SUMMARY
Formation of the vertebrate neuromuscular junction requires, among others proteins, Agrin, a neuronally derived ligand, and the following muscle proteins: LRP4, the receptor for Agrin; MuSK, a receptor tyrosine kinase (RTK); and Dok7, a cytoplasmic adapter protein. Dok7 comprises a pleckstrin-homology (PH) domain, a phosphotyrosine-binding (PTB) domain, and C-terminal sites of tyrosine phosphorylation. Unique among adapter proteins recruited to RTKs, Dok7 is not only a substrate of MuSK but also an activator of MuSK’s kinase activity. Here, we present the crystal structure of the Dok7 PH-PTB domains in complex with a phosphopeptide representing the Dok7 binding site on MuSK. The structure and biochemical data reveal a dimeric arrangement of Dok7 PH-PTB that facilitates trans-autophosphorylation of the kinase activation loop. The structure provides the molecular basis for MuSK activation by Dok7 and for rationalizing several Dok7 loss-of-function mutations found in patients with congenital myasthenic syndromes.
INTRODUCTION
Neuromuscular junction (NMJ) formation requires an elaborate exchange of signals between motor neurons and muscle fibers, culminating in a highly specialized postsynaptic muscle membrane and a differentiated nerve terminal (Burden, 2002; Kummer et al., 2006). One key signal is Agrin, a ligand supplied by motor neurons. Agrin binds to LRP4 (low-density lipoprotein receptor-related protein-4) expressed in muscle (Kim et al., 2008; Zhang et al., 2008), leading to activation of MuSK (muscle-specific kinase), a receptor tyrosine kinase (RTK) that plays a central signaling role in NMJ formation (Glass and Yancopoulos, 1997). Activated MuSK results in clustering of essential postsynaptic proteins, including acetylcholine receptors (AChRs) and the AChR-associated protein Rapsyn; synapse-specific transcription; and the generation of critical retrograde signals that regulate presynaptic differentiation. MuSK, LRP4, Rapsyn, and AChRs are pre-clustered in the muscle endplate prior to innervation, and their clustering is refined and sharpened upon release of Agrin from the developing nerve terminal (Kummer et al., 2006). Failure to properly cluster AChRs is responsible for multiple disorders in neuromuscular transmission, including myasthenia gravis and congenital myasthenic syndromes (CMS) (Beeson et al., 2008; Engel and Sine, 2005).
The cytoplasmic adapter-protein Dok7 is also essential for NMJ formation. Dok7−/− mice die at birth, as do Agrin−/− and MuSK−/− mice, because they lack NMJs and are unable to move or breathe (Okada et al., 2006). Dok7 is required for both MuSK-mediated pre-clustering of AChRs and for Agrin-stimulated AChR clustering upon innervation (Inoue et al., 2009; Okada et al., 2006).
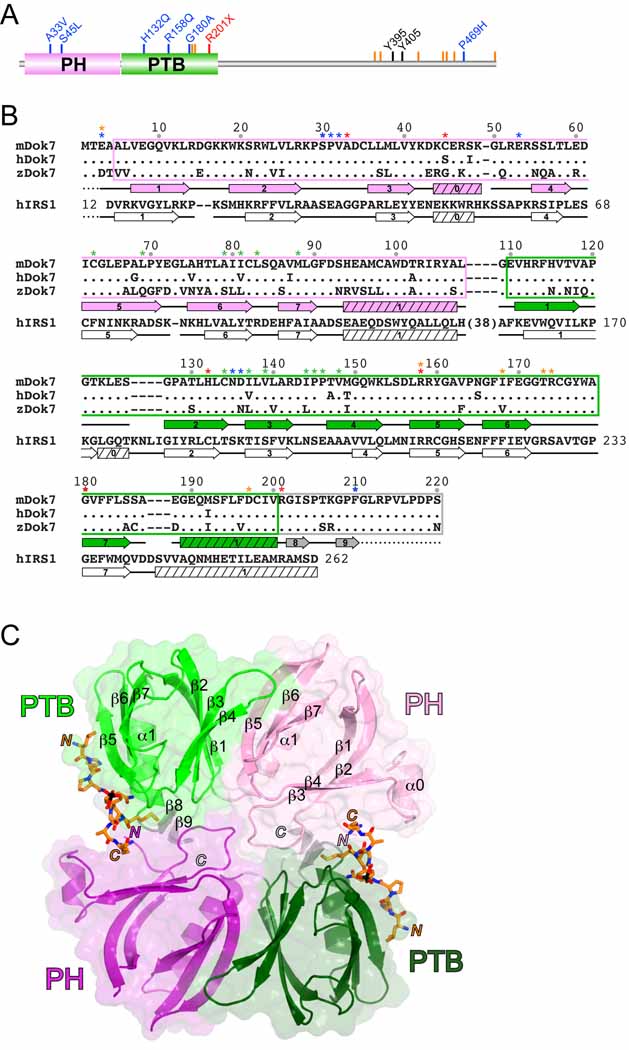
Dok7 (504 residues, human) is a member of a family of adapter proteins that includes Dok1-7 and insulin receptor substrates 1–4 (IRS1-4) (Cai et al., 2003). Members of this protein family are characterized by N-terminal pleckstrin-homology (PH) and phosphotyrosine-binding (PTB) domains, followed by an extended C-terminal region with multiple sites of tyrosine phosphorylation. Recessive mutations throughout Dok7 are causative for CMS (Beeson et al., 2008) (Figure 1A).
Figure 1. Crystal Structure of the Dok7(PH-PTB)-MuSK(pTyr553) Complex.
(A) Domain architecture of human Dok7 drawn to linear scale (504 residues). The abbreviations are: PH, pleckstrin-homology (domain), and PTB, phosphotyrosine-binding (domain). The positions of CMS mutations are shown by blue (missense), red (nonsense), or orange (frame shift) lines and labels. The tyrosine phosphorylation sites are shown in black.
(B) The sequences for the PH and PTB domains of mouse, human, and zebrafish Dok7 are shown along with the corresponding sequence for human IRS1. Residues in human and zebrafish Dok7 that are identical to those in mouse Dok7 are represented by a period. The PH domain is boxed in pink, the PTB domain is boxed in green, and the C-terminal extension to the PTB domain is boxed in gray, consistent with the coloring in Figure 1. Residue numbering is identical for mouse, human, and zebrafish Dok7. Residue numbering for human IRS1 is in-line with the sequence. Secondary-structure elements (α helices and β strands) for mouse Dok7(PH-PTB), as determined by PROCHECK (Laskowski et al., 1993), appear below the Dok7 sequences, and those for human IRS1(PH-PTB) (PDB code 1QQG) (Dhe-Paganon et al., 1999) appear below the IRS1 sequence. A dashed line indicates a disordered region. Residues denoted with a blue asterisk mediate Dok7(PH-PTB) dimerization, with a green asterisk mediate PH-PTB interactions, with an orange asterisk mediate MuSK pTyr553 binding, and with a red asterisk are CMS mutations.
(C) Structure of the Dok7(PH-PTB)-MuSK(pTyr553) complex. The two Dok7(PH-PTB) protomers in the asymmetric unit are shown in ribbon representation, with one protomer colored pink (PH domain) and light green (PTB domain) and the second protomer colored purple (PH domain) and dark green (PTB domain). The C-terminal extension is colored gray in both copies. The molecular two-fold axis passes through the center, perpendicular to the plane of the figure. A semi-transparent molecular surface is shown for each PH-PTB protomer. The MuSK pTyr553 phosphopeptide is shown in stick representation with carbon atoms colored orange, oxygen atoms red, nitrogen atoms blue, sulfur atoms yellow, and phosphorus atoms black. Secondary-structure elements (α helices and β strands) for one of the PH-PTB protomers are labeled. The N- and C-termini of the two copies of the peptide and PH-PTB are labeled by ‘N’ and ‘C’ in the appropriate colors.
MuSK undergoes autophosphorylation (in trans) on Tyr553 in the juxtamembrane region (between the transmembrane helix and the tyrosine kinase domain) and on three tyrosines in the activation loop of the kinase domain: Tyr750, Tyr754, and Tyr755 (Till et al., 2002; Watty et al., 2000). Autophosphorylation of the MuSK activation loop is critical for increased catalytic efficiency (kinase activation) and biological function (Herbst and Burden, 2000; Till et al., 2002). Autophosphorylation of Tyr553 in the juxtamembrane region of MuSK is also critical for kinase activation: a MuSK Y553F mutant does not undergo activation-loop autophosphorylation in muscle cells (Herbst and Burden, 2000).
