role in covalent modification of the enzyme by F2CDP (12). To identify the site of covalent modification by the sugar derived from F2CDP, [1'-3H] F2CDP was incubated with active site mutants of human α: C429S/A, E431Q/D, C787S/A, C790S/A, C444S/A, C218S/A (the equivalent of C225 in E. coli) and β or β' in the presence of DTT. Each mutant was purified as described for wt-α with similar recovery. The extent of covalent labeling measured subsequent to Sephadex G50 chromatograpy (in the absence of denaturant) was determined by scintillation counting and compared to the labeling of wt-RNR. The results are summarized in Table 2.
Table 2.
Quantitation of sugar covalently attached to hRNR mutants incubated with [1'-3H]-F2CDP analyzed by SEC.
| protein | [3H]/α2β'2 | [3H]/α2β2 |
|---|---|---|
| wt α | 0.5 (0.8)b | 0.8–0.9 |
| C429S-α (C439)a | 0 | 0.008 |
| C429A-α (C439) | 0 | 0.01 |
| E431Q-α (E441) | 0 | 0.01 |
| E431D-α (E441) | 0 | 0.08 |
| C787S-α (C754) | 0.3 (0.5) | 0.8 |
| C787A-α (C754) | 0.26 (0.4) | 0.3 |
| C790S-α (C759) | 0.3 (0.5) | 0.6 |
| C790A-α (C759) | 0 | 0.1 |
| C444S-α (C462) | 0.03 (0.05) | 0.4 |
| C444A-α (C462) | 0.2 (0.3) | 0.7 |
| C218S-α (C225) | 0 | 0.1 |
| C218A-α (C225) | 0 | 0.1 |
| Y138F- β' (Y122) | 0 |
The equivalent residues of E. coli RNR are given in ( ).
The values are normalized to 1 Y•/β′2 are given in ( ).
As expected no label is associated with RNR when the radical initiator on α (C429), essential for removal of the 3'-H for all mechanism based inhibitors, is changed to S or A. Two other mutants E431Q/D-α and C218S/A-α also showed no labeling. The E431 mutant is likely not labeled as this glutamate plays an important role (although not essential role) in the first two steps of the reduction process by removal of proton from the 3'-OH of F2CDP (28, 42, 43). Thus the rate of chemistry on any nucleotide is likely to be greatly reduced when the E is changed to a Q/D. In addition a recent structure of the S. cerevisiae α soaked with F2CDP showed that E431 is 6.2 Å from the 3'-HO of the sugar and that a water molecule intervenes (44). Thus, it is unlikely that this E is the site of labeling.
C225 on E. coli α, on the other hand, has been implicated as playing a key role in the inactivation pathway involving covalent modification of the E. coli RNR (12). Our previous studies with the C225S mutant of E. coli have demonstrated that first few steps in catalysis are not impaired by this mutation (45, 46). Thus lack of labeling with C218S/A-hα, make this a likely candidate for alkylation. Consistent with this proposal, the S. cerevisiae structure shows that this is the only nucleophilic residue close to the sugar (44). The caveat is however, that the second subunit, essential for catalysis, is missing in the structure. Thus our favored model is that C218 is covalently modified by a sugar moiety derived from F2CDP. In line with this proposal, the SDS-PAGE gel (Figure 2 and Figure S2 of the Supporting Information), revealed that with the C218S-α mutant and either hβ or β', that no α with modified migratory properties is observed.
The C-terminal tail mutants of α, C787S/A and C790S/A retain variable amounts of label associated with hRNR (with either β or β') (Table 2). These cysteines are involved in re-reduction of the active site disulfide generated during the synthesis of dNDP (47, 48). This C terminus must be able to enter the active site to reduce the disulfide between C218 and C444 and then must swing out of the active site to react with the protein reductant TR. The variability in labeling suggests that one of these cysteines, may be involved in covalent interaction with the sugar moiety as well. Labeling of the active site C218 and a C (787 or 790) in the tail might give rise to the observed conformational change found by SDS PAGE (Figure 2 and Figure S2 of the Supporting Information).
