Abstract
Objective
Ath11, an atherosclerosis susceptibility locus on proximal chromosome 10 (0–21cM) revealed in a cross between apolipoprotein E deficient C57BL/6 (B6) and FVB mice, was recently confirmed in congenic mice. The objectives of this study were to assess how Ath11 affects lesion development and morphology, to determine aortic gene expression in congenics, and to narrow the congenic interval.
Methods and Results
Assessing lesion area over time in congenic mice showed that homozygosity for the FVB allele increased lesion area at 6 weeks persisting through to 24 weeks of age. Staining of aortic root sections at 16 weeks did not reveal obvious differences between congenics. Aortic expression-array analysis at 6 weeks revealed 97 >2 fold regulated genes, including one gene in the QTL interval, Aldh8a1, and two gene clusters regulated by Hnf4α and Esr1. Analysis of lesion area in 11 subcongenic strains revealed two narrowed regions, 10a (21 genes) acting in females and 10b (7 genes) acting in both genders.
Conclusions
Ath11 appears to act early in lesion formation with significant effects on aortic gene expression. This QTL is genetically complex containing a female specific region 10a from 0 to 7.3 Mb, and a gender independent region 10b from 20.1 to 21.9 Mb.
Keywords: Atherosclerosis, QTL, Ath11, congenics, subcongenics
Atherosclerosis is a complex disorder involving many genes and environmental factors. Animal models have been used to sort out these complexities. Particularly with regard to genetic factors, one approach has been to alter candidate gene expression or function and observe effects on atherosclerotic lesions. Another approach is to use reverse genetics to identify regions of the genome co-inherited with atherosclerosis susceptibility and determine the culprit gene(s). One way to do this is quantitative trait locus (QTL) mapping.1 A mapped region must be confirmed by creation of congenics, and congenics can then be used to create subcongenics to narrow the region and ultimately identify the culprit gene(s). Following this strategy, in previous work we carried out an intercross between atherosclerosis susceptible C57BL6/J (B6) and atherosclerosis resistant FVB/N (FVB) mice on the Apoe−/− and Ldlr−/−backgrounds.2,3 QTL mapping revealed an atherosclerosis susceptibility locus on mouse proximal chromosome 10 (Chr10), designated Ath11.4,5 This is the only QTL reported thus far that is present in two atherosclerotic sensitizing backgrounds for the same intercross and is independent of the animal’s gender and lineage. The Chr10 QTL suggested a dominant B6 allele that lowered atherosclerotic lesion area, despite the fact that B6 is the atherosclerosis susceptible strain in this intercross. Ath11 was validated using interval-specific congenic strains on the F1 background with either the Apoe−/− or Ldlr−/− sensitizing background.6 The congenic interval defining the Ath11 locus extends from 0 (D10Mit49) to 21.9 cM (D10Mit60) (58.3 Mb) on Chr10 and contains 382 annotated genes. The F1 background was used because it provided a clearer confirmation of the interval than either the B6 or FVB backgrounds, perhaps because it permitted genetic interactions not present on either inbred background.
The current study shows that gene(s) in the QTL region act early in lesion formation with altered expression of clusters of genes regulated by the transcription factors Hnf4α and Esr1. Analysis of subcongenic strains revealed a proximal region, 10a, that is female specific and contains 21 genes, and a distal region, 10b, in both genders containing 7 genes.
Methods
Generation of congenic and subcongenic mice
Apoe−/− deficient mice on the C57BL/6 (B6.Apoe−/−) and FVB/N (FVB.Apoe−/−) backgrounds were taken from our own colony.2 Mice were named as follows: first the strain and the overall background are indicated, e.g. F1.Apoe−/− for mice that were heterozygous (B6xFVB) on the Apoe−/− background and second the genotype of the Chr10 interval is depicted, e.g. Chr10B6/FVB for mice that were heterozygous at Chr10. Congenic littermates of all three genotypes on the F1 background were used for the time course, lesion morphology and gene expression studies.6 To narrow Ath11, subcongenic strains containing a reduced portion of the original interval were generated by crossing B6.Apoe−/−Chr10B6/FVB with B6.Apoe−/−. Recombinants missing part of the original FVB interval were identified and bred to FVB.Apoe−/− mice to generate littermates that were either F1.Apoe−/−Chr10FVB/FVB* or F1.Apoe−/−Chr10B6/FVB (the new shortened interval is indicated by a *). Animal care and experimental procedures conformed to the guidelines of the American Heart Association. Research animals were housed in the Rockefeller University’s Comparative Bioscience Center in a specific pathogen-free environment in rooms with a 7AM to 7PM light/dark cycle. The Rockefeller University’s Institutional Animal Care and Use Committee approved all procedures.
Genotyping
Genomic DNA was isolated and genotyped as described.6 The boundaries of each subcongenic strain were fine mapped by sequencing regions containing polymorphic SNPs between B6 and FVB. Primers were purchased from Sigma.