Tyr553 resides in an NPXY (X, any residue) sequence motif that, when phosphorylated, binds the PTB domains of Dok7 (Okada et al., 2006) and ShcD (Jones et al., 2007). In contrast to all other known PTB domain-containing proteins recruited to phosphorylated RTKs, Dok7 serves not only as a substrate of MuSK, on Tyr395/405 (Hamuro et al., 2008), but also as an activator of MuSK’s catalytic (kinase) activity—in effect, a cytoplasmic ligand. MuSK is poorly activated by Agrin in myotubes lacking Dok7, and co-transfection of Dok7 and MuSK in non-muscle cells leads to robust MuSK phosphorylation independent of Agrin and LRP4 (Inoue et al., 2009; Okada et al., 2006).
To understand the molecular mechanism by which Dok7 activates MuSK from the “inside” (cytoplasmically), we determined the crystal structure of the Dok7 PH and PTB domains in complex with a 13-residue phosphopeptide representing pTyr553 in MuSK. The structure reveals that Dok7 PH-PTB forms an integrated, dimeric structural unit, which dimerizes MuSK and facilitates trans-autophosphorylation of tyrosines in the MuSK activation loop.
RESULTS
Overall Structure of Dok7(PH-PTB)-MuSK(pTyr553)
Based on sequence conservation among Dok7 proteins from different species (Figure 1B), we expressed in E. coli mouse Dok7, residues 1–220 (referred to below as Dok7[PH-PTB]), which includes the PH domain, the PTB domain, and a Dok7-conserved region C-terminal to the PTB domain (residues 200–220). We crystallized Dok7(PH-PTB) with a 13-residue phosphopeptide representing pTyr553 in MuSK (juxtamembrane region), determined the structure by single anomalous diffraction (SAD) phasing of selenomethionyl-substituted protein crystals, and refined the structure at 2.6 Å resolution. Data collection and refinement statistics appear in Table 1.
Table 1.
X-ray Data Collection and Refinement Statistics
| Data collection | |
|---|---|
| Resolution (Å) | 50-2.6 |
| Observations | 890,683 |
| Unique reflections | 75,848 |
| Redundancy | 11.7 |
| Completeness (%) | 100.0 (100.0)1 |
| Rsym2 (%) | 9.8 (35.7)1 |
| <I/σI> | 10.0 (4.8)1 |
| Refinement | |
| Resolution (Å) | 50-2.6 |
| Reflections | 37,533 |
| Rcryst3 / Rfree (%) | 25.6 / 30.3 |
| Rmsd bond lengths (Å) | 0.007 |
| Rmsd bond angles (°) | 1.09 |
| Rmsd B-factors4 (Å2) (main/side chain) | 0.5 / 0.8 |
| Average B-factors (Å2) | |
| Protein (6322 atoms) | 37.4 |
| Phosphopeptide (320 atoms) | 40.0 |
| Solvent (78 atoms) | 35.3 |
Value in parentheses is for the highest resolution shell: 2.69-2.60 Å.
Rsym = 100 × Σ |I – 〈I〉| / Σ I.
Rcryst = 100 × Σ ∥Fo| – |Fc∥ / Σ |Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree determined from 5% of the data.
For bonded protein atoms.
The asymmetric unit contains four Dok7(PH-PTB)-phosphopeptide complexes, arranged as a dimer of dimers (D2 symmetry). Each of the two dimer interfaces buries ~2,000 Å2 of total surface area, whereas the dimer-dimer interface buries ~1,200 Å2. Because the two (presumptive) phosphoinositide binding sites in the PH domains face the same direction in each dimer, but the two pairs of sites face opposite directions in the tetramer, we conclude that the dimer (Figure 1C) and not the tetramer is of potential physiological significance.
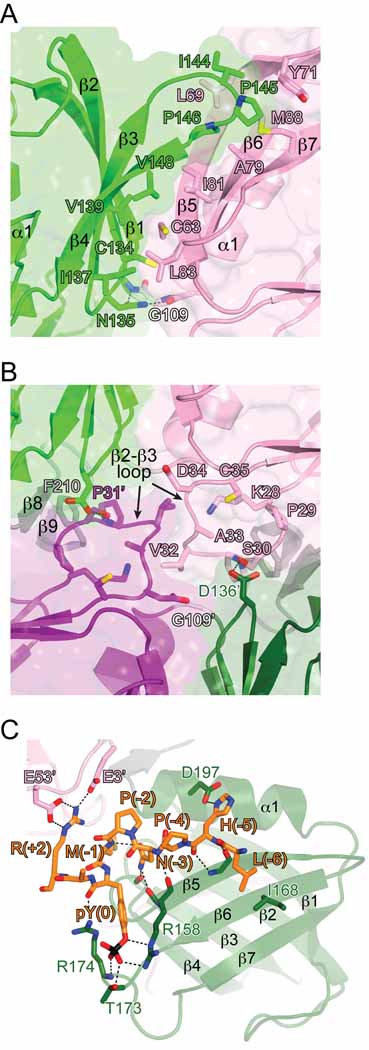
The Dok7 PH and PTB domains adopt the canonical domain architecture shared between these two signaling modules, each comprising a seven-stranded β sandwich and a C-terminal α helix (Figure 1C). The Dok7 PH domain contains an additional, short α helix between β-strand 3 (β3) and β4, similar to the PH domain of IRS1 (Eck et al., 1996). The linker between the PH and PTB domains of Dok7 is anomalously short for tandem PH and PTB domains, essentially a single residue (Gly109), compared to ≥23 residues in Dok1-6 and IRS1-4 (Figures 1B and C). At the end of the C-terminal helix (α1) in the PH domain, the polypeptide chain makes a tight turn, centered at Gly109 (conserved in Dok7), and the first residue of β1 in the PTB domain is Val111. The turn is stabilized by conserved Asn135 in the β2-β3 loop of the PTB domain, which is hydrogen-bonded to backbone atoms on both sides of Gly109 (Figure 2A).
Figure 2. Molecular Interactions in the Dok7(PH-PTB)-MuSK(pTyr553) Structure.
(A) View of the interface between the Dok7 PH and PTB domains. The viewing angle (down the molecular dyad axis) is the same as in Figure 1C. Side chains that mediate the interaction between the two domains are shown in stick representation. Carbon atoms are colored either pink (PH) or green (PTB), oxygen atoms are colored red, nitrogen atoms are colored blue, and sulfur atoms are colored yellow. Hydrogen bonds are represented by black dashed lines. Select secondary elements are labeled.
(B) View of the Dok7 dimerization interface. The viewing angle is the same as in Figure 1C (two-fold axis is perpendicular to the plane of the figure), and the coloring scheme is the same as in (A). Residues in the β2-β3 loop of the PH domain and select other residues are shown in stick representation. Residues in the second protomer (dark colors) are labeled with an apostrophe.
(C) Mode of MuSK pTyr553 binding to Dok7. The pTyr553 phosphopeptide is shown in stick representation, as are select residues from the PTB domain (dark green) and the PH domain (pink) from the other protomer (labeled with an apostrophe). Hydrogen bonds/salt bridges are represented by black dashed lines. See also Figure S1. Figures 1, 2, and 6 were rendered with PyMOL (http://pymol.sourceforge.net).
The PH and PTB domains share an extensive interface, burying ~2,300 Å2 of total surface area. Hydrophobic residues in the interface include Cys63, Leu69, Ala79, Ile81, Leu83, and Met88 from the PH domain, and Cys134, Ile137, Val139, Ile144, Pro145, Pro146, and Val148 from the PTB domain (Figure 2A). In other Dok/IRS family members, many of the corresponding residues are hydrophilic, in particular, the equivalents of Leu69, Leu83, and Ile144. The only other available structure of tandem PH and PTB domains is that of IRS1 (Dhe-Paganon et al., 1999). In the IRS1 PH-PTB crystal structure, the domains also form an interface, but the residues involved are, for the most part, non-overlapping with those in Dok7.