SEC to examine the quaternary structure of hRNR (with β′) inactivated by F2CDP
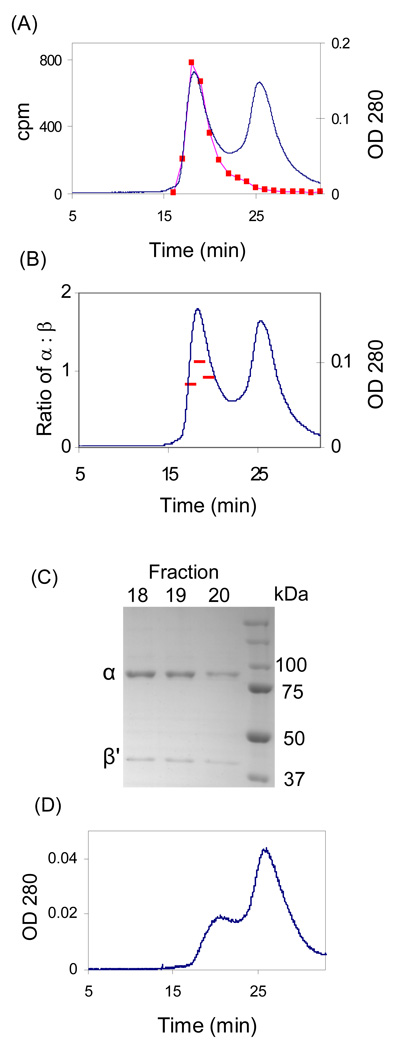
Our previous studies have shown that a tight α6β6 complex is generated when human RNR is inactivated by F2CDP. To examine the quaternary structure of αn(β′2)m subsequent to inactivation with [1'-3H]-F2CDP, the inactivated complex was examined by SEC with 0.5 mM ATP in the elution buffer. The A280nm revealed protein elution at 18.6 min and at 25.6 min (Figure 3). Comparison of the retention times with a standard curve generated with globular proteins of known molecular weight (Figure S3 of the Supporting Information) suggested that the protein eluting at 18.6 min has a mass (794 kDa) more consistent with α6β'6 than α6β′2 (798 vs 633 kDa). Control experiments with α (88 kDa), α2 (189 kDa with TTP), β′2 (116 kDa) (Table 3 and Figure S4 of the Supporting Information) suggest that the peak eluting at 25.6 min may be composed of β′2 and α. Each fraction from the SEC column was examined by SDS-PAGE with the amount of α and β′ determined from a standard curve made with known concentrations of E. coli α and β (Figure S5 of the Supporting Information). Analysis of protein in fractions 18, 19 and 20, gave a ratio of α: β' as 1.2, 1.0, 1.0 respectively and an average of 1.06 ± 0.15 (Figure 3B and 3C). This analysis supports the formation of an α6β'6 complex in the presence of F2CDP and ATP. Each fraction was also analyzed for radioactivity and fractions 18 and 19 contained 0.57 and 0.54 labels /α2 respectively.
Figure 3.
SEC on a S200 column to detect complex formation of α/β' inactivated by [1'-3H] F2CDP (37.5 µM). The elution buffer contained 0.5 mM ATP. A. The elution profile monitored by A280nm and scintillation counting ( ); B. Analysis of the ratio of α:β (
); B. Analysis of the ratio of α:β ( ) using standard curves generated from known amounts of E. coli α and β; C. SDS PAGE of fractions 18–20 from A; D. The elution profile in the absence of F2CDP.
) using standard curves generated from known amounts of E. coli α and β; C. SDS PAGE of fractions 18–20 from A; D. The elution profile in the absence of F2CDP.
Table 3.
Molecular weight determination of inactivated hRNR by SEC.
| Protein (effector) |
Retention time (min) |
Apparent mass (kDa) |
Caculated mass (kDa) |
Oligomeric state |
|---|---|---|---|---|
| β' | 26 | 116 | 81 | β'2 |
| α | 25.6a | 88 | 92 | α |
| α (100µM TTP) | 24a | 189 | 184 | α2 |
| α, β' (ATP) | 20.9,26.2 | 543, 109 | 552, 184, 81 |
α6, α2, β'2 |
| α, β' inactivated by F2CDP |
18.6, 25.6 |
794, 116 |
798 | α6β'6 complexb |
| α, Y138F-β' inactivated by F2CDP |
20–22.5, 26.9 |
389–712, 88 |
633, 552, 184, 81 |
α6β'2, α6 α2, β'2 |
Superose 12 column was used.
Un-reacted α2, β'2 were also present.
