Abstract
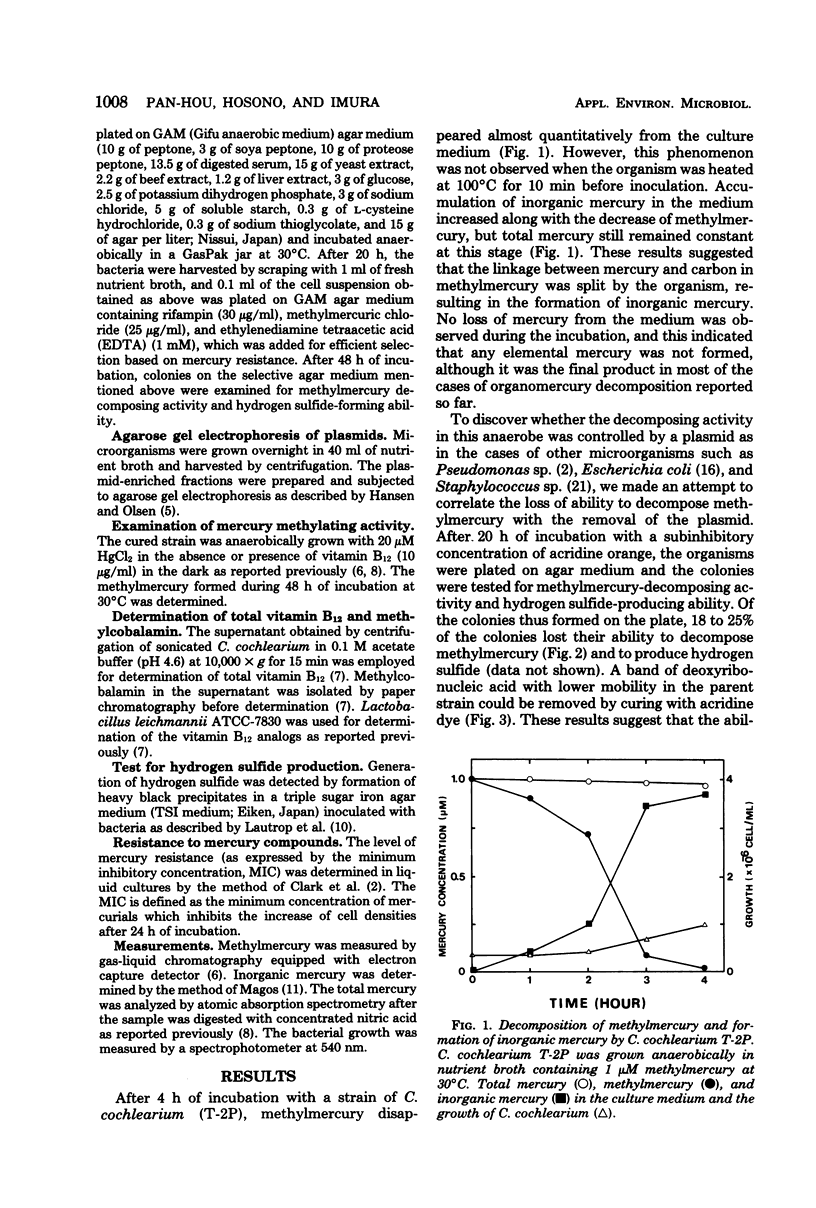
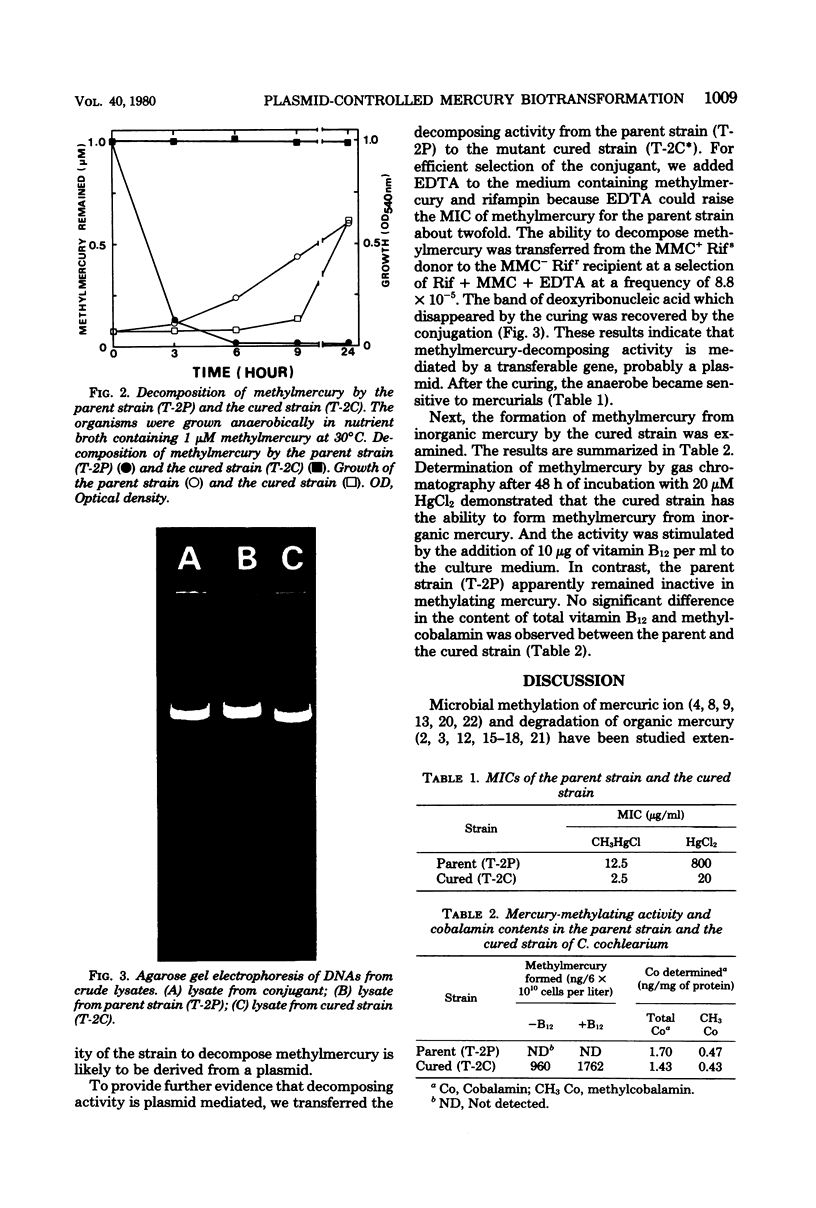
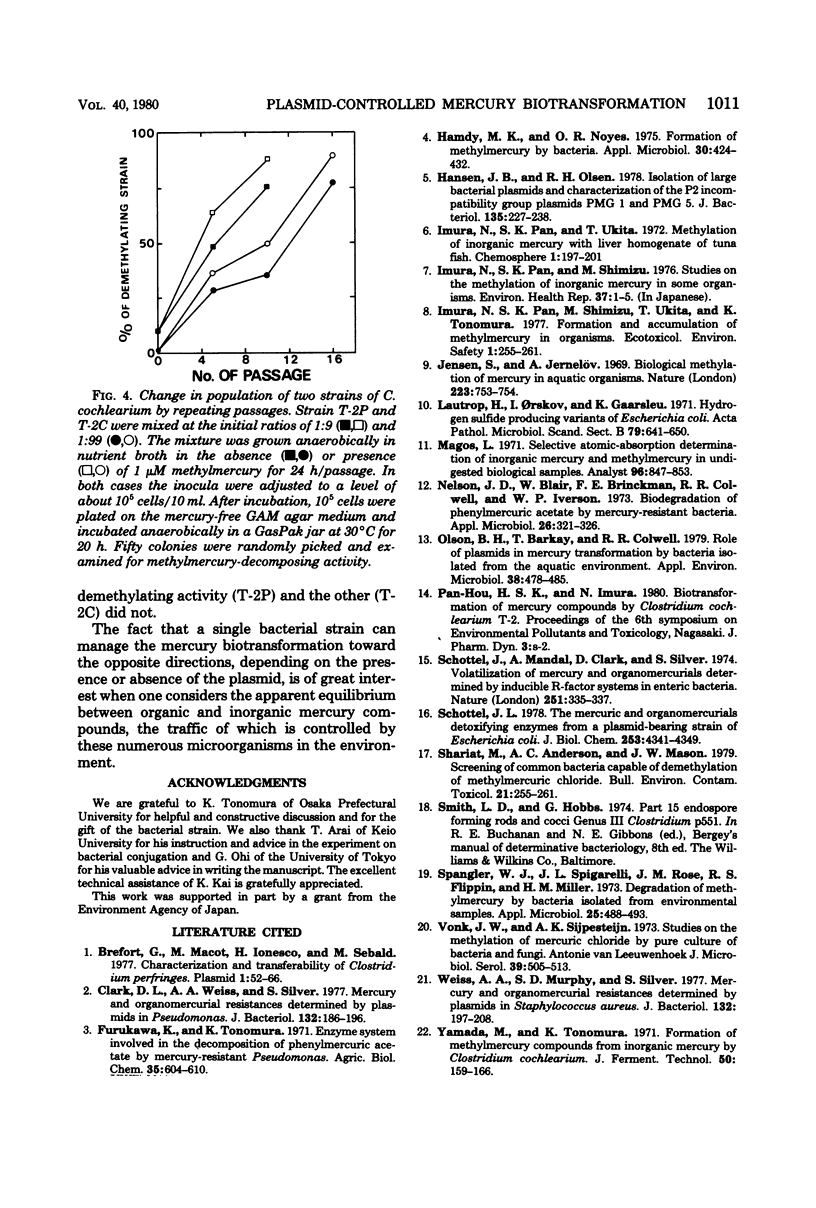
A strain of Clostridium cochlearium having methylmercury-decomposing ability was isolated. The ability was cured by the treatment with acridine dye and recovered by the conjugation of the cured strain with the parent strain. The cured strain then showed the activity to methylate mercuric ion as previously reported (M. Yamada and K. Tonomura, J. Ferment. Technol. 50:159-166, 1971). These results and the agarose gel electrophoretic pattern of the deoxyribonucleic acids from the lysates indicate a possible role of plasmids in controlling the mercury biotransformation of the two opposite directions in a single bacterial strain: methylation in the absence of the plasmid and demethylation in the presence of it. A possible mechanism for mercury resistance involving hydrogen sulfide is discussed.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brefort G., Magot M., Ionesco H., Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977 Nov;1(1):52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- Clark D. L., Weiss A. A., Silver S. Mercury and organomercurial resistances determined by plasmids in Pseudomonas. J Bacteriol. 