The C-terminal α helix (α1) of the Dok7 PTB domain ends at residue 200. There is interpretable electron density through Phe210 in the C-terminal extension to the PTB domain, although the sequence conservation extends to residue 220 (Figure 1B); residues 211 through 220 are disordered in the structure. Residues in the C-terminal extension form two short interacting β strands (β8, β9) (Figure 2B). This secondary structure may be important for positioning Phe210 in the dimer interface (see below).
Dimerization of Dok7
As stated above, the Dok7(PH-PTB) dimer interface buries ~2000 Å2 of total surface area with a shape complementarity value of 0.69 (Lawrence and Colman, 1993). These interface metrics are consistent with biologically relevant protein-protein interactions. The Dok7(PH-PTB) dimer is mediated primarily by residues in the β2-β3 loop of the PH domain, interacting across the molecular two-fold axis with their counterparts in the other protomer and with residues in the PH-PTB linker and in the region C-terminal to the PTB domain (Figure 2B). The eight residues in the β2-β3 loop (28–35) are invariant throughout vertebrate species (Figure 1B). In the structure, Ser30 is hydrogen-bonded to Asp136’ (an apostrophe denotes a residue in the second protomer) in the PTB domain (Figure 2B), and Pro31 is in van der Waals contact with conserved Phe210’ in the C-terminal extension to the PTB domain. Val32, at the tip of the loop, occupies the “linchpin” position in the dimer interface, and is in van der Waals contact with its counterpart (Val32’). Val32 is also in van der Waals contact with Gly109’, the PH-PTB linker residue in the other protomer (Figure 2B). For steric reasons, a linker of greater length, or a residue other than glycine, would prevent formation of this dimer.
Two crystal structures of Dok7(PH-PTB) (mouse and human) were determined and partially refined in the absence of the MuSK pTyr553 phosphopeptide (data not shown). In both of these structures, the PH-PTB dimer (Figure 1C) is absent. Furthermore, for one structure, of mouse Dok7(PH-PTB) at 2.3 Å resolution, the PTB domain was readily placed, but the position of the PH domain is not evident. These additional structural data indicate that the PH-PTB intramolecular interaction, as well as Dok7 dimerization, may depend on PTB-domain engagement of the MuSK juxtamembrane region.
Mode of Phosphotyrosine Binding
In the crystal structure, the 13-residue phosphopeptide is bound in the phosphopeptide binding groove of the PTB domain, between β5 and α1. The phosphate group of pTyr553 is coordinated by Arg158 (β5) and by Thr173 and the backbone amide nitrogen of Arg174 (β6-β7 loop) (Figure 2C). The guanidinium group of Arg174 is proximal to the phosphate group (<4 Å) and is hydrogen-bonded to the backbone carbonyl oxygen of pTyr553. As in other PTB domain-NPXpY structures, Asn550 of the NPXpY motif stabilizes the β turn in the phosphopeptide by hydrogen-bonding to the backbone amide nitrogen of Met552 (phosphopeptide) and to backbone carbonyl oxygens of Leu154 and Leu157 (β4-β5 loop, PTB domain) (Figure 2C). Using isothermal titration calorimetry, we measured the binding affinity (Kd) of the pTyr553 phosphopeptide to Dok7 PH-PTB as 4.7 µM (Supplemental Data; Figure S1), a fairly typical value for a PTB domain-NPXpY interaction.
For PTB domains in general, specificity for particular NPXpY sequences derives from residue differences in α1 and in β5 and β6. This is true for the Dok7 PTB domain as well (Figure 2C), with specificity at the P-6 position (relative to pTyr553), Leu547, conferred by Ile168 (β6), and at the P-5 position, His548, by Asp197 (α1). Non-canonical mechanisms dictate specificity at the P-1 position, Met552, and at the P+2 position, Arg555. The C-terminal α helix (α1) of the Dok7 PTB domain is six and four residues shorter than the corresponding helices in the related IRS1 (Eck et al., 1996) and Dok1 (Shi et al., 2004) PTB domains, respectively, which affords the necessary space to accommodate the long side chain of Met552 (P-1) (Figures 1B and 2C). The specificity for Arg555 (P+2) comes not from the PTB domain, but rather from the PH domain of the other PH-PTB protomer in the dimer. Arg555 is salt-bridged to Glu3 (pre-β1) and Glu53 (α0-β4 loop) of the “trans” PH domain (Figure 2C). Glu3 is either glutamic or aspartic acid across Dok7 species, and Glu53 is strictly conserved (Figure 1B), as is Arg555 of MuSK.
Solution Studies of the Dok7-MuSK Interaction
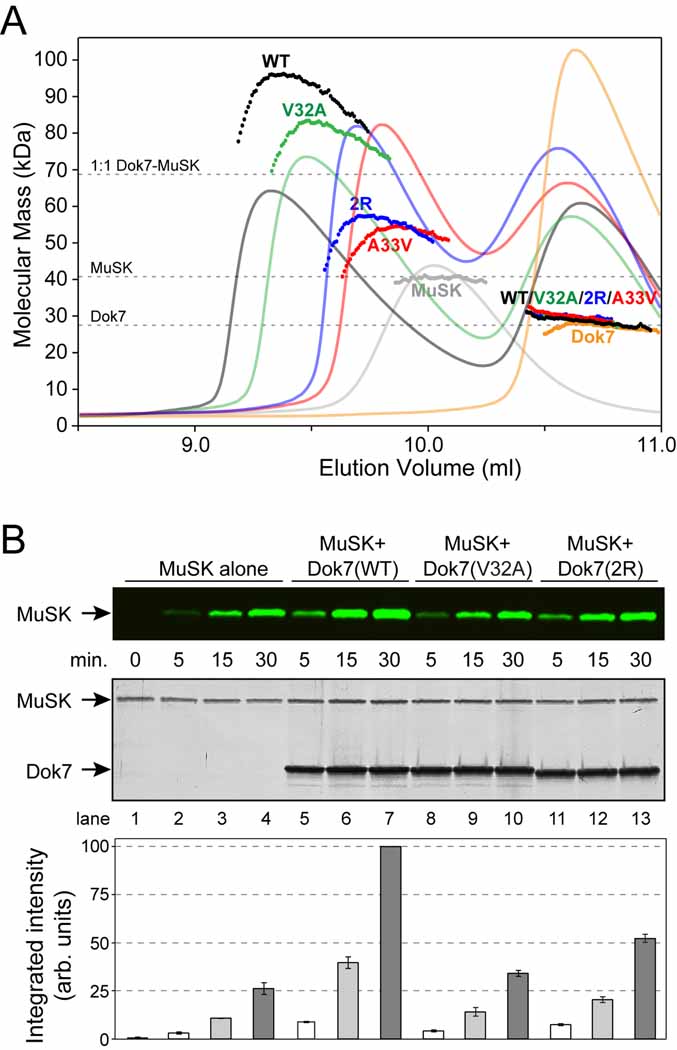
To monitor the oligomeric state of Dok7(PH-PTB) and MuSK in solution, we performed size-exclusion chromatography (SEC) coupled to static multi-angle light scattering (MALS), which yields absolute particle masses. Dok7(PH-PTB) alone eluted in a single peak, with a measured molecular mass of 27 kDa (24.6 kDa calculated), indicative of a monomeric species (Figure 3A). The phosphorylated cytoplasmic domain of MuSK (pMuSK[cyto]) also eluted in a single peak, with a measured mass of 41 kDa (39.0 kDa calculated), consistent with a monomer. A mixture of Dok7(PH-PTB) and pMuSK(cyto), with the former in molar excess, eluted in two peaks. The first peak contained both Dok7(PH-PTB) and pMuSK(cyto), and the second peak contained Dok7(PH-PTB) only. The concentration-dependent molecular-mass profile across the first peak (Figure 3A) indicates that the proteins are undergoing a fast, reversible oligomerization. At the apex of the first peak (highest protein concentration), MALS analysis yielded a molecular mass of 98 kDa, which is between the molecular masses for 1:1 (68 kDa) and 2:2 (136 kDa) Dok7-MuSK complexes. These data demonstrate that Dok7(PH-PTB) binds to pMuSK(cyto) to form 1:1 and higher-order (2:2) complexes.