As discussed above, β'2 contains only 0.6 Y• and thus gives rise to protein that does not form a complex. The relative amounts of the two peaks (Figure 3) support the proposal that the percentage of active quaternary structure correlates with the amount of the Y•. Together with the observations of 0.5 eq. sugar/α2 attached, and 0.6 eq. cytosine released after inactivation, these results support a model where 1 eq. of F2CDP inactivates the two active sites of α2 by enhancing subunit interactions.
Given the poor resolution of the SEC method, a control experiment in which F2CDP was omitted was carried out under identical conditions (Figure 3D). The mixture analyzed by SEC gave two protein peaks: one with a retention time of 20.9 min and an apparent molecular weight of 543 kDa and the second with a retention time of 26.2 min and an apparent molecular weight of 109 kDa. The SDS-PAGE analysis of the first peak showed only the presence of α, which correspond to α6, and the second peak showed the presence of both α and β' corresponding to α and β'2 based on the apparent MW.
An additional control: SEC to examine the quaternary structure of hRNRs with Y138F-β' in the presence of F2CDP and ATP
To gain further insight into the quaternary structure of hRNR without covalent modification in the presence of F2CDP, Y138F-β' in which the essential Y• of β'2 was replaced with an F, was studied. The reaction mixture was identical to that described above except that Y138F-β' replaced wt-β'. Analysis of the reaction mixture by SEC showed that a small portion of protein migrated between 20–22.5 min, with apparent MWs ranging from 389–712 kDa followed by a second protein peak with a retention time at 26.9 min and an apparent MW of 88 kDa (Figure 4, Table 3). No radioactivity eluted with either peak when [1'-3H] F2CDP was used. SDS-PAGE analysis revealed that both α and β' were present in the peak that eluted from 20–22.5 min, but the ratio of α: β' ranged from 5.8–9.1 by comparison with standards (Figure S5 of the Supporting Information). The results suggest a complex equilibrium between nucleotide analog/ATP and subunits, consistent with the existence of α6β2, α6 and other species prior to the chemistry of inactivation.
Figure 4.
SEC on a S200 column to detect complex formation of α/Y138F-β' incubated with [1'-3H] F2CDP. The elution buffer contained 0.5 mM ATP. A. The elution profile monitored by A280nm and scintillation counting shows no radiolabel. B. Fractions 20–23 in A were monitored by 10% SDS PAGE. Lane 1–8 are E. coli α and β (0.05- 0.4 µM) standards at different concentrations analyzed by SDS-PAGE to determine the ratio of α, Y138F-β′.
DISCUSSION
The mechanism of inactivation of RNRs by F2CDP is obviously very complex. However, despite this complexity, the RNRs from E. coli, L. leichmannii and the two RNRs from humans share common and mechanistically/structurally informative features. Stoichiometric amounts of inhibitor are sufficient for complete inactivation (10, 11). In the case of the class I RNRs, this inhibition prevents the second α/β pair from reaction because of the tight and unusual subunit interactions (11). In all cases, the protein becomes covalently modified in a chemically fragile state by a sugar moiety generated from F2CDP and the cofactor (Y• or adenosylcobalamin) is partially inactivated. In all cases, if one waits long enough, two fluoride ions and one cytosine are released, requiring ketone formation at the C3' position of the nucleotide during the inactivation process. Unexpectedly, the α subunit is observed to have two conformations in about an approximately 1:1 ratio on SDS PAGE analysis in the absence of heat. Mutagenesis studies suggest that the second conformation is likely the result of a crosslink between a cysteine in the C-terminus of α and its active site C218, directly involved in water loss in the normal reduction process. The key to the function of RNR is understanding the gymnastics of the structurally floppy, and thus structurally inaccessible, C-termini of β/β' and C-termini of α.
From our present observations information about the subunit architecture/interactions result. The altered α is likely to be formed from an intramolecular crosslink, rather than a cross-link from the C-terminus of the adjacent α in the α2 dimer. If the crosslink were intermolecular, α should migrate on the SDS-PAGE gel as a dimer and would likely be sensitive to β-ME treatment. Re-reduction of the active site disulfide in one α, by the C-terminal tail of the second α in the α2 complex had previously been suggested by pre-steady state in vitro studies on E. coli RNR by Ericksson (49) and in vivo studies of S. cerevisiae RNR by Huang and coworkers (50). Our previous pre-steady state experiments (51) with mutations of the C-terminal cysteines were not inconsistent with the Ericksson proposal. In the Huang studies, the kinetics of re-reduction of the active site disulfide is not measured due to the in vivo nature of the experiments. We have previously shown that the two cysteines in the C-terminal tail of E. coli RNR can function as a thioredoxin and this observation is also likely to be applicable to the S. cerevisiae RNR and could account for their in vivo results (47).