Mouse feeding and sacrifice, blood drawing and analysis, and atherosclerosis assessment
For atherosclerosis studies, mice were weaned at 28 days of age and fed a semi-synthetic modified AIN76a diet containing 0.02% cholesterol. Animals were sacrificed at 6, 12, 16, 20 and 24 weeks of age for the time course study, at 6 weeks of age for gene expression analysis and at 16 weeks of age for all other studies. Mice were sacrificed and plasma isolated for lipoprotein analysis as described.6 The isolated hearts were stored frozen in Tissue-Tek OCT compound (time course study, immunostaining) or in buffered formalin (subcongenic strains). To quantify atherosclerosis at the aortic root, hearts were sectioned and stained with oil red O (ORO) as described.6, 7
Histochemistry and immunostaining of aortic root sections
Histochemistry and immunostaining were performed as previously described.7 Movat pentachrome stain was used to determine lesion composition. To identify cellular components, lipids, and apoptotic cells, lesions were fixed and stained for CD-68 (anti CD68/MAC 1957 GA (rat), Serotec), CD-31 (anti CD31 (PECAM) (rat), BD Pharma), α-actin (anti actin IgG (rabbit), Biomedical Technologies), lipids (ORO) and TUNEL (In Situ Cell Death Detection Kit, POD from Roche) according to the manufacturer’s instructions.
Gene expression analysis
RNA was isolated from 6 week old female mice (F1.Apoe−/−Chr10FVB/FVB, F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10B6/B6). Aortic tissue was homogenized and pipetted onto a Qiashredder prior to RNA isolation with the RNeasy fibrous tissue Kit (Qiagen). Contaminating DNA was removed (DNA-free, Ambion) and 5 μgRNA, pooled from five 6 week old female mice per strain, were reverse transcribed using Superscript II (Invitrogen) and a poly-dT primer containing the T7 RNA polymerase bindingsite (Genset Corporation). Double-stranded cDNA was made and purified (Genechip Sample Cleanup Module, Qiagen/Affymetrix). cRNA was synthesized using biotin-labeled ribonucleotidesand T7 RNA polymerase (Enzo Bioarray) and purifiedwith the Genechip Sample Cleanup Module (Qiagen/Affymetrix) prior to fragmentation. Fragmented cRNA samples were hybridized to Affymetrix mouse expression array MOE430 A and B sets. Gene expression differences between both F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10FVB/FVB mice and F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10FVB/FVB mice were identified with the GeneSpring 7 software (Silicon Genetics). The GO slimmer tool (http://amigo.geneontology.org/cgi-bin/amigo/slimmer) was used to identify the number of regulated genes for level-1 GO terms, GO-2376, GO-8219 and GO-3018. Greater than 2-fold differentially regulated genes were uploaded into MetaCore™ Analytical suite (GeneGo Inc., USA) to generate sub-networks centered on transcription factors. Calculation of statistical significance of networks is based on p-values, which are defined as the probability of the network’s assembly from a random set of nodes (genes) the same size as the input list.8 A p-value of < 10−30 was considered significant.
Statistical Analysis
All data are expressed as mean ± STDEV unless indicated otherwise. Distributions were tested for normality and statistical analysis was done by t-test (2 groups) and analysis of variance (ANOVA) (3 groups) for normally distributed data and Mann-Whitney (2 groups) and the Kruskal-Wallis test (3 groups) for non-normally distributed data using the Prism software, version 4.0.
Results
Time course of atherosclerotic lesion development in congenic mice
The time course of lesion progression for the Ath11 locus was determined. F1.Apoe−/− mice that were homozygous B6 (Chr10B6/B6), heterozygous (Chr10B6/FVB), and homozygous FVB (Chr10FVB/FVB) in the congenic interval were weaned at 4 weeks of age onto the semi-synthetic AIN76a diet containing 0.02 % cholesterol, and sacrificed for atherosclerotic lesion quantification at 6, 12, 16, 20 and 24 weeks of age (figure 1a, males and 1b, females). In general, atherosclerotic lesion area did not differ at any time point between F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10B6/FVB mice. In contrast, at each time point atherosclerotic lesion area was increased in F1.Apoe−/−Chr10FVB/FVB compared to F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10B6/FVB mice (for most comparisons p<0.001). This pertained even at 6 weeks of age, where lesion formation had begun at the aortic root in F1.Apoe−/−Chr10FVB/FVB mice but not in F1.Apoe−/−Chr10B6/B6 or F1.Apoe−/−Chr10B6/FVB mice. There were only minor, and probably not biologically significant, genotypic effects on plasma total- or HDL-cholesterol levels at any of the time points (Supplemental Table I). This time course study indicated that the gene(s) in Ath11 that differ between B6 and FVB operate very early in lesion development at the beginning of the foam cell or fatty streak stage, and that the effect persists through to 24 weeks of age when the lesions are much more complex.
Figure 1.