Figure 3. In Vitro Characterization of the Dok7(PH-PTB)-MuSK(cyto) Interaction.
(A) A280 traces from the SEC runs are shown as colored solid lines, plotted on the same relative scale. The MALS-derived molecular mass distributions are plotted as individual points in the colors corresponding to the A280 traces, with the scale shown on the left-hand side. The samples analyzed were (Dok7=Dok7[PH-PTB] and MuSK=pMuSK[cyto]): MuSK + wild-type Dok7 (‘WT’, black), MuSK + Dok7 V32A (‘V32A’, green), MuSK + Dok7 A33V (‘A33V’, red), MuSK + Dok7 double-arginine mutant (‘2R’, blue), MuSK alone (‘MuSK’, gray), and wild-type Dok7 alone (‘Dok7’ orange). The dashed line at 68 kDa indicates the molecular mass of a 1:1 Dok7-MuSK complex. The molecular mass distributions in the second peak (labeled ‘WT/V32A/2R/A33V’), which represents excess Dok7, are nearly superimposable for the four Dok7-MuSK samples.
(B) MuSK autophosphorylation assay. MuSK(cyto) was incubated with Mg-ATP in the absence (lanes 1–4) or presence of Dok7(PH-PTB) wild-type (lanes 5–7), V32A (lanes 8–10), or the R158Q/R174A mutant (2R, lanes 11–13). The top panel is an immunoblot with anti-MuSK pTyr754 antibodies visualized on an infrared imaging system. The middle panel shows the silver-stained gel of the reaction mixtures (loading controls), and the lower panel shows the quantification of the immunoblot in the top panel. The error bars represent the standard error from experiments done in triplicate. For each experiment, the highest intensity (lane 7) was scaled to 100 (hence, no error bar), then the data were averaged. The zero time point (lane 1) is prior to the addition of Mg-ATP and is only shown for the MuSK-alone sample. See also Figure S2.
We then tested the ability of three Dok7(PH-PTB) mutants to form complexes with MuSK: V32A (β2-β3 loop), a key Dok7 dimer-interface residue in the crystal structure (Figure 2B), A33V, the adjacent residue and a CMS mutation, and a double-arginine mutant (R158Q [CMS mutation] + R174A), which should abrogate PTB-domain binding to pTyr553 in MuSK (Figure 2C). When we performed SEC on Dok7 V32A mixed with pMuSK(cyto) (same protein concentrations as before), the complex eluted later than the complex containing wild-type Dok7, and MALS analysis gave a molecular mass of 82 kDa at the highest protein concentration (again, the profile was concentration-dependent). For the double-arginine Dok7 mutant, the peak containing the complex was shifted further to the right, with a molecular-mass maximum of 57 kDa. Finally, for A33V, the complex eluted later still, with a molecular-mass maximum of 54 kDa. From these SEC-MALS data, we conclude that Dok7 V32A impairs formation of the 2:2 but not the 1:1 Dok7-MuSK complex, as predicted from the crystal structure, and that A33V and the double-arginine mutant impair formation of the 1:1 complex.
Dok7-Stimulated MuSK Autophosphorylation in Vitro
The crystal structure of Dok7(PH-PTB)-MuSK(pTyr553) and the SEC results suggest a mechanism by which Dok7 might stimulate the kinase activity of MuSK: Dimerization of Dok7 bound to the juxtamembrane region of MuSK juxtaposes the two kinase domains for trans-autophosphorylation on the activation loop. To test this hypothesis, we performed autophosphorylation reactions with MuSK(cyto) in the presence or absence of Dok7(PH-PTB). We showed previously that incubation of MuSK(cyto) with Mg-ATP results in trans-autophosphorylation of Tyr553 (juxtamembrane region) and Tyr750, Tyr754, and Tyr755 (activation loop) (Till et al., 2002). With MuSK alone, incubation with Mg-ATP results in a relatively slow rate of autophosphorylation as measured by immunoblotting with anti-pTyr754 (activation loop) phosphospecific antibodies (Figure 3B, lanes 1–4). In the presence of Dok7, the rate of MuSK Tyr754 autophosphorylation is substantially increased, approximately 4-fold under these reaction conditions (Figure 3B, lanes 5–7).
To test whether dimerization of Dok7(PH-PTB) as observed in the crystal structure is necessary for Dok7 stimulation of MuSK autophosphorylation, we performed the MuSK autophosphorylation reaction in the presence of the dimer-interface mutant V32A, which impairs 2:2 Dok7-MuSK complex formation (Figure 3A). With this mutant, the stimulatory effect on MuSK autophosphorylation is largely absent (Figure 3B, lanes 8–10).
The double-arginine mutant (R158Q/R174A), which is impaired in its ability to form a 1:1 complex with MuSK (Figure 3A), was also impaired in its ability to stimulate MuSK autophosphorylation, as expected (Figure 3B, lanes 11–13). Interestingly, a ~2-fold stimulatory effect was still present, evidently because the dimerization interface of this Dok7 mutant is intact, and the mutant retains some MuSK binding capacity (Figure 3A). We also tested A33V (CMS mutation) and S30Q, another dimer-interface residue (Figure 2B). Both of these mutations abrogated the ability of Dok7 to stimulate MuSK autophosphorylation (Supplemental Data; Figure S2).
Dok7-Mediated MuSK Activation in Cells
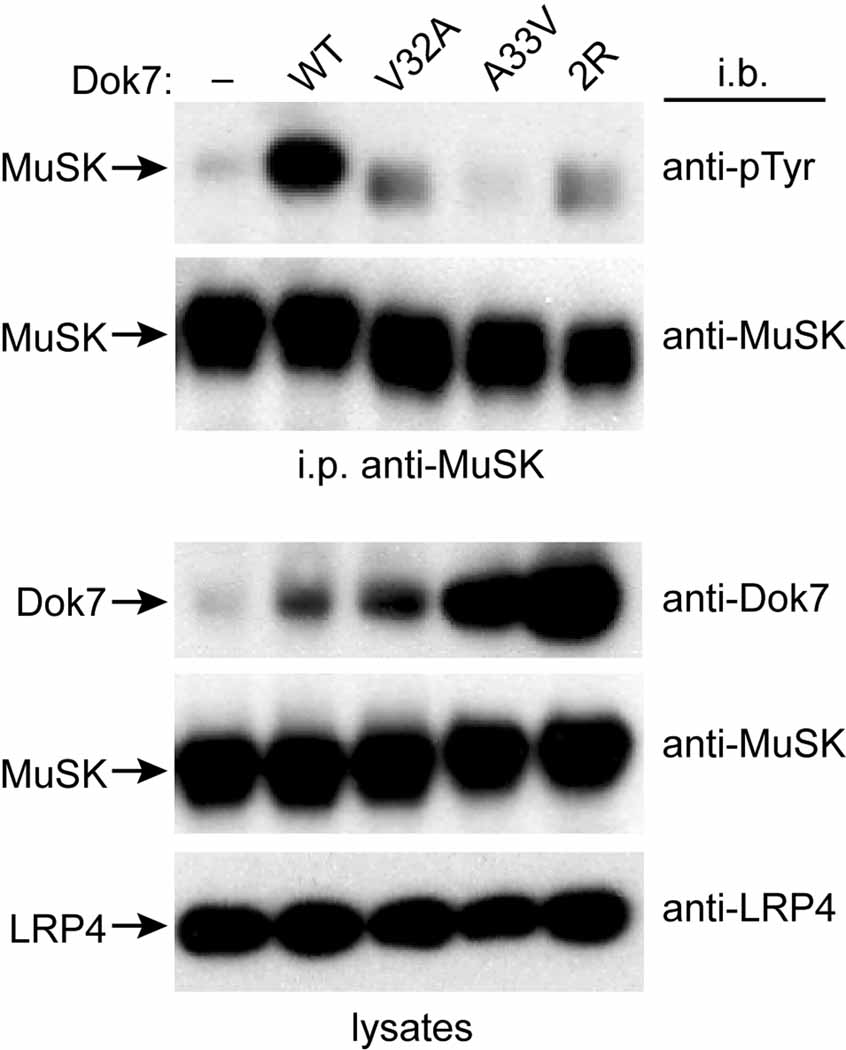
To determine whether Dok7 dimerization regulates MuSK activation in vivo, we expressed wild-type or mutant forms of Dok7, together with MuSK and LRP4, in NIH 3T3 cells and measured Agrin-stimulated MuSK phosphorylation. Because Dok7 overexpression can strongly stimulate MuSK phosphorylation in the absence of Agrin/LRP4, and can obscure the effects of partial loss-of-function mutations in Dok7 (data not shown and [Okada et al., 2006]), we expressed low levels of full-length Dok7 using a weak c-Fos basal promoter cassette (Gilman et al., 1986; Simon and Burden, 1993). We treated parental 3T3 cells or cells expressing wild-type Dok7, V32A, A33V, or the double-arginine mutant (R158Q/R174A) with Agrin, immunoprecipitated MuSK, and probed Western blots with antibodies to phosphotyrosine. As shown in Figure 4, Agrin stimulated robust MuSK phosphorylation in cells expressing wild-type Dok7, but weak phosphorylation in cells expressing Dok7 V32A, A33V, or the double-arginine mutant. These data demonstrate that residues that are important for Dok7 to activate MuSK in vitro, in particular, Val32 in the dimer interface, are likewise critical for Dok7 to activate MuSK in vivo.