A second observation of interest is that the C-terminus of α must have access to the active site of the same α, while β remains bound. If our interpretation of the altered migratory properties of α is correct, a recent structure of a complex of a class Ib RNR from Salmonella typhimurium at 4.5 Å resolution may be informative in this regard (52). In this structure, the two subunits are present with a 360 Å2 interface. The interface however, is very informative with respect to the function of the C-termini of each subunit. One can observe the end of β (residue 285), a gap of 20 amino acids and the last eleven amino acids of β bound in a hydrophobic patch formed by 3 helices (α 10, α I and αD) within α. One can also locate the C-terminus (residue 699 missing the last 15 amino acids) of α within the structure and its position relative to β. One β is interacting with one α in a fashion that, with tightening, could align the radical transfer pathway between the two subunits in a chemically competent fashion. In this αβ the active site cysteines are oxidized and some electron density exists for the ADP substrate. The second α has no substrate bound and no contact with the second β. Examination of the locations of the C-termini of the subunits suggest that the tails of α and β could be intertwined and that complete dissociation of the subunits might require a push from an interaction with a second α2 interacting with the dangling β2. This model would be consistent with the kinetics of inactivation by F2CDP in all the class I RNRs where a 1:1 complex of subunits is inactive, while assaying for activity of one subunit in the presence of an excess of the other subunit leads to some recovery of β2 activity as all the Y• has not been destroyed.
Uhlin has suggested that the structure of the class Ib RNR complex is a snapshot of one of the steps during the RNR-mediated reduction: either an initial step in subunit interaction or a final step after the nucleotide reduction process. The unusual conformation of α in the F2CDP inactivated RNR complex suggests that it might be trapping the enzyme in an asymmetrical configuration as well. The tight interaction between the subunits subsequent to inactivation by F2CDP suggests that crystallization of the complex may be possible. Recently we have obtained crystals of the E. coli RNR that diffract to 6 Å resolution (Stock, Drennan, unpublished work). Obviously higher resolution structures are essential to figure out the key role of the tails of the subunits in the mechanism of ribonucleotide reduction. Despite the absence of molecular understanding, our studies suggest that both human RNRs can be targeted by F2CDP, and that the consequences of inactivation may differ depending on growth conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Yun Yen (City of Hope National Medical Center) for providing phRRM1 and phRRM2, Dr. Lars Thelander (Umeå University, Sweden) for providing the p53R2, and Eli Lilly for providing [5-3H] F2C and 2-deoxy-3,5-di-O-benzoyl-3,3-difluororibonolactone. We thank Ellen Minnihan in our lab for her thoughtful comments.
Abbreviations
- RNR
ribonucleotide reductases
- α
ribonucleotide reductase large subunit
- β
ribonucleotide reductase small subunit, also called R2
- β′
the second ribonucleotide reductase small subunit, also called p53R2
- h
human
- hRNR
human RNR
- hβ
human β
- dNTP
deoxynucleoside 5′-triphosphate
- dNDP
deoxynucleoside 5′-diphosphate
- NDP
nucleoside 5′-diphosphate
- eq.
equivalent
- DTT
dithiothreitol
- β-ME
β-mercaptoethanol
- IPTG
isopropyl-β-D-thiogalactopyranoside
- wt
wild type
- Y•
tyrosyl radical
- SEC
size exclusion chromatography
- Amp
ampicillin
- CM
chloramphenicol
- PMSF
phenylmethanesulphonylfluoride
- F2C
2', 2'-dideoxy-difluorocytidine or gemcitabine
- F2CDP
5′-diphosphate of F2C
- F2CTP
5′-triphosphate of F2C
- TR
thioredoxin
- TRR
thioredoxin reductase
- ATM
ataxia telangiectasia mutated
- ERK
extracellular signal-regulated kinase
- assay buffer
50 mM Hepes (pH 7.6), 15 mM MgCl2, 1 mM EDTA
- buffer A
25 mM Tris pH 7.6, 10% glycerol 30% w/v (NH4)2SO4
- buffer B
25 mM Tris (pH 7.6), 10% glycerol 5% w/v (NH4)2SO4
- buffer C
50 mM Tris pH7.6, 5% glycerol 1 mM EDTA, 1mM PMSF
Footnotes
This work was supported by the NIH grant (GM29595 to J.S.)