Effect of the proximal Chr10 interval (0–21 cM) on atherosclerosis development over time in F1.Apoe−/− congenic mice (a: males and b: females). Atherosclerotic lesion size was measured at 6, 12, 16, 20 and 24 weeks of age. The genetics of the proximal 0–21.9 cM of Chr10 are F1.Apoe−/−Chr10B6/B6, F1.Apoe−/−Chr10B6/FVB, and F1.Apoe−/−Chr10FVB/FVB indicated in the figure as B/B (black bars), B/F (grey bars), and F/F (white bars), respectively. The numbers of mice for each genotype and for each time point are shown at the bottom of the bars. Values represent means ± SEM.
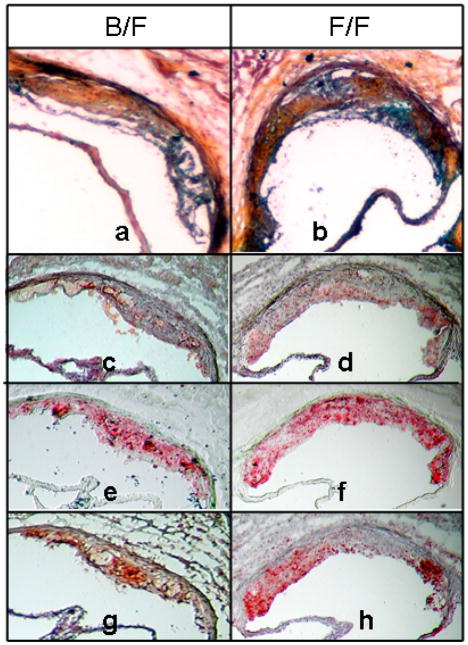
Morphology of atherosclerotic lesions in congenic mice
Aortic root lesion morphology was compared between F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10FVB/FVB mice sacrificed at 16 weeks of age. The results for male and female mice were similar and representative sections are shown for male mice in figure 2. Frozen aortic root sections were stained with Movat’s pentachrome to assess the general morphology of the lesions (figures 2a and 2b), immunostained with anti-CD68 to detect macrophages (figures 2c and 2d), stained with Oil Red O (ORO) to detect lipid (figures 2e and 2f), TUNEL stained for apoptotic cells (figures 2g and 2h), and immunostained with anti-CD31 and anti-α-actin to detect endothelial cells (ECs) and smooth muscle cells (SMCs) (data not shown). Lesion composition was quite similar between genotypes and consisted mainly of foam cells with occasional cholesterol clefts and/or necrotic core areas and beginning fibrous caps. TUNEL staining revealed apoptotic cells in macrophage rich regions in aortic root and brachiocephalic artery (data not shown) sections from mice with both genotypes.
Figure 2.
Frozen aortic root sections of male F1.Apoe−/−Chr10B6/FVB (left panels) and F1.Apoe−/−Chr10FVB/FVB congenic mice (right panels) sacrificed at 16 weeks of age. Panels a and b: Movats stain (100x), panels c and d: CD68 staining for macrophages (red), panels e and f: Oil Red-O staining for lipid (red), panels g and h: TUNEL staining for apoptosis (red). Panels c, e, and g show serial sections as do panels d, f, and h.
Gene expression analysis in the aorta of congenic mice
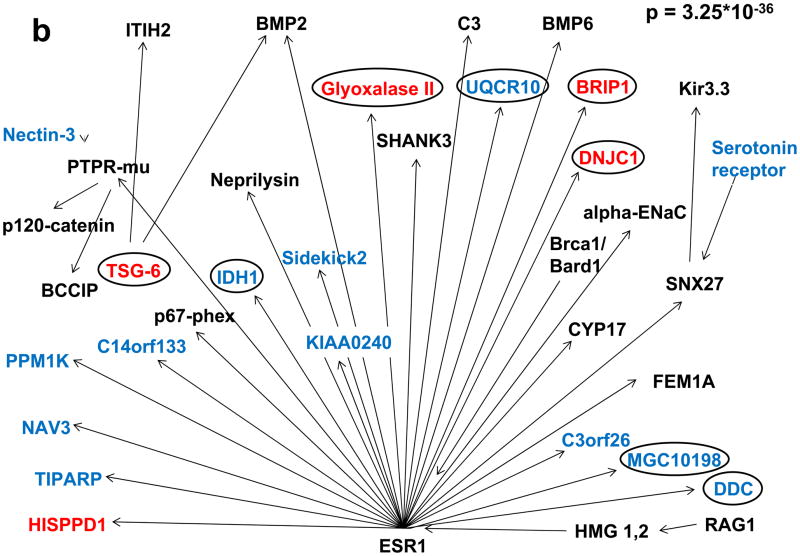
The gene expression pattern of aortas from 6 week old female congenic F1.Apoe−/−Chr10B6/B6, F1.Apoe−/−Chr10B6/FVB, and F1.Apoe−/−Chr10FVB/FVB mice were compared in a gene expression array. A comparison between F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10FVB/FVB mice revealed 354 genes with greater than two-fold expression differences, and between F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10FVB/FVB mice 253 genes. 97 genes were regulated in both comparisons and of these only Aldh8a1 maps within the congenic interval. The complete list of differentially regulated genes is provided in Supplemental Table IIa. Radioactive RT-PCR (n=6 per congenic genotype) with aortic tissue was used to confirm the microarray results for a subset of the 97 genes. The results of this confirmation study are presented in Supplemental Table IIb. Using the GO Slimmer Tool for both comparisons, the categories with the highest fraction of regulated genes were cellular and metabolic processes, regulation of biological processes, and developmental processes (see Supplemental Table IIc). This suggests that similar biological processes are underlying the decrease in atherosclerosis in both F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10B6/FVB mice compared to F1.Apoe−/−Chr10FVB/FVB mice. Due to the nature of the atherosclerotic process, regulated genes in the GO term categories for immune system processes, cell death pathways, and vascular processes in the circulatory system were identified comparing both