Figure 4. Dok7-Mediated Activation of MuSK in NIH 3T3 Cells.
Shown are immunoblots from Agrin-treated NIH 3T3 cells stably transfected with MuSK, LRP4, and wild-type Dok7 (WT), V32A, A33V, the double-arginine mutant (2R), or no Dok7 (−). The top two blots are from anti-MuSK immunoprecipitates (i.p.), and the bottom three blots are from lysates. The antibodies used for immunoblotting (i.b.) are listed on the right sides of the blots, and the protein identifications are listed on the left sides. As can be seen in the anti-Dok7 immunoblot of the lysates (top lysate blot), the levels of Dok7 vary in the different stably transfected cell lines, but in all cases the mutant Dok7 level is greater than or equal to the wild-type level.
Phosphoinositide Binding to the Dok7 PH domain
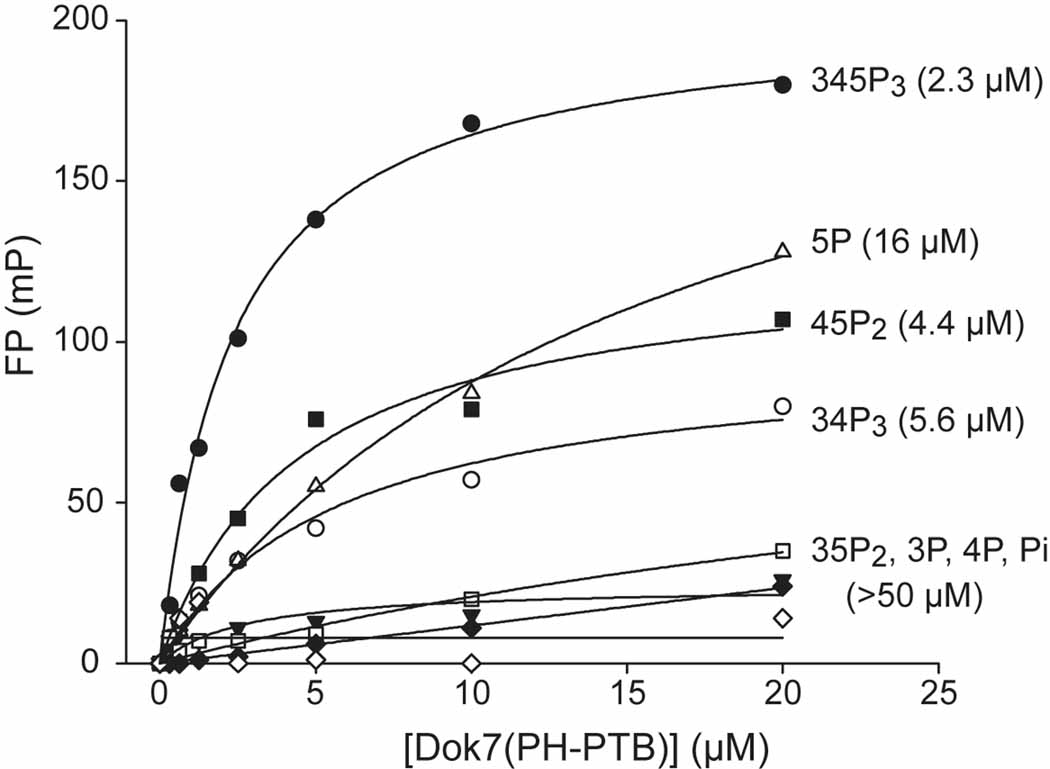
Because the majority of PH domains in phosphotyrosine signaling pathways bind to membrane phosphoinositides, we measured the phosphoinositide binding affinities to Dok7(PH-PTB) in vitro using a fluorescence-polarization assay (Depetris et al., 2009). PI(3,4,5)P3, PI(4,5)P2, and PI(3,4)P2 bound to Dok7(PH-PTB) with affinities (Kd values) in the range 2 to 6 µM, whereas PI(3,5)P2 and the mono-phosphorylated phosphoinositides bound with much lower affinities (Figure 5). These data indicate that the Dok7 PH domain binds relatively non-specifically to phosphoinositides and with modest (low micromolar) affinities, characteristics that are typical for PH domains (Lemmon and Ferguson, 2000). Although the crystals of Dok7(PH-PTB) were obtained only in the presence of the headgroup Ins(1,3,4,5)P4, there is no discernable electron density for it.
Figure 5. Phosphoinositide Binding to Dok7(PH-PTB).
Fluorescence-polarization measurements (millipolarization [mP] versus protein concentration) of phosphoinositide binding to mouse Dok7(PH-PTB). Fits are based on a saturable, single-site binding model. Kd values (in µM) extracted from the fits are given in parentheses to the right of the headgroup labels.
Based on structure-function studies performed on other PH domains, several residues in the Dok7 PH domain, mainly in the β1-β2 region, are probably responsible for phosphoinositide headgroup binding, including Lys18 (β1-β2 loop), Lys20 (β2), Arg22 (β2), and Arg54(α0-β4 loop). As measured in the fluorescence polarization assay, mutation of Arg54 to alanine resulted in a significant loss (>5-fold) of phosphoinositide binding (data not shown). It should be noted that the β2-β3 loop, implicated in Dok7 dimerization, is distal to the β1-β2 loop (Figure 1C), i.e., binding of phosphoinositides by the PH domain is compatible with PH-PTB dimerization and also with PTB-domain binding to the juxtamembrane region (pTyr553) of MuSK.
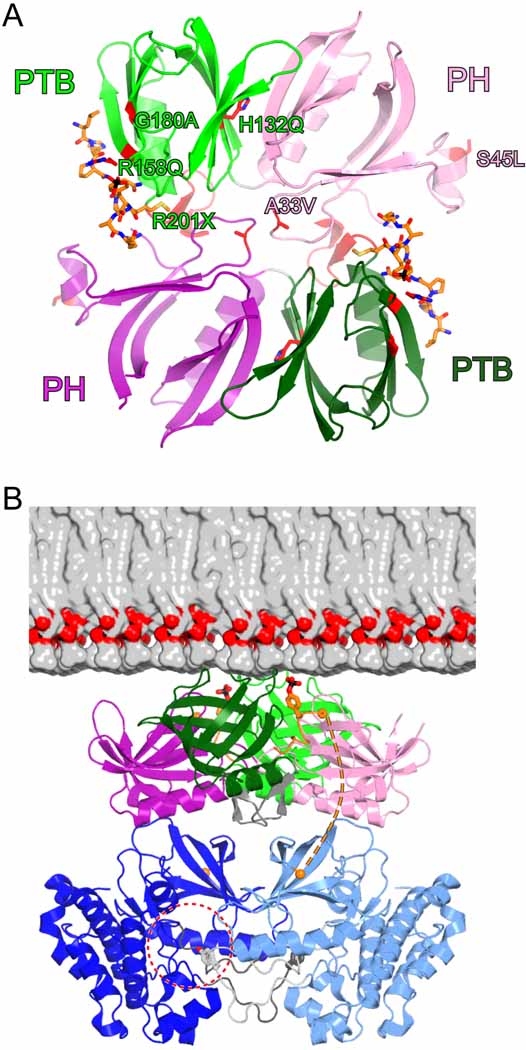
CMS Mutations
Recessive, loss-of-function mutations in Dok7 that are causative for CMS are distributed throughout the Dok7 sequence (Figure 1A). In the PH and PTB domains, the missense mutations are A33V, S45L, H132Q, R158Q, and G180A. These PH-PTB mutations have not been found in combination in both Dok7 alleles, suggesting that, individually, they significantly impair Dok7 function. Our structure and biochemical studies of Dok7(PH-PTB) provide a molecular basis for understanding the deleterious effects of many of these mutations, which are mapped onto the structure in Figure 6A. Ala33 is situated in the β2-β3 loop of the PH domain, the critical structural element for Dok7 PH-PTB dimerization. As the SEC-MALS data show (Figure 3A), A33V results in an impairment of 1:1 Dok7-MuSK complex formation, leading to loss of MuSK activation [Figures 3B and 4 and Supplemental Data; Figure S2]). Interestingly, mutation of the adjacent residue, V32A, does not impair 1:1 complex formation, only formation of the 2:2 complex (Figure 3A and data not shown). Given the tight structural coupling between the PH and PTB domains (Figure 1C), it is conceivable that a major perturbation in the conformation of the β2-β3 loop, which could be caused by A33V (much less so by V32A), could be transmitted to the MuSK-binding PTB domain.