SUPPORTING INFORMATION AVAILABLE
Tables 1 listed the primers for mutants of β′ and α. Figure S1 shows the analysis of products generated during inactivation of αn(β′2)m by [5-3H]-F2CDP. Figure S2 shows the SDS-PAGE analysis of the α/β inactivated by F2CDP and ATP. Figure S3 shows the gel filtration molecular weight standards, and figure S4 shows SEC controls with β'2, α and α2 with TTP. Figure S5 shows the E. coli α and β standards at different concentrations analyzed by SDS-PAGE to determine the ratio of α, β′ in Figure 3B. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Stubbe J, van der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 2.Hertel LW, Boder GB, Kroin JS, Rinzel SM, Poore GA, Todd GC, Grindey GB. Evaluation of the antitumor-activity of gemcitabine (2',2'-difluoro-2'-deoxycytidine) Cancer Res. 1990;50:4417–4422. [PubMed] [Google Scholar]
- 3.Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2',2'-difluorodeoxycytidine on DNA-synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- 4.Plunkett W, Huang P, Gandhi V. Gemcitabine: actions and interactions. Nucleosides & nucleotides. 1997;16:1261–1270. [Google Scholar]
- 5.Rivera F, López-Tarruella S, Vega-Villegas ME, Salcedo M. Treatment of advanced pancreatic cancer: from gemcitabine single agent to combinations and targeted therapy. Cancer Treat Rev. 2009;35:335–339. doi: 10.1016/j.ctrv.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Danesi R, Altavilla G, Giovannetti E, Rosell R. Pharmacogenomics of gemcitabine in non-small-cell lung cancer and other solid tumors. Pharmacogenomics. 2009;10:69–80. doi: 10.2217/14622416.10.1.69. [DOI] [PubMed] [Google Scholar]
- 7.Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2',2'-difluorodeoxycytidine (gemcitabine) Drug Resistance Updates. 2002;5:19–33. doi: 10.1016/s1368-7646(02)00002-x. [DOI] [PubMed] [Google Scholar]
- 8.Nordlund N, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 9.Baker CH, Banzon J, Bollinger JM, Stubbe J, Samano V, Robins MJ, Lippert B, Jarvi E, Resvick R. 2'-deoxy-2'-methylenecytidine and 2'-deoxy-2',2'-difluorocytidine 5'-diphosphates - potent mechanism-based inhibitors of ribonucleotide reductase. J. Med. Chem. 1991;34:1879–1884. doi: 10.1021/jm00110a019. [DOI] [PubMed] [Google Scholar]
- 10.van der Donk WA, Yu GX, Perez L, Sanchez RJ, Stubbe J, Samano V, Robins MJ. Detection of a new substrate-derived radical during inactivation of ribonucleotide reductase from Escherichia coli by gemcitabine 5'-diphosphate. Biochemistry. 1998;37:6419–6426. doi: 10.1021/bi9729357. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5'-diphosphate inhibit ribonucleotide reductases. Proc. Natl. Acad. Sci. U S A. 2007:14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artin E, Wang J, Lohman G, Yu G, Griffin G, Barr G, Stubbe J. Insight into the mechanism of inactivation of ribonucleotide reductase by gemcitabine 5'-diphosphate in the presence and absence of reductant. submitted to Biochemistry. 2009 doi: 10.1021/bi901590q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42:1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- 14.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha(6)beta(2) octamer. J. Biol. Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 15.Sjöberg BM, Reichard P. Nature of free-radical in ribonucleotide reductase from Escherichia-coli. J. Biol. Chem. 1977;252:536–541. [PubMed] [Google Scholar]
- 16.Sjöberg BM, Reichard P, Gräslund A, Ehrenberg A. The tyrosine free radical in ribonucleotide reductase from Escherichia coli. J. Biol. Chem. 1978;253:6863–6865. [PubMed] [Google Scholar]
- 17.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 18.Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 19.Pontarin G, Fijolek A, Pizzo P, Ferraro P, Rampazzo C, Pozzan T, Thelander L, Reichard PA, Bianchi V. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17801–17806. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi T, Matsuda K, Saqiya Y, Iwadate M, Fujino MA, Nakamura Y, Arakawa H. p53R2-dependent pathway for DNA synthesis in a p53-regulated cell cycle checkpoint. Cancer Res. 2001;61:8256–8262. [PubMed] [Google Scholar]
