Abstract
Objective
To quantitate risk and study heterogeneity at high resolution for HLA in the most common JIA subtypes, IgM RF negative polyarticular and oligoarticular. Four digit comprehensive HLA typing enabled great precision and a large cohort allowed for consideration of both age of onset and subtype.
Methods
PCR-based high resolution HLA typing for Class I and Class II loci was accomplished for 802 JIA patients and 273 controls. Specific HLA epitopes, potential interactions of alleles at specific loci and between loci, (accounting for linkage disequilibrium and haplotypic associations), and an assessment of the current ILAR classification were considered.
Results
An HLA DRB1-DQB1 effect was shown to be exclusively due to DRB1 and showed similarity between oligoarticular patients and a younger subgroup of polyarticular patients. Furthermore, polyarticular patients showed age-specific related effects with susceptibility in the >6 year group limited to the HLA–DRB1*08 haplotype, a marked distinction from the additional susceptibility haplotypes, HLA-DRB1*1103/1104, found in the oligoarticular cohort and the younger polyarticular group. Also in contrast to findings for oligoarticular JIA, polyarticular patients had no evidence of an HLA class I effect. Markers associated with reduced risk for disease include DRB1*1501, DRB1*0401 and DRB1*0701. The DRB1*1501 effect was shown to reduce risk across the whole cohort, whereas DRB1*0401 and DRB1*0701 were protective for selected subtypes. Surprisingly, the susceptibility mediated by DPB1*0201, even in the absence of DR susceptibility haplotypes, overcame the protective effect of DRB1*1501.
Conclusions
Inherited HLA factors in JIA show similarities overall as well as differences between subtypes.
INTRODUCTION
The term Juvenile Idiopathic Arthritis (JIA), refers to multiple clinically defined arthropathies which are likely attributable to a series of complex genetic traits, some of which are autoimmune. The genetic risk factors are known to include genes in the Human Leukocyte Antigen (HLA) complex on chromosome 6, which carry the more substantial component of risk, as well as non-HLA genes or regions. As with other autoimmune diseases, the non-HLA genetic component appears to be largely mediated by multiple low risk polymorphisms. Some of these polymorphisms are likely to be JIA-specific and some shared with other autoimmune diseases, such as reported recently for celiac disease and type 1 diabetes mellitus (1).
Extensive and well-documented associations with multiple HLA alleles in JIA have been described (2-4) for at least 25 years. It has been long realized that HLA associations in children are largely distinct from those in adult rheumatoid arthritis (RA), with HLA-DRB1*08, *11 and *13 reported in children with inflammatory arthritis (5-7). Furthermore, the shared epitope strongly associated with adult RA (8), consisting of selected HLA-DRB1*01/*04 alleles, is minimally important in JIA; HLA-DRB1*04 (and HLA-DRB1*07) are in fact protective depending on clinical subtype. Interestingly, when adult RA is stratified by the presence or absence of anti-CCP antibodies, HLA-DRB1*13 is protective for anti-citrullinated protein antibody-positive RA and is a risk factor for anti-citrullinated protein antibody-negative RA (9).
Another early finding included an association with a HLA- DP2 variant which when combined with HLA-DR3, DR5 (DR11), or DR6 (DR13) gave an enhanced level of risk, but not when combined with HLA-DR8. This suggested that genes from both loci are involved in generating predisposition to disease (7, 10). Class I involvement has also been demonstrated, with evidence of association with HLA-A2 (11, 12).
An additional level of complexity exists in JIA with associations related to age at disease onset and gender; not fully explained by age and gender representation in clinical phenotypes. Several of the class II associations including HLA-DR11 and HLA-DR13 are more often observed with younger age at onset, with 6 years of age used as the dividing point; others, including HLA-B27 and HLA-DR4 alleles, are associated with protection early in life but increase risk for disease later in childhood (13).