Figure 6. CMS Mutations and Model for Dok7-Facililated MuSK Activation.
(A) CMS missense and nonsense mutations are mapped onto the Dok7(PH-PTB) structure, shown as a ribbon diagram. The side chains of the wild-type protein affected by mutation are shown in stick representation, with carbon atoms colored red. The residues in only one protomer are labeled. The ribbon is colored red for residues 201–210, which are deleted as a consequence of the R201X nonsense mutation.
(B) The structure of the 2:2 Dok7(PH-PTB):MuSK(pTyr553) complex is colored as in Figure 1B, with pTyr553 shown in stick representation. The view is approximately 90° from the view in (A). Half of a bilayer consisting of 1-palmitoyl-2-oleoyl-phosphatidylcholine is shown to scale. Two MuSK kinase domains (light and dark blue) are represented by the dimer of trans-autophosphorylating kinase domains of the insulin-like growth factor-1 receptor (IGF1R) (Wu et al., 2008). In this symmetric dimeric configuration (two-fold axis is vertical, perpendicular to the bilayer plane), Tyr1135 in the IGF1R activation loop, corresponding to Tyr754 in MuSK, is bound in the active site of the other kinase domain (dashed-red circle). The activation loop is colored either light gray (corresponding to the light-blue kinase) or dark gray (corresponding to the dark-blue kinase), with Tyr1135 (Tyr754) shown in stick representation. The orange spheres denote the Cα positions at the end of the pTyr553 peptide (Arg555) and at the beginning of the MuSK kinase domain (Tyr569). In this manually assembled model, the distance between these two residues is 34 Å.
The deleterious consequences of the S45L mutation are not readily predicted from the crystal structure, but it is possible that an exposed hydrophobic residue in α0 in the PH domain (Figure 6A) could result in protein aggregation. His132 is present in the interface between the PH and PTB domains, and mutation to glutamine could disrupt the PH-PTB interface. We attempted to express Dok7(PH-PTB) H132Q to measure its ability to activate MuSK in vitro. The protein was poorly expressed in E. coli and was ill-behaved by SEC (data not shown). Thus, H132Q, by disrupting the PH-PTB interface, appears to adversely affect the structural integrity of the protein.
Arg158 is salt-bridged to the phosphate group of pTyr553 (Figure 2C), and is therefore directly involved in MuSK binding. As predicted, mutation of this arginine (combined with R174A) impaired MuSK binding (Figure 3A) and consequently MuSK activation (Figures 3B and 4). Gly180 is conserved in Dok1-7 and IRS1-4, and a non-glycl residue at this position would possess unfavorable backbone torsion angles. A point mutation found in a subset of CMS patients introduces a stop codon at Arg201, near the end of the C-terminal α helix in the PTB domain. Expression of Dok7 residues 1–200 resulted in a poorly behaved protein, suggesting that this mutation/truncation compromises the structural integrity of the PTB domain.
DISCUSSION
For most ligand-RTK systems, signal transduction occurs via a relatively straightforward process: A polypeptide ligand (often dimeric) binds directly to the ectodomain of its cognate receptor, leading to receptor oligomerization (often dimerization) and juxtaposition of the cytoplasmic kinase domains for trans-autophosphorylation (Hubbard and Miller, 2007; Schlessinger, 2000). Tyrosine phosphorylation stimulates kinase activity and provides recruitment sites for downstream substrates. Activation of MuSK is more complicated in at least two ways. First, the ligand, Agrin, does not bind directly to MuSK, but rather to LRP4, which acts in conjunction with Agrin to stimulate MuSK trans-autophosphorylation (Kim et al., 2008; Zhang et al., 2008). Second, a cytoplasmic adapter protein, Dok7, is required for MuSK activation, in addition to Agrin/LRP4 (Okada et al., 2006).
Our structural and biochemical studies are consistent with a Dok7-mediated MuSK activation mechanism in which Dok7 binds to the phosphorylated juxtamembrane region of MuSK and, through dimerization of its PH-PTB domains, juxtaposes two MuSK kinase domains for trans-autophosphorylation on their activation loops (Figure 6B). The PTB domain of Dok7 binds selectively to pTyr553 (and adjacent residues) in the juxtamembrane region (Figure 2C), and the PH domain serves two roles: main mediator of Dok7 dimerization (Figure 2B) and plasma-membrane localization via phosphoinositide binding (Figure 5). This juxtamembrane activation mechanism (heterologous, through Dok7) contrasts with the mechanism by which other RTKs such as Ephs and c-Kit are activated through juxtamembrane auto-phosphorylation—release of autoinhibition (Hubbard, 2004).
Why does MuSK require a cytoplasmic activator? The dual activation mechanism for MuSK—from the “inside” (Dok7) as well as from the “outside” (Agrin/LRP4)—could have arisen to supply two levels of MuSK activity: a basal (non-Agrin-stimulated) activity, which is necessary for pre-patterning of MuSK and AChRs, and an Agrin-stimulated activity upon muscle innervation. In general for RTKs, only the ligand-stimulated activity of an RTK is required for biological function, and any basal activity is not thought to have an important functional role, other than to enable trans-autophosphorylation upon ligand-induced dimerization.
The requirement for MuSK clustering and signaling in the pre-patterning stage could explain, on the one hand, why MuSK is highly autoinhibited, and, on the other hand, why a cytoplasmic activator is required. The crystal structure of the MuSK kinase domain, along with kinetics studies (Till et al., 2002), revealed that unphosphorylated MuSK is autoinhibited by the activation loop through a mechanism similar to that of the insulin receptor kinase (Hubbard et al., 1994), but to an even greater degree. It is plausible that this level of autoinhibition, which may be necessary to restrict the extent of MuSK activation and AChR pre-clustering, requires a cytoplasmic activator to overcome it.
How does pTyr553 in the MuSK juxtamembrane region become phosphorylated to create a docking site for Dok7, while the activation loop remains unphosphorylated or partially phosphorylated? The unphosphorylated activation loop of MuSK must be transiently displaced from its own active site to phosphorylate any substrate, including another MuSK molecule. Evidence that a heterologous kinase is not involved in Tyr553 phosphorylation comes from mutation of the conserved catalytic lysine (Lys608) in the MuSK kinase domain, which results in a loss of all tyrosine phosphorylation on MuSK (Zhou et al., 1999). By analogy with the insulin receptor kinase, only when the MuSK activation loop is phosphorylated on both Tyr754 and Tyr755 (the second and third tyrosines in the loop) will the receptor be fully activated catalytically (Hubbard, 1997).