1977 Oct;132(1):186–196. doi: 10.1128/jb.132.1.186-196.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy M. K., Noyes O. R. Formation of methyl mercury by bacteria. Appl Microbiol. 1975 Sep;30(3):424–432. doi: 10.1128/am.30.3.424-432.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura N., Pan S. K., Shimizu M., Ukita T., Tonomura K. Formation and accumulation of methylmercury in organisms. Ecotoxicol Environ Saf. 1977 Sep;1(2):255–261. doi: 10.1016/0147-6513(77)90040-9. [DOI] [PubMed] [Google Scholar]
- Jensen S., Jernelöv A. Biological methylation of mercury in aquatic organisms. Nature. 1969 Aug 16;223(5207):753–754. doi: 10.1038/223753a0. [DOI] [PubMed] [Google Scholar]
- Lautrop H., Orskov I., Gaarslev K. Hydrogensulphide producing variants of Escherichia coli. Acta Pathol Microbiol Scand B Microbiol Immunol. 1971;79(5):641–650. doi: 10.1111/j.1699-0463.1971.tb00092.x. [DOI] [PubMed] [Google Scholar]
- Magos L. Selective atomic-absorption determination of inorganic mercury and methylmercury in undigested biological samples. Analyst. 1971 Dec;96(149):847–853. doi: 10.1039/an9719600847. [DOI] [PubMed] [Google Scholar]