- 21.Xue L, Zhou B, Liu X, Qiu W, Jin Z, Yen Y. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–986. [PubMed] [Google Scholar]
- 22.Bourdon A, Minai L, Serre V, Jais J, Sarzi E, Aubert S, Chretien D, Lonlay P, Paquis-fluckinger V, Arakawa H, Nakamura Y, Munnich A, Rötig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 23.Chang L, Zhou B, Hu S, Guo R, Liu X, Jones SN, Yen Y. ATM-mediated serine 72 phosphorylation stabilizes ribonucleotide reductase small subunit p53R2 protein against MDM2 to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18519–18524. doi: 10.1073/pnas.0803313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piao C, Jin M, Kim HB, Lee SM, Amatya PN, Hyun JW, Chang IY, You HJ. Ribonucleotide reductase small subunit p53R2 suppresses MEK-EPK activity by binding to ERK kinase 2. Oncogene. 2009;28:2173–2184. doi: 10.1038/onc.2009.84. [DOI] [PubMed] [Google Scholar]
- 25.Shao JM, Zhou BS, Zhu LJ, Qiu WH, Yuan YC, Xi BX, Yen Y. In vitro characterization of enzymatic properties and inhibition of the p53R2 subunit of human ribonucleotide reductase. Cancer Res. 2004;64:1–6. doi: 10.1158/0008-5472.can-03-3048. [DOI] [PubMed] [Google Scholar]
- 26.Guittet O, Håkansson P, Voevodskaya N, Fridd S, Gräslund A, Arakawa H, Nakamura Y, Thelander L. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J. Biol. Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Xue L, Yen Y. Redox property of ribonucleotide reductase small subunit M2 and p53R2. Methods Mol Biol. 2008;477:195–206. doi: 10.1007/978-1-60327-517-0_15. [DOI] [PubMed] [Google Scholar]
- 28.Silva DJ, Stubbe J, Samano V, Robins MJ. Gemcitabine 5'-triphosphate is a stoichiometric mechanism-based inhibitor of Lactobacillus leichmannii ribonucleoside triphosphate reductase: Evidence for thiyl radical-mediated nucleotide radical formation. Biochemistry. 1998;37:5528–5535. doi: 10.1021/bi972934e. [DOI] [PubMed] [Google Scholar]
- 29.Lohman GJS, Stubbe J. Inactivation of Lactobacillus leichmannii ribonucleotide reductase by F2CTP: covalent modification (part I) manuscript in preparation for Biochemistry. 2009 doi: 10.1021/bi902132u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohman GJS, Gerfen GJ, Stubbe J. Inactivation of L. leichmannii ribonucleotide reductase by F2CTP: adenosylcobalamin destruction and formation of a nucleotide based radical. manuscript in preparation for Biochemistry. 2009 doi: 10.1021/bi9021318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stubbe J, van der Donk WA. Ribonucleotide reductases: radical enzymes with suicidal tendencies. Chem. Biol. 1995;12:793–801. doi: 10.1016/1074-5521(95)90084-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Zhenchuk A, Wiman KG, Albertioni F. Regulation of p53R2 and its role as potential target for cancer therapy. Cancer Lett. 2009;276:1–7. doi: 10.1016/j.canlet.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Lunn CA, Kathju S, Wallace BJ, Kushner SR, Pigiet V. Amplification and purification of plasmid-encoded thioredoxin from Escherichia coli K12. J. Biol. Chem. 1984;259:469–474. [PubMed] [Google Scholar]
- 34.Russel M, Model P. Direct cloning of the trxB gene that encodes thioredoxin reductase. J. Bacteriol. 1985;163:238–242. doi: 10.1128/jb.163.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkin SE, Chen S, Ley BA, Mangravite L, Edmondson DE, Huynh BH, Bollinger JM., Jr Electron injection through a specific pathway determines the outcome of oxygen activation at the diiron cluster in the F208Y mutant of Escherichia coli ribonucleotide reductase protein R2. Biochemistry. 1998;37:1124–1130. doi: 10.1021/bi9723717. [DOI] [PubMed] [Google Scholar]
- 36.Berglund O, Eckstein F. ATP- and dATP-substituted agaroses and the purification of ribonucleotide reductases. Methods Enzymol. 1974;34:253–261. doi: 10.1016/s0076-6879(74)34021-9. [DOI] [PubMed] [Google Scholar]