F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10B6/FVB with F1.Apoe−/−Chr10FVB/FVB mice (Supplemental Table IId). Genes regulated in both comparisons that are present on a list of over 4000 human genes associated with cardiovascular processes compiled by the Cardiovascular GO Annotation Initiative are shown in table 1. Finally, the 354 genes regulated between F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10FVB/FVB mice and the 253 genes regulated between F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10FVB/FVB were analyzed using the GeneGO MetaCore database to identify gene networks linked to a single transcription factor. This analysis revealed two clusters of regulated genes; one driven by the transcription factor Hepatocyte Nuclear Factor 4 alpha (Hnf4α) and the other by the Estrogen Receptor alpha (Esr1) for both comparisons. The former is not encoded in the congenic interval, but the latter lies within the interval on chromosome 10 at 5.3–5.7 Mb. Figure 3 shows the networks retrieved from the comparison of F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10FVB/FVB mice. The respective networks from the comparison of F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10FVB/FVB (data not shown) were qualitatively similar with p-values of 2.11*10−61 and 4.42*10−21 for Hnf4α and Esr1 respectively.
Table 1.
Regulated Genes present on Cardiovascular GO Annotation initiative list
| Ensemble Gene ID | BB vs FF | BF vs FF | MGI symbol | Description |
|---|---|---|---|---|
| ENSMUSG00000027350 | −8.6 | −2.5 | Chgb | chromogranin B Gene |
| ENSMUSG00000025479 | −3.9 | −3.8 | Cyp2e1 | cytochrome P450, fam 2, subfam e, polypeptide 1 Gene |
| ENSMUSG00000071356 | 2.7 | 5.0 | Reg3b | regenerating islet-derived 3 beta Gene |
| ENSMUSG00000028245 | 2.1 | 2.6 | Nsmaf | neutral sphingomyelinase (N-SMase) activation associated factor Gene |
| ENSMUSG00000031710 | −2.1 | −3.0 | Ucp1 | uncoupling protein 1 (mitochondrial, proton carrier) Gene |
| ENSMUSG00000053475 | 3.4 | 4.3 | Tnfaip6 | tumor necrosis factor alpha induced protein 6 Gene |
| ENSMUSG00000029819 | −3.1 | −2.8 | Npy | neuropeptide Y Gene |
| ENSMUSG00000037705 | 4.1 | 4.1 | Tecta | tectorin alpha Gene |
| ENSMUSG00000043089 | 2.2 | 2.5 | Mmp1a | matrix metallopeptidase 1a (interstitial collagenase) Gene |
| ENSMUSG00000025950 | −2.3 | −2.0 | Idh1 | isocitrate dehydrogenase 1 (NADP+). soluble Gene |
| ENSMUSG00000022878 | −2.7 | −3.2 | Adipoq | adiponectin, C1Q and collagen domain containing Gene |
| ENSMUSG00000022952 | 7.9 | 5.1 | Runx1 | runt related transcription factor 1 Gene |
| ENSMUSG00000027513 | −2.2 | −4.1 | Pck1 | phosphoenolpyruvate carboxykinase 1, cytosolic Gene |
| ENSMUSG00000020182 | −5.0 | −10.8 | Ddc | dopa decarboxylase Gene |
| ENSMUSG00000035232 | 2.8 | 2.8 | Pdk3 | pyruvate dehydrogenase kinase, isoenzyme 3 Gene |
| ENSMUSG00000037542 | −2.6 | −10.2 | Aldh8a1 | aldehyde dehydrogenase 8 family, member A1 Gene |
| ENSMUSG00000041607 | −2.1 | −2.7 | Mbp | myelin basic protein Gene |
| ENSMUSG00000025855 | −2.2 | −2.4 | Prkar1b | protein kinase, cAMP dependent regulatory, type I beta |
| ENSMUSG00000027984 | 5.9 | 6.6 | Hadh | hydroxyacyl-Coenzyme A dehydrogenase Gene |
| ENSMUSG00000031392 | 2.4 | 2.0 | Irak1 | interleukin-1 receptor-associated kinase 1 Gene |
| ENSMUSG00000031722 | −4.0 | −4.8 | Hp | haptoglobin Gene |
| ENSMUSG00000052962 | 6.1 | 6.3 | Mrpl35 | mitochondrial ribosomal protein L35 Gene |
| ENSMUSG00000000889 | −2.5 | −2.0 | Dbh | dopamine beta hydroxylase Gene |
| ENSMUSG00000029486 | 3.4 | 3.4 | Mrpl1 | mitochondrial ribosomal protein L1 Gene |
| ENSMUSG00000058258 | −2.2 | −2.2 | Idi1 | isopentenyl-diphosphate delta isomerase Gene |
| ENSMUSG00000038641 | 13.3 | 3.3 | Akr1d1 | aldo-keto reductase family 1, member D1 Gene |
Figure 3.