In the context of the soluble cytoplasmic domain of MuSK, Tyr553 and Tyr754 are the preferred sites of autophosphorylation (in trans), and Tyr750 and Tyr755 are autophosphorylated more slowly (Till et al., 2002). We posit that, independent of Dok7 and Agrin/LRP4, adventitious interactions of MuSK in the muscle cell membrane result in a low, sub-stoichiometric phosphorylation of Tyr553. Formation of a pTyr553-mediated 2:2 Dok7-MuSK complex will facilitate trans-autophosphorylation of the activation loop to activate the kinase. Through this process, Dok7 could activate receptors “catalytically”, that is, a Dok7-activated receptor could potentially autophosphorylate a non-activated receptor independent of whether Dok7 is bound to the pair. Indeed, relatively low levels of Dok7 (compared to MuSK) are sufficient to saturate MuSK autophosphorylation in heterologous cells (data not shown). Upon kinase activation, phosphorylation of downstream signaling proteins, in particular, Dok7 itself, will ensue. Now that the basic molecular mechanism by which Dok7 mediates MuSK activation is in hand, the next challenge will be to determine how Agrin and LRP4 work in concert with Dok7 to activate MuSK during neuromuscular synaptogenesis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA encoding residues 1–220 (PH and PTB domains) of mouse and human Dok7 were subcloned into an E. coli expression vector that includes a TEV-cleavable, N-terminal 6xHis tag. The vector encoding Dok7(PH-PTB) was transformed into E. coli strain BL21(DE3) (for mouse Dok7) or Rosetta2(DE3) (for human Dok7), and cultures were grown in Luria broth media at 37°C to an OD600 of 0.6. Protein expression was induced by the addition of IPTG (0.2 mM) for 12 hr at 19°C. Cells are harvested, resuspended in lysis buffer (50 mM Tris [pH 8.0], 300 mM NaCl, 0.1% (v/v) Triton X-100, 5 mM 2-mercaptoethanol, and complete EDTA-free protease inhibitor cocktail [Roche Diagnostics]), and lysed using a cell disrupter (Emulsiflex-C2, Avestin). The lysate was centrifuged at 37,000×g for 1 hr, and the supernatant was collected. The soluble fraction was purified by Ni2+-affinity chromatography (Sigma), followed by size-exclusion chromatography (Superdex 75, GE Healthcare). The 6xHis tag is then cleaved with TEV, and the un-tagged protein was further purified by Source S ion-exchange chromatography (GE Heathcare).
To produce selenomethionyl-labeled Dok7(PH-PTB), transformed BL21(DE3) cells from 1 ml of an overnight culture in LB medium were spun down in 1 ml of M9 minimal medium with glucose (4 g/l) as carbon source, and then added to one liter of the same pre-warmed medium. Cells were grown to mid-log phase before addition of the following amino acids: lysine, phenylalanine and threonine at 100 mg/l; isoleucine, leucine and valine at 50 mg/l; and L-selenomethionine at 60 mg/l. Protein expression was induced with the addition of 0.2 mM IPTG at 19°C 15 min after addition of the amino acids. Purification of the selenomethionyl-labeled Dok7(PH-PTB) was performed as for native Dok7(PH-PTB). The incorporation of selenium was confirmed by MALDI mass spectrometry.
MuSK(cyto) (rat, residues 529–868) was expressed in baculovirus-infected Sf9 insect cells and purified as described previously (Till et al., 2002). A 13-residue phosphopeptide representing the region encompassing MuSK Tyr553, Ac-LDRLHPNPMpYQRM, was synthesized (Genemed Synthesis) and solubilized in 100 mM Tri-HCl (pH 8.0) and 150 mM NaCl.
X-ray Crystallography
Crystals of mouse Dok7(PH-PTB) in complex with the MuSK pTyr553 phosphopeptide were grown at 17°C in hanging drops. 3 µl of a stock solution containing Dok7(PH-PTB) at 4 mg/ml, a 5:1 molar excess of phosphopeptide, and a 2:1 molar excess of Ins(1,3,4,5)P4 (Echelon) was mixed with 1 µl of reservoir buffer containing 0.7 M of sodium/potassium tartrate. Selenomethionyl (SeMet)-derivatized crystals were obtained under the same conditions. Crystals of native and SeMet Dok7(PH-PTB) belong to space group P3221 with unit cell parameters of a=b=134.6 Å, c=121.6 Å. There are four complexes per asymmetric unit. Before flash-freezing in liquid nitrogen, crystals were equilibrated in a series of stabilizing solutions containing reservoir buffer plus 10%, 15%, 20%, then 25% ethylene glycol.
Data for a single SeMet crystal was collected on beam line X4C at the National Synchrotron Light Source, Brookhaven National Laboratory at 0.9785 Å, the wavelength corresponding to the peak of the K-absorption edge for selenium. Diffraction data were processed using HKL2000 (Otwinowski and Minor, 1997). The Dok7(PH-PTB)-MuSK(pTyr553) structure was determined by SAD (single anomalous diffraction) phasing using SOLVE/RESOLVE (Terwilliger and Berendzen, 1999) (figure of merit=0.31 from SOLVE, 0.76 from RESOLVE; 30-2.6 Å). A total of 20 ordered Se atoms were found in the four copies of Dok7(PH-PTB) in the asymmetric unit, representing all possible SeMet residues other than the ones at the N-termini. Model building was performed in Coot (Emsley and Cowtan, 2004). Structure refinement (to 2.6 Å) was performed using CNS (Brünger et al., 1998) and REFMAC (Murshudov et al., 1997). The final atomic model contains residues 2–220 for the four copies of Dok7(PH-PTB), excluding residues 15–17 (β1-β2 loop) in copies A and B, and residues 16–17 in copy D. Residues 547–555 of the MuSK phosphopeptide are built in all four copies. The inositol headgroup, present in the crystallization solution, is not visible in the electron density but was required for obtaining crystals.
In Vitro MuSK Autophosphorylation
Dephosphorylated MuSK(cyto) was produced by treating the purified protein (30 µM) with calf intestinal alkaline phosphatase (1 unit per µg MuSK) (Promega) for 1 hr at room temperature. Phosphatase-treated MuSK(cyto) was purified by Source Q ion-exchange chromatography (GE Healthcare), and the dephosphorylated state was confirmed by immunoblotting with anti-phosphotyrosine antibodies (4G10, Upstate). Dephosphorylated MuSK(cyto) (1 µM) was incubated or not with 5 µM of mouse Dok7(PH-PTB), wild-type or mutant, at room temperature in the reaction buffer (5 mM ATP, 50 mM MgCl2, 25 mM Tris-HCl [pH 7.5], 0.5 mM sodium vanadate, and 10 mM TCEP) into a final volume of 40 µl. Aliquots of 8 µl were removed at 5, 15 and 30 min, and the reaction was stopped by adding 36 µl of 2× Laemmli sample buffer to each aliquot and boiling. The samples were separated by SDS-PAGE (15%) and transferred to a nitrocellulose membrane (GE Healthcare). The phosphorylation level of the MuSK activation loop was detected by immunoblotting with anti-MuSK pY754 phosphospecific antibodies (Jones et al., 2007). The relative signal intensity at each time point was measured on an Odyssey infrared imaging system (Li-Cor Biosciences).
SEC and MALS Analysis
To obtain phosphorylated MuSK(cyto) for binding with Dok7(PH-PTB), MuSK(cyto) at 8 mg/ml was autophosphorylated in the presence of 5 mM ATP and 50 mM MgCl2 for 6 hr at room temperature. The phosphorylation reaction was stopped with the addition of EDTA to a final concentration of 100 mM. For SEC combined with in-line MALS analysis, 50 µl of pMuSK(cyto) at 6 mg/ml was combined with 50 µl of wild-type or mutant human Dok7(PH-PTB) at 8 mg/ml. 100 µl of each Dok7(PH-PTB)-pMuSK(cyto) complex, of pMuSK(cyto) alone (at 3 mg/ml), or of Dok7(PH-PTB) alone (at 4 mg/ml) was subjected to SEC using a Shodex KW-803 column (JM Science, Inc.) equilibrated in 20 mM MES pH 6.5, 200 mM NaCl, and 1 mM DTT. The HPLC system (Waters) was connected in-line to a three-angle light-scattering detector (miniDAWN TREOS) and a refractive-index detector (Optilab rEX) (Wyatt Technology). Data were collected every second at a flow rate of 0.5 ml/min. Data analysis was carried out using the program ASTRA (Wyatt Technology).