- 37.Perlstein DL, Ge J, Ortigosa AD, Robblee JH, Zhang Z, Huang M, Stubbe J. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry. 2005;44:15366–15377. doi: 10.1021/bi051616+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steeper JR, Steuart CC. A rapid assay for CDP reductase activity in mammalian cell extracts. Anal. Biochem. 1970;34:123–130. doi: 10.1016/0003-2697(70)90092-8. [DOI] [PubMed] [Google Scholar]
- 39.Salowe SP, Stubbe J. Cloning, overproduction, and purification of the B2 subunit of ribonucleoside-diphosphate reductase. J Bacteriol. 1986;165:363–366. doi: 10.1128/jb.165.2.363-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortigosa AD, Hristova D, Perlstein DL, Zhang Z, Huang MX, Stubbe J. Determination of the in vivo stoichiometry of tyrosyl radical per beta beta ' in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry. 2006;45:12282–12294. doi: 10.1021/bi0610404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hristova D, Wu CH, Jiang W, Krebs C, Stubbe J. Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry. 2008;47:3989–3999. doi: 10.1021/bi702408k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Donk WA, Yu G, Silva DJ, Stubbe J. Inactivation of ribonucleotide reductase by (E)-2'-fluorromethylene-2'-deoxycytidine 5′-diphosphate: a paradigm for nucleotide mechanism based inhibitors. Biochemistry. 1996;35:8381–8391. doi: 10.1021/bi960190j. [DOI] [PubMed] [Google Scholar]
- 43.Zipse H, Artin E, Wnuk S, Lohman GJ, Martino D, Griffin RG, Kacprzak S, Kaupp M, Hoffman B, Bennati M, Stubbe J, Lees N. Stucture of the nucleotide radical formed during reaction of CDP/TTP with the E441Q-α2β2 of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2009;131:200–211. doi: 10.1021/ja806693s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu HFC, Uchiki T, Racca J, Dealwis C. Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4028–4033. doi: 10.1073/pnas.0600440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao SS, Johnston MI, Bollinger JM, Stubbe J. Mechanism-based inhibition of a mutant Escherichia coli ribonucleotide reductase (cysteine-225-serine) by its substrate CDP. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1485–1489. doi: 10.1073/pnas.86.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao SS, Holler TP, Bollinger JM, Jr, Yu GX, Johnston MI, Stubbe J. Interaction of C225SR1 mutant subunit of ribonucleotide reductase with R2 and nucleoside diphosphates: tales of a suicidal enzyme. Biochemistry. 1992;31:9744–9751. doi: 10.1021/bi00155a030. [DOI] [PubMed] [Google Scholar]
- 47.Mao SS, Holler TP, Yu GX, Bollinger JM, Booker S, Johnston MI, Stubbe J. A model for the role of multiple cysteine residues involved in ribonucleotide reduction - amazing and still confusing. Biochemistry. 1992;31:9733–9743. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- 48.Lin ANI, Ashley GW, Stubbe J. Location of the redox-active thiols of ribonucleotide reductase: sequence similarity between the Escherichia coli and Lactobacillus leichmannii enzymes. Biochemistry. 1987;26:6905–6909. doi: 10.1021/bi00396a006. [DOI] [PubMed] [Google Scholar]
- 49.Eriksson HK. Kinetics in the pre-steady state of the formation of cystines in ribonucleoside diphosphate reductase: evidence for an asymmetric complex. Biochemistry. 2001;40:9631–9637. doi: 10.1021/bi010651n. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Yang K, Chen C, Feser J, Huang M. Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2217–2222. doi: 10.1073/pnas.0611095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ge J, Yu GX, Ator MA, Stubbe J. Pre-steady-state and steady-state kinetic analysis of E. coli class I ribonucleotide reductase. Biochemistry. 2003;42:10071–10083. doi: 10.1021/bi034374r. [DOI] [PubMed] [Google Scholar]
- 52.Uppsten M, Färnegårdh M, Domkin V, Uhlin U. The first holocomplex structure of ribonucleotide reductase gives new insight into its mechanism of action. J. Mol. Biol. 2006;359:365–377. doi: 10.1016/j.jmb.2006.03.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.