Transcription factor analysis of genes for which expression in aortic tissue differed by more than 2 fold between F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/− Chr10FVB/FVB congenic mice, utilizing the GeneGo MetaCore database, detected two significant clusters of differentially regulated genes driven by (a) Hnf4α and (b) Esr1. Genes colored in blue were down- and genes colored in red up-regulated in F1.Apoe−/−Chr10B6/B6 vs. F1.Apoe−/−Chr10FVB/FVB congenic mice. Unregulated genes of the network are shown in black. Circles mark genes that were regulated in a similar manner when F1.Apoe−/−Chr10B6/FVB and F1.Apoe−/−Chr10FVB/FVB congenic mice were compared.
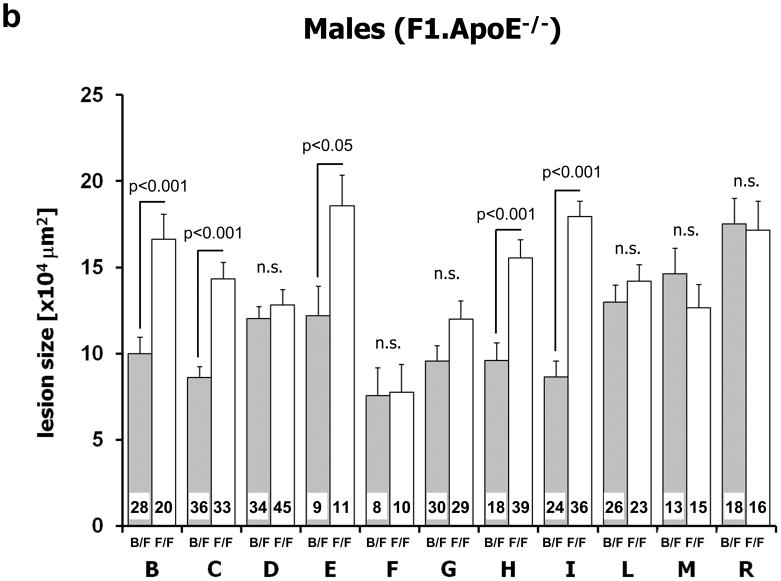
Narrowing the congenic interval by studies of subcongenic mice
Ath11 (0 to 21.9 cM interval), which was confirmed in congenic mice and for which the B6 allele displays a dominant atheroprotective effect, contains ~380 genes. To narrow this interval 11 subcongenic strains were created and aortic root atherosclerotic lesion area assessed in 957 mice. As shown in figure 4, the interval is complex. The proximal portion (region 10a) indicates a B6 atheroprotective gene detectable only in female mice and the distal portion (region 10b) another B6 atheroprotective gene detectable in both genders. The 10a region is defined by subcongenics D (0-rs50156646) and G (0-rs4228112), which showed decreased atherosclerosis lesion area in F1.Apoe−/−Chr10B6/FVB vs. F1.Apoe−/−Chr10FVB/FVB* female mice, and subcongenics L (rs29349441-rs50156646) and R (rs29349441-rs13480506), which showed equal lesion area in F1.Apoe−/−Chr10B6/FVB vs. F1.Apoe−/−Chr10FVB/FVB* mice of both genders. This places the culprit gene in the 10a region between 0 and rs29349441 (see figure 4a), a region spanning 7.3 Mb and containing 21 genes including Esr1 (Table 2a). As shown in figure 4, the 10b region is defined by subcongenic strain I (JMR2001 rs52653661), which showed decreased atherosclerosis lesion area in F1.Apoe−/−Chr10B6/FVB vs. F1.Apoe−/−Chr10FVB/FVB* mice of both genders. Thus the culprit gene in the 10b region resides in a considerably narrowed 1.8 Mb interval containing 7 genes (Table 2b). The interval between regions 10a and 10b was excluded because of equal lesion area in F1.Apoe−/−Chr10B6/FVB vs. F1.Apoe−/−Chr10FVB/FVB* male and female mice in subcongenic strains L (rs29349441-rs50156646), M (rs4228112-rs50156646), and R (rs29349441-rs13480506). The interval distal to subcongenic strain I was excluded as carrying a locus with a major lesion size effect because of equal lesion area in subcongenic strain F (12–21.9cM).
Figure 4.
Analysis of atherosclerosis lesion size in subcongenic strains. Panel a: Schematic illustration of the subcongenic strains on an F1 background. The horizontal bar indicates the extent of the genomic interval for each subcongenic strain. The dotted lines at either side of a bar indicate the region in which the recombination occurred. A black bar describes a subcongenic strain that showed increased lesion area in FF vs. BF mice, a white bar a strain that did not, and a gray bar a strain that showed increased lesion area only when female FF vs BF mice were compared. Panels b (males) and c (females): Effect of different proximal Chr10 intervals on atherosclerosis lesion size. For each subcongenic strain the gray column indicates heterozygousity (B/F) and the white column homozygousity (F/F) in the interval. At the bottom of each column the numbers of mice studied with that genotype are indicated. Values represent means ± SEM.
Table 2.
Table 2a: Genes in Chromosome 10a region
Table 2b: Genes in Chromosome 10b region
| Description | symbol | position on chr10 | Ori |
|---|---|---|---|
| Cnksr family member 3 | Cnksr3 | 3,134,304–3,227,479 | (+) |
| interaction protein for cytohesin exchange factors 1 | Ipcef1 | 3,294,060–3,460,745 | (+) |
| opioid receptor, mu 1 | Oprm1 | 3,308,332–3,587,948 | (−) |
| regulator of G-protein signaling 17 | Rgs17 | 4,424,175–4,514,372 | (+) |
| mitochondrial translational release factor 1-like | Mtrf1l | 4,522,631–4,533,459 | (+) |
| F-box protein 5 Gene | Fbxo5 | 4,541,119–4,546,998 | (+) |
| vasoactive intestinal polypeptide | Vip | 4,698,927–4,707,323 | (−) |
| myc target 1 | Myct1 | 4,740,217–4,752,745 | (−) |
| synaptic nuclear envelope 1 | Syne1 | 4,795,849–5,326,338 | (+) |
| estrogen receptor 1 (alpha) | Esr1 | 5,340,927–5,734,552 | (−) |
| AC158617.9 | AC158617.9 | 5,785,430–5,833,846 | (−) |
| RIKEN cDNA 1700052N19 | 1700052N19Rik | 5,891,401–5,914,028 | (−) |
| required for meiotic nuclear division 1 homolog (S. cerevisiae) | Rmnd1 | 5,914,207–5,943,366 | (+) |
| zinc finger and BTB domain containing 2 | Zbtb2 | 5,958,433–5,979,459 | (+) |
| A kinase (PRKA) anchor protein (gravin) 12 | Akap12 | 5,987,073–6,080,178 | (−) |
| methylenetetrahydrofolate dehydrogenase (NADP+ dep) 1-like | Mthfd1l | 6,179,460–6,373,428 | (−) |
| pleckstrin homology domain containing, family G member 1 | Plekhg1 | 6,379,238–6,606,177 | (−) |
| iodotyrosine deiodinase Gene | Iyd | 6,791,664–6,806,285 | (−) |
| protein phosphatase 1, regulatory (inhibitor) subunit 14c | Ppp1r14c | 6,881,570–6,980,484 | (−) |
| RIKEN cDNA 9230019H11 | 9230019H11Rik | 7,216,995–7,226,702 | (−) |
| low density lipoprotein receptor-related protein 11 | Lrp11 | 7,309,659–7,345,263 | (+) |
| Description | symbol | position on chr10 | Ori |
|---|---|---|---|
| phosphodiesterase 7B Gene | Pde7b | 20,117,810–20,444,884 | (−) |
| Abelson helper integration site 1 | Ahi1 | 20,672,465–20,800,234 | (+) |
| myeloblastosis oncogene | Myb | 20,844,736–20,880,790 | (−) |
| Hbs1-like (S. cerevisiae) | Hbs1l | 21,015,823–21,088,689 | (+) |
| aldehyde dehydrogenase 8 family, member A1 | Aldh8a1 | 21,097,106–21,116,384 | (+) |
| serum/glucocorticoid regulated kinase 1 | Sgk1 | 21,601,990–21,719,704 | (+) |
| retinoic acid early transcript 1E | Raet1e | 21,878,375–22,093,945 | (+) |
Genes highlighted in gray are present on the Cardiovascular GO Annotation initiative’s list
Total and HDL cholesterol levels were determined for all of the subcongenic strains and there were no significant differences between F1.Apoe−/−Chr10B6/FVB vs. F1.Apoe−/−Chr10FVB/FVB* male or female mice (Supplemental Table III).
SNP analysis
The actual amount of functional protein present of a certain gene does not only depend on expression but also on efficacy of splicing and translation as well as coding region differences. Therefore the amount of functional protein can be altered by SNPs in critical regions of a gene such as intronic splice sites, 3- or 5-prime UTRs and the coding region. We searched the Ensembl database for polymorphic (B6 vs. FVB) SNPs in the 10a (coding region only) and 10b region genes. This analysis identified coding SNPs in Oprm1, Myct1, Syne1, 9230019H11Rik and Esr1 (Supplemental Table IVa) in the 10a region. Polymorphic SNPs in the 10b region are found in regions that can affect expression of Ahi1, Myb, Hbsl1, Aldh8a1, Sgk1 and Raet1 including one potential non-synonymous SNP each in Myb and Hbs1l. A detailed list of all 10b region SNPs is given in Supplemental Table IVb.
Discussion
Previously we identified and confirmed in congenic mice Ath11, a region on proximal Chr10 (0 to 21.9 cM) containing 382 genes, in which heterozygosity or homozygosity for the B6 allele decreased and homozygosity for the FVB allele increased atherosclerosis. Here we present the time course of lesion development, lesion morphology, and aortic gene expression microarray analysis for F1.Apoe−/− congenic mice. We also present aortic root lesion area analysis for 11 subcongenic strains, which revealed the Ath11 QTL to be complex with two atherosclerosis susceptibility regions; 10a from 0 to 7.3 Mb containing 21 genes present only in females, and 10b from 20.1 to 21.9 Mb containing 7 genes present in both genders.
Analysis of lesion development at 6, 12, 16, 20 and 24 weeks of age in F1.Apoe−/−mice that were Chr10B6/B6, Chr10B6/FVB, and Chr10FVB/FVB in the congenic interval revealed significant differences in lesion area even at 6 weeks between Chr10B6/B6 and Chr10B6/FVB congenics versus Chr10FVB/FVB congenics that persisted to 24 weeks. This suggests that the culprit gene(s) act early in lesion development at stages that might include activation of the endothelium, monocyte attachment to the endothelium, monocyte migration to the subendothelial space, and subendothelial foam cell formation, rather than later events such as foam cell death, vascular smooth muscle cell migration, or fibrous cap formation. Since there were no biologically significant differences in total and HDL cholesterol between the congenics throughout the time course, this suggests the culprit gene(s) are involved in vessel wall biology rather than cholesterol metabolism.
The survey of aortic gene expression at 6 weeks of age in female congenic mice revealed 97 genes with >2-fold expression differences between both F1.Apoe−/−Chr10B6/B6 and F1.Apoe−/−Chr10B6/FVB vs. F1.Apoe−/−Chr10FVB/FVB mice (Supplemental Table IIa). Searching for the presence of these 97 regulated genes in the Cardiovascular GO Annotation Initiative list revealed 26 genes (Table 1), including Aldh8a1, that is present in the 10b interval (Tables 1 and 2b). Finally, analysis of the regulated genes using the GeneGO MetaCore database revealed two clusters of regulated genes; one driven by Hnf4α, which is not encoded in the congenic interval, and the other by Esr1 encoded in the 10a region.
Hnf4α belongs to the nuclear hormone receptor superfamily and is involved in metabolism, diabetes and liver development.9 It does not appear to be ligand activated, but activity can be affected by phosphorylation, co-activator recruitment, and interaction with other nuclear hormone receptors.9 It is possible that the culprit gene of Ath11 modifies Hnf4α in this manner.
Esr1 is a ligand activated nuclear hormone receptor that mediates the transcriptional effects of estrogen. Epidemiological studies strongly suggest that estrogens protect premenopausal women from coronary heart disease.10 In mice the atheroprotective effect of estradiol was demonstrated in Apoe−/−11 and Ldlr−/−mice.12 There are two estrogen receptors, Esr1 and Esr2, but the major protective effect is mediated through Esr1, as shown by the failure of estradiol to diminish atherosclerosis lesion size in Esr1 knockout ovariectomized Apoe−/−mice.13 Esr1 is present in many important cell types involved in atherosclerosis including ECs, VSMCs, macrophages and T lymphocytes. Since Esr1 is encoded in the 10a region, it becomes an interesting candidate gene.
The classical genetic strategy of narrowing the original congenic interval (0 to 21.9 cM) by creating subcongenic strains was used. Previous studies showed the effect of the congenic interval was easiest to discern on the F1 background.6 Therefore, all eleven subcongenic strains, each containing a portion of the congenic interval, were phenotyped on the F1 background. Utilizing this strategy we elucidated two non-overlapping regions, 10a and 10b. The gene for A20, which is a prominent regulator of NFkB mediated inflammation and located at 5.5 cM, was originally considered a promising candidate gene.7 However, it lies between the 10a and 10b regions and is ruled out by the subcongenic analysis.
The 10a region is female specific and contains 21 genes, including Esr1. The finding that Esr1 was not differentially expressed in aortas from female mice, does not exclude it as the culprit gene in the 10a region. There are many SNPs, including two synonymous coding SNPs (listed in Supplemental Table 4a) in and near this gene that differ between B6 and FVB and could modify Esr1 function for example by influencing translational efficacy. It is also possible that the criterion of 2-fold expression difference between the strains was too stringent, and lesser differences in expression of Esr1 might be physiologically significant. In fact, Esr1 is 1.8 fold up regulated in F1.Apoe−/−Chr10B6/B6 compared to F1.Apoe−/−Chr10FVB/FVB mice. Other genes in the 10a region must also be considered. Besides Esr1 the Cardiovascular GO Annotation Initiative lists six other 10a region genes (highlighted in gray in Table 2a) and there are coding SNPs in Oprm1, Myct1, Syne1 and 9230019H11Rik. Even though Esr1 seems to be a very promising candidate, further analysis, including sequencing, expression and functional studies of the genes in the 10a region, will be required to ultimately determine the culprit gene.
The 10b region is gender independent, spans 1.8 Mb, and contains 7 genes, from proximal to distal Pde7b, Ahi1, Myb, Hbs1l, Aldh8a1, Sgk1, and Raet1 (see Table 2b). Based on aortic microarray expression data, Aldh8a1 is the only one of these genes that is differentially expressed between congenic strains, whereas Myb and Hbs1l are the only genes that contain potential non-synonymous SNPs. However, there are SNPs present in important regions of other genes as well that could alter their functionality. Additional studies, confirming the presence of these SNPs and determining which are functional and causative in atherosclerosis will be needed for each of these genes to identify the culprit gene of the 10b region.
In summary, the congenic region representing Ath11 appears to act early in lesion formation with altered aortic expression of clusters of genes regulated by the transcription factors Hnf4α and Esr1. Moreover, subcongenic strain analysis of Ath11 reveals two regions influencing atherosclerosis susceptibility, one in females containing 21 genes (10a) and the other gender independent containing 7 genes (10b).
Supplementary Material
Acknowledgments
b) This study was supported by the NIH PO1HL54591 Program Project 4/07/2006-3/31/2011 Project 1 (to J. L. B.) and a grant of the Deutsche Forschungsgemeinschaft (to D. T.; DFG Th374/1-1).
Footnotes
c) All authors have no disclosures.
References
- 1.Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 2.Dansky HM, Charlton SA, Sikes JL, Heath SC, Simantov R, Levin LF, Shu P, Moore KJ, Breslow JL, Smith JD. Genetic background determines the extent of atherosclerosis in ApoE-deficient mice. Arterioscler Thromb Vasc Biol. 1999;19:1960–1968. doi: 10.1161/01.atv.19.8.1960. [DOI] [PubMed] [Google Scholar]
- 3.Teupser D, Persky AD, Breslow JL. Induction of atherosclerosis by low-fat, semisynthetic diets in LDL receptor-deficient C57BL/6J and FVB/NJ mice: comparison of lesions of the aortic root, brachiocephalic artery, and whole aorta (en face measurement) Arterioscler Thromb Vasc Biol. 2003;23:1907–1913. doi: 10.1161/01.ATV.0000090126.34881.B1. [DOI] [PubMed] [Google Scholar]
- 4.Dansky HM, Shu P, Donavan M, Montagno J, Nagle DL, Smutko JS, Roy N, Whiteing S, Barrios J, McBride TJ, Smith JD, Duyk G, Breslow JL, Moore KJ. A phenotype-sensitizing Apoe-deficient genetic background reveals novel atherosclerosis predisposition loci in the mouse. Genetics. 2002;160:1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teupser D, Tan M, Persky AD, Breslow JL. Atherosclerosis quantitative trait loci are sex- and lineage-dependent in an intercross of C57BL/6 and FVB/N low-density lipoprotein receptor-/- mice. Proc Natl Acad Sci U S A. 2006;103:123–128. doi: 10.1073/pnas.0509570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teupser D, Wolfrum S, Tan M, Persky AD, Dansky HM, Breslow JL. Novel strategy using F1-congenic mice for validation of QTLs: studies at the proximal chromosome 10 atherosclerosis susceptibility locus. Arterioscler Thromb Vasc Biol. 2009;29:678–683. doi: 10.1161/ATVBAHA.108.179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfrum S, Teupser D, Tan M, Chen KY, Breslow JL. The protective effect of A20 on atherosclerosis in apolipoprotein E-deficient mice is associated with reduced expression of NF-kappaB target genes. Proc Natl Acad Sci U S A. 2007;104:18601–18606. doi: 10.1073/pnas.0709011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet. 2008;23:2–7. doi: 10.2133/dmpk.23.2. [DOI] [PubMed] [Google Scholar]
- 10.Hsia J, Criqui MH, Herrington DM, Manson JE, Wu L, Heckbert SR, Allison M, McDermott MM, Robinson J, Masaki K. Conjugated equine estrogens and peripheral arterial disease risk: the Women’s Health Initiative. Am Heart J. 2006;152:170–176. doi: 10.1016/j.ahj.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Elhage R, Arnal JF, Pieraggi MT, Duverger N, Fievet C, Faye JC, Bayard F. 17 beta-estradiol prevents fatty streak formation in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 1997;17:2679–2684. doi: 10.1161/01.atv.17.11.2679. [DOI] [PubMed] [Google Scholar]
- 12.Marsh MM, Walker VR, Curtiss LK, Banka CL. Protection against atherosclerosis by estrogen is independent of plasma cholesterol levels in LDL receptor-deficient mice. J Lipid Res. 1999;40:893–900. [PubMed] [Google Scholar]
- 13.Hodgin JB, Krege JH, Reddick RL, Korach KS, Smithies O, Maeda N. Estrogen receptor alpha is a major mediator of 17beta-estradiol’s atheroprotective effects on lesion size in Apoe−/− mice. J Clin Invest. 2001;107:333–340. doi: 10.1172/JCI11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.