Phosphoinositide Binding
Purified mouse Dok7(PH-PTB) at concentrations ranging from 0.15 µM to 20 µM were mixed with 12.5 nM (final) of BODIPY TMR-labeled phosphoinositides (Echelon) in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.05% Tween-20. After 5 min of incubation, 20 µl of the reaction mix was added to individual wells of a 384-well assay plate (Corning). Fluorescence polarization was measured at room temperature using a TECAN Infinite F500 plate reader. A 535/25 nm filter and a pair of 590/20 nm filters were used as excitation and emission polarization filters, respectively. Curve fitting to a single-site (saturating) binding model was performed with SigmaPlot (Systat Software).
Dok7-Mediated MuSK Activation in NIH 3T3 Cells
Surface MuSK/LRP4-positive pools of NIH 3T3 cells were generated by retroviral-mediated transduction using pMXs Flag-LRP4-mCherry and pMXs Myc-MuSK-GFP, and sorted for membrane surface expression by FACS, using antibodies to extracellular epitope tags Flag and Myc (Cell Signaling Technology). Similarly, wild-type or mutant Dok7 (mouse) expression was reconstituted in MuSK/LRP4-positive 3T3 cells using pBabe ECFP-cFosDok7-HA as previously described (Kim et al., 2008). LRP4/MuSK/Dok7-expressing cells were stimulated with 5 nM of neural rat Agrin (R&D Systems) for 45 min, then lysed in buffer containing 30 mM triethanolamine, 50 mM NaF, 2 mM Na orthovanadate, 1 mM Na tetrathionate, 5 mM EDTA, 5 mM EGTA, 1 mM N-ethylmaleimide, 50 mM NaCl, 1% NP-40, and complete protease cocktail inhibitor tablet (Roche Diagnostics). The clarified lysates were resolved by SDS-PAGE and immunoblotted with antibodies against MuSK (R&D Systems), Flag (for LRP4; Sigma), or HA (for Dok7; Santa Cruz). To probe the phosphotyrosine content of MuSK, the clarified lysates were incubated overnight with an anti-MuSK antibody (Herbst and Burden, 2000) then incubated with protein-A-agarose beads (Roche) for 1 h at 4 °C. The beads were centrifuged and extensively washed with fresh lysis buffer before solubilization in SDS-PAGE buffer. Bound proteins were resolved by SDS-PAGE and immunoblotted using anti-MuSK antibodies (R&D Systems) or anti-phosphotyrosine antibodies (Millipore).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health, NS053414 (to S.R.H.) and NS036193 (to S.J.B.). We thank Dr. R. Ghirlando for sedimentation velocity experiments, J. Wu for help with the fluorescence-polarization experiments, J. Burrill for technical support, N. Yumoto for NIH 3T3 cells stably expressing MuSK and LRP4, and A. Stiegler and A. Blais for helpful discussions. Beamline X4C at the National Synchrotron Light Source, Brookhaven National Laboratory, a DOE facility, is supported by the New York Structural Biology Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes. Atomic coordinates and structure factors for Dok7(PH-PTB)-MuSK(pTyr553) have been deposited in the Protein Data Bank with accession code XXXX.
REFERENCES
- Beeson D, Webster R, Cossins J, Lashley D, Spearman H, Maxwell S, Slater CR, Newsom-Davis J, Palace J, Vincent A. Congenital myasthenic syndromes and the formation of the neuromuscular junction. Ann. NY Acad. Sci. 2008;1132:99–103. doi: 10.1196/annals.1405.049. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burden SJ. Building the vertebrate neuromuscular synapse. J. Neurobiol. 2002;53:501–511. doi: 10.1002/neu.10137. [DOI] [PubMed] [Google Scholar]
- Cai D, Dhe-Paganon S, Melendez PA, Lee J, Shoelson SE. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J. Biol. Chem. 2003;278:25323–25330. doi: 10.1074/jbc.M212430200. [DOI] [PubMed] [Google Scholar]
- Depetris RS, Wu J, Hubbard SR. Structural and functional studies of the Ras-associating and pleckstrin-homology domains of Grb10 and Grb14. Nat. Struct. Mol. Biol. 2009;16:833–839. doi: 10.1038/nsmb.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Ottinger EA, Nolte RT, Eck MJ, Shoelson SE. Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc. Natl. Acad. Sci. USA. 1999;96:8378–8383. doi: 10.1073/pnas.96.15.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Engel AG, Sine SM. Current understanding of congenital myasthenic syndromes. Curr. Opin. Pharmacol. 2005;5:308–321. doi: 10.1016/j.coph.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Gilman MZ, Wilson RN, Weinberg RA. Multiple protein-binding sites in the 5'-flanking region regulate c-fos expression. Mol Cell Biol. 1986;6:4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ, Yancopoulos GD. Sequential roles of agrin, MuSK and rapsyn during neuromuscular junction formation. Curr Opin Neurobiol. 1997;7:379–384. doi: 10.1016/s0959-4388(97)80066-9. [DOI] [PubMed] [Google Scholar]
- Hamuro J, Higuchi O, Okada K, Ueno M, Iemura S, Natsume T, Spearman H, Beeson D, Yamanashi Y. Mutations causing DOK7 congenital myasthenia ablate functional motifs in Dok-7. J Biol Chem. 2008;283:5518–5524. doi: 10.1074/jbc.M708607200. [DOI] [PubMed] [Google Scholar]
- Herbst R, Burden SJ. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J. 2000;19:67–77. doi: 10.1093/emboj/19.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell. Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr. Opin. Cell Biol. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- Inoue A, Setoguchi K, Matsubara Y, Okada K, Sato N, Iwakura Y, Higuchi O, Yamanashi Y. Dok-7 activates the muscle receptor kinase MuSK and shapes synapse formation. Sci. Signal. 2009;2:ra7. doi: 10.1126/scisignal.2000113. [DOI] [PubMed] [Google Scholar]
- Jones N, Hardy WR, Friese MB, Jorgensen C, Smith MJ, Woody NM, Burden SJ, Pawson T. Analysis of a Shc family adaptor protein, ShcD/Shc4, that associates with muscle-specific kinase. Mol. Cell. Biol. 2007;27:4759–4773. doi: 10.1128/MCB.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer TT, Misgeld T, Sanes JR. Assembly of the postsynaptic membrane at the neuromuscular junction: paradigm lost. Curr. Opin. Neurobiol. 2006;16:74–82. doi: 10.1016/j.conb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- Müller JS, Herczegfalvi A, Vilchez JJ, Colomer J, Bachinski LL, Mihaylova V, Santos M, Schara U, Deschauer M, Shevell M, et al. Phenotypical spectrum of DOK7 mutations in congenital myasthenic syndromes. Brain. 2007;130:1497–1506. doi: 10.1093/brain/awm068. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Okada K, Inoue A, Okada M, Murata Y, Kakuta S, Jigami T, Kubo S, Shiraishi H, Eguchi K, Motomura M, et al. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Shi N, Ye S, Bartlam M, Yang M, Wu J, Liu Y, Sun F, Han X, Peng X, Qiang B, et al. Structural basis for the specific recognition of RET by the Dok1 phosphotyrosine binding domain. J. Biol. Chem. 2004;279:4962–4969. doi: 10.1074/jbc.M311030200. [DOI] [PubMed] [Google Scholar]
- Simon AM, Burden SJ. An E box mediates activation and repression of the acetylcholine receptor delta-subunit gene during myogenesis. Mol. Cell. Biol. 1993;13:5133–5140. doi: 10.1128/mcb.13.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till JH, Becerra M, Watty W, Lu Y, Ma Y, Neubert TA, Burden SJ, Hubbard SR. Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. Structure. 2002;10:1187–1196. doi: 10.1016/s0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- Watty A, Neubauer G, Dreger M, Zimmer M, Wilm M, Burden SJ. The in vitro and in vivo phosphotyrosine map of activated MuSK. Proc Natl Acad Sci U S A. 2000;97:4585–4590. doi: 10.1073/pnas.080061997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li W, Craddock BP, Foreman KW, Mulvihill MJ, Ji QS, Miller WT, Hubbard SR. Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO J. 2008;27:1985–1994. doi: 10.1038/emboj.2008.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Glass DJ, Yancopoulos GD, Sanes JR. Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J. Cell Biol. 1999;146:1133–1146. doi: 10.1083/jcb.146.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.