The HLA literature for JIA (JRA), summarized above, was limited by multiple variables which included international differences in nomenclature, the lack of diagnostic biomarkers (nomenclature still based largely on active joint counts), changes in disease phenotype during the course of disease, available cohorts (most considerably underpowered), and not least, the low resolution of the HLA typing. Given this complexity, it has been difficult to arrive at firm conclusions. In addition, efforts to define critical epitopes or motifs within the associated HLA alleles have been limited in part for the same reasons. A sequence motif encoded in exon 2 of certain DQA1 alleles has been associated with susceptibility in young onset oligoarticular disease (14). Likewise a DRB1 shared epitope encompassing critical amino-acid residues in the third hypervariable region has been identified for oligoarticular patients, but the strong association does not extend to proximal markers as it does in polyarticular patients (15).
A more recent large study (16) did not report interactions or define epitopes, and was somewhat limited in resolution of HLA typing. In contrast as reported here, high resolution HLA class-I and class-II typing in a substantial and well- studied population allows a more detailed evaluation of HLA associations in JIA. The comprehensive nature of this DNA-based study allows consideration of specific HLA epitopes, potential interactions of alleles of the same locus as well as between loci, while accounting for linkage disequilibrium and haplotype effects, and an assessment of the validity of the current classification.
SUBJECTS AND METHODS
Description of Cohorts
A case control study design was employed. Approximately 95% of the samples were recruited at the Cincinnati Children’s Hospital Medical Center (CCHMC). The remaining samples were provided by collaborating centers which included Children’s Hospital of Wisconsin, Schneider Children’s Hospital and Children’s Hospital of Philadelphia, or as part of a NIAMS supported JIA affected sibpair registry. The medical and clinical data relating to samples were collected for the affected sibpair NIAMS registry in standardized case report forms including ILAR criteria, or in the Research Registry maintained within the CCHMC Division of Rheumatology. This study was approved by the Institutional Review Board of CCHMC and collaborating centers. Given that the sample size for Hispanic and African American individuals was insufficient for independent analysis, the study was limited to non-Hispanic Caucasians to avoid confounding related to underlying population substructure. Most cases and all controls correspond to an ongoing genome-wide association study cohort, allowing review of ancestry informative SNPs to confirm population homogeneity.
The ILAR revised criteria for juvenile idiopathic arthritis (17) were the criteria of choice. The cohort was limited to the two most common subtypes, IgM Rheumatoid Factor (RF) negative polyarticular and oligoarticular JIA. Oligoarthritis was further classified as persistent (4 or fewer joints) or extended (more than 4 joints) at 1 year after disease onset.
Patients recruited before ILAR criteria were published were originally classified using the American College of Rheumatology criteria for Juvenile Rheumatoid Arthritis (18) or the EULAR criteria for Juvenile Chronic Arthritis (19) and subsequently reclassified for this study by ILAR criteria when possible. For this study, a patient was considered rheumatoid factor negative on the basis of a single test. When data was available for multiple polyarticular RF negative or oligoarticular arthritis patients within a pedigree, a single JIA patient was randomly selected.
Controls
A control cohort of 273 Caucasian individuals evenly dispersed through 3-18 years of age and gender was chosen from a larger cohort selected in a regionally representative manner to ensure comparability to the population of patients within the general area of CCHMC. This was done to avoid the typical bias associated with recruitment from tertiary medical centers or large group physician practices and are healthy individuals without known major health conditions. Participants answered questions regarding health and medical histories for themselves and their families including their grandparents, and have undergone a physical examination.
HLA Typing
PCR-based HLA typing was carried out with a panel of immobilized sequence-specific oligonucleotide probes for the HLA class I (A, B, and C) and class II (DRB1, DQA1, DQB1, DPA1, and DPB1) loci for 820 JIA patients and 273 controls. The PCR products were labeled using biotinylated primers, hybridized to an immobilized probe panel, and the labeled amplicons bound to specific probes were detected using Streptavidin-HRP and a chromogenic substrate (20). The probe reactivity patterns were scanned and the genotypes assigned using an in-house (RMS) software program, StripScan. This system examines polymorphisms in exon 2 for the class II loci and in exons 2 and 3 for the class I loci.
Statistical Analysis
The data were analyzed by gender; age at disease onset (classified as greater or less than 6 years of age, based on previous studies (13)); clinical subtype, limited to polyarticular RF negative and oligoarticular (persistent or extended); and combinations of these when sample size permitted. Sample sizes for the various groupings of the patients are given in table 1. The data were examined at the allele, genotype, and haplotype level. Haplotype estimations and tests for fit to expectations under Hardy-Weinberg Equlibrium (HWE) were performed using the Pypop package (21) (www.pypop.org), which is able to handle the high levels of polymorphism characteristic of the HLA loci.
Table 1.
JIA cohort sizes by subtype, gender and age at disease onset
| Male |
Female |
All total |
|||||
|---|---|---|---|---|---|---|---|
| <6 yrs* | >6 yrs | total | <6 yrs | >6 yrs | total | ||
| RF neg Poly | 30 | 44 | 74 | 124 | 135 | 259 | 333 |
| Oligo-Persistent | 42 | 36 | 78 | 203 | 79 | 282 | 360 |
| Oligo-Extended | 13 | 7 | 20 | 81 | 26 | 107 | 127 |
| All JIA | 85 | 87 | 172 | 408 | 240 | 648 | 820 |
| Control | 139 | 134 | 273 | ||||
Age at disease onset
Tests for heterogeneity between specific groups and association analyses were performed using contingency table testing and a standard Chi-square measure. We stress that we must identify all heterogeneity in disease risk at the primary disease predisposing gene, including relatively weak effects, before proceeding with analyses to detect additional genetic effects. The relative predispositional effect (RPE) method (22) was used to identify all heterogeneity in disease risk at the primary disease gene; alleles, haplotypes, or genotypes with the strongest predisposing or protective effects were sequentially removed from the analysis until no further heterogeneity in risk effects was seen. Determining the order in which haplotypes or genotypes are sequentially removed is not trivial, and requires interplay between the contribution to the Chi-square heterogeneity test, the ORs or Patient/Control (P/C) ratio (23), the OR and P/C ratio are often close in value) and the control frequencies of the allele, haplotype, or genotype. For the same control frequency and equivalent strength of effects, a positive association will contribute more to the overall Chi-square than a negative association. Also, less frequent classes even with stronger predisposing or protective effects can contribute less to the overall Chi-square.
In order to account for linkage disequilibrium within the HLA region, stratification by the disease-associated alleles using the conditional haplotype method (CHM) was employed (24). If all HLA region genes directly involved in disease susceptibility have been identified, then the relative frequencies of alleles at the other HLA loci on high-risk haplotypes should be the same in cases and controls; similarly for neutral and protective haplotypes. While fit to these expectations does not exclude the possibility that other genes in the HLA complex are involved in disease, lack of fit unequivocally shows that all disease-predisposing genes in the region have not been identified.
Survival curves were generated using the Kaplan-Meier method in GraphPad Prism version 5.0 for Macintosh, GraphPad Software, San Diego California USA, www.graphpad.com; significance testing was accomplished with the Mantel-Haenszel method. Principal component analysis was performed using the GenAlEx 6 package (25).
RESULTS
Disease heterogeneity
Contingency table analysis of HLA DR-DQ allele frequency distributions reveals significant (p<0.05) heterogeneity with regard to age of onset and clinical subtypes. Close examination of between-disease group variation suggests that clinical subtype and age of onset generally serve as proxies for one another, although it appears that there is additional variation with regard to age of onset within the polyarticular clinical subtype. In contrast, within the oligoarticular subtypes, little heterogeneity with respect to age of onset is observed.
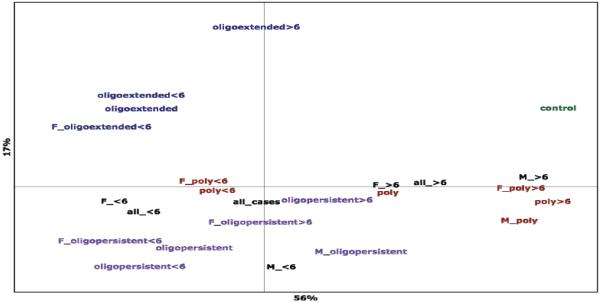
Overall HLA variation in the relevant subgroups of the data are summarized in a Principal Component Analysis (PCA); a plot of coordinates one and two, which account for 56% and 17% respectively of the total variation observed for all eight HLA loci typed (figure 1). Examination of the PCA plot reveals that with respect to HLA variation, three general clusters, each roughly inhabiting a different quadrant of the plot, can be related to clinical subtype. The exception is within the polyarticular subgroup, where earlier age of onset appears to be related to HLA variation more similar to that observed for individuals diagnosed with oligoarticular disease, in keeping with the findings for the association analyses. However, this group clusters very close to the mean for all cases, suggesting that it may be comprised of individuals with a mix of true disease subtypes. Although it appears that a significant proportion of the total variation observed in the HLA loci can be attributed to age of onset differences, substantial variation remains with regard to clinical subtype. To verify that the associations observed are not due to confounding with population substructure, we reanalyzed the primary associations adjusting for the top 10 principal components derived from Affymetrix SNP6.0 GeneChip data. These analyses yielded comparable statistical evidence and magnitudes of the odds ratios (not shown). Thus, these associations are robust to the potential confounding of population substructure.
Figure 1.
Principal component analysis for HLA class I and II loci showing clustering of important subsets of the patient population. The first two coordinates are shown, comprising 56% and 17% respectively of the total variation observed.
Key: oligoextended = oligoarticular extended disease; oligopersistent = oligoarticular persistent disease; poly = IgM RF negative polyarticular disease; <6 = age of onset prior to six years of age; >6 = age of onset after to six years of age; F = female patients; M = male patients.
HLA class II
DR-DQ haplotypic associations
Based on these observations of disease heterogeneity, the data were subdivided to examine the differences between disease subtypes and age of onset. Patient and control frequencies for these comparisons for all DRB1-DQA1-DQB1 haplotypes observed three or more times in the patient population are provided in Table 2. DRB1*1103 and *1104 are combined for this analysis since each is found on the identical DQA1-DQB1 haplotypes. Furthermore DRB1*1103 is not observed among the control population; it is a relatively rare allele and there is no evidence for heterogeneity in its effect relative to that for DRB1*1104.
Table 2.
Common (count ≥ 3) DRB1-DQA1-DQB1 estimated haplotypes in study cohorts
| RF negative polyarticular | Oligoarticular persistent | Oligoarticular extended |
Combined | Controls | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All | <6 yrs* | ≥6 yrs | All | <6 yrs | ≥6 yrs | All | <6 yrs | ≥6 yrs | ||
| Haplotype | freq (n) | freq (n) | freq (n) | freq (n) | freq (n) | freq (n) | freq (n) | freq (n) | freq (n) | freq (n) |
| 0101-101-0501 | 0.128 (85) | 0.112 (34) | 0.152 (50) | 0.105 (74) | 0.111 (52) | 0.090 (19) | 0.099 (25) | 0.112 (109) | 0.116 (75) | 0.09 (49) |
| 0102-0101-0501 | 0.012 (8) | 0.007 (2) | 0.018 (6) | 0.013 (9) | 0.011 (5) | 0.019 (4) | 0.004 (1) | 0.008 (8) | 0.015 (10) | 0.009 (5) |
| 0103-0101-501 | 0.006 (4) | 0.003 (1) | 0.009 (3) | 0.004 (3) | 0.004 (2) | 0.005 (1) | 0.008 (2) | 0.005 (5) | 0.006 (4) | 0.018 (10) |
| 0301-0400-0201 | 0.002 (1) | 0.003 (1) | 0 | 0.004 (3) | 0.006 (3) | 0 | 0.008 (2) | 0.006 (6) | 0 | 0 |
| 0301-0500-0201 | 0.107 (71) | 0.099 (30) | 0.115 (38) | 0.116 (82) | 0.126 (59) | 0.099(21) | 0.075 (19) | 0.107 (104) | 0.105 (68) | 0.112 (61) |
| 0401-0300-0301 | 0.033 (22) | 0.030 (9) | 0.036 (12) | 0.018 (13) | 0.011 (5) | 0.028 (6) | 0.020 (5) | 0.017 (17) | 0.036 (23) | 0.049 (27) |
| 0401-0300-0302 | 0.029 (19) | 0.023 (7) | 0.033 (11) | 0.011 (8) | 0.006 (3) | 0.019 (4) | 0.044 (11) | 0.017 (17) | 0.033 (21) | 0.035 (19) |
| 0402-0300-0302 | 0.002 (1) | 0 | 0.003 (1) | 0.004 (3) | 0.006 (3) | 0 | 0.004 (1) | 0.003 (3) | 0.003 (2) | 0.011 (6) |
| 0403-0300-0302 | 0.006 (4) | 0.007 (2) | 0.006 (2) | 0.008 (6) | 0.009 (4) | 0.009 (2) | 0.012 (3) | 0.009 (9) | 0.006 (4) | 0.004 (2) |
| 0404-0300-0302 | 0.024 (16) | 0.020 (6) | 0.027 (9) | 0.008 (6) | 0.002 91) | 0.024 (5) | 0.012 (3) | 0.009 (9) | 0.025 (16) | 0.029 (16) |
| 0701-0201-0201 | 0.054 (36) | 0.049 (15) | 0.061 (20) | 0.028 (20) | 0.017 (8) | 0.052 (11) | 0.067 (17) | 0.036 (35) | 0.059 (38) | 0.081 (44) |
| 0701-0201-0303 | 0.026 (17) | 0.020 (6) | 0.03 (10) | 0.014 (10) | 0.013 (6) | 0.014 (3) | 0.004 (1) | 0.013 (13) | 0.023 (15) | 0.031 (17) |
| 0801-0400-0402 | 0.08 (53) | 0.102 (31) | 0.064 (21) | 0.137 (97) | 0.152 (71) | 0.099(21) | 0.099 (25) | 0.128 (125) | 0.077 (50) | 0.02 (11) |
| 0801-0600-0301 | 0.003 (2) | 0.007 (2) | 0 | 0.004 (3) | 0.006 (3) | 0 | 0 | 0.005 (5) | 0 | 0 |
| 0805-0400-0402 | 0.003 (2) | 0.003 (1) | 0.003 (1) | 0.004 (3) | 0.006 (3) | 0 | 0.004 (1) | 0.004 (4) | 0.003 (2) | 0 |
| 0901-0300-0303 | 0.012 (8) | 0.016 (5) | 0.009 (3) | 0.010 (7) | 0.009 (4) | 0.014 (3) | 0.012 (3) | 0.012 (12) | 0.009 (6) | 0.007 (4) |
| 1001-0101-0501 | 0.009 (6) | 0.007 (2) | 0.012 (4) | 0.003 (2) | 0 | 0.005 (1) | 0 | 0.003 (3) | 0.009 (6) | 0.007 (4) |
| 1101-0500-0301 | 0.057 (38) | 0.069 (21) | 0.036 (12) | 0.078 (55) | 0.081 (38) | 0.075(16) | 0.079 (20) | 0.076 (74) | 0.06 (39) | 0.064 (35) |
| 1103-0500-0301 | 0.026 (17) | 0.033 (10) | 0.021 (7) | 0.017 (12) | 0.017 (8) | 0.014 (3) | 0.008 (2) | 0.025 (24) | 0.02 (13) | 0 |
| 1104-0103-0603 | 0 | 0 | 0 | 0.005 (4) | 0.006 (3) | 0 | 0 | 0.003 (3) | 0 | 0 |
| 1104-0500-0301 | 0.059 (39) | 0.092 (28) | 0.024 (8) | 0.073 (52) | 0.083 (39) | 0.057 (12) | 0.087 (22) | 0.085 (83) | 0.048 (31) | 0.02 (11) |
| 1201-0500-0301 | 0.012 (8) | 0.016 (5) | 0.009 (3) | 0.010 (7) | 0.006 (3) | 0.019 (4) | 0.029 (7) | 0.012 (12) | 0.015 (10) | 0.015 (8) |
| 1301-0103-0603 | 0.074 (49) | 0.095 (29) | 0.055 (18) | 0.121 (86) | 0.130 (61) | 0.104 (22) | 0.075 (19) | 0.11 (107) | 0.073 (47) | 0.057 (31) |
| 1302-0102-0604 | 0.020 (13) | 0.026 (8) | 0.015 (5) | 0.035 (25) | 0.041 (19) | 0.019 (4) | 0.02 (5) | 0.032 (31) | 0.019 (12) | 0.027 