- Nelson J. D., Blair W., Brinckman F. E., Colwell R. R., Iverson W. P. Biodegradation of phenylmercuric acetate by mercury-resistant bacteria. Appl Microbiol. 1973 Sep;26(3):321–326. doi: 10.1128/am.26.3.321-326.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson B. H., Barkay T., Colwell R. R. Role of plasmids in mercury transformation by bacteria isolated from the aquatic environment. Appl Environ Microbiol. 1979 Sep;38(3):478–485. doi: 10.1128/aem.38.3.478-485.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Shariat M., Anderson A. C., Mason J. W. Screening of common bacteria capable of demethylation of methylmercuric chloride. Bull Environ Contam Toxicol. 1979 Jan;21(1-2):255–261. doi: 10.1007/BF01685420. [DOI] [PubMed] [Google Scholar]
- Spangler W. J., Spigarelli J. L., Rose J. M., Flippin R. S., Miller H. H. Degradation of methylmercury by bacteria isolated from environmental samples. Appl Microbiol. 1973 Apr;25(4):488–493. doi: 10.1128/am.25.4.488-493.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk J. W., Sijpesteijn A. K. Studies on the methylation of mercuric chloride by pure cultures of bacteria and fungi. Antonie Van Leeuwenhoek. 1973;39(3):505–513. doi: 10.1007/BF02578894. [DOI] [PubMed] [Google Scholar]
- Weiss A. A., Murphy S. D., Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977 Oct;132(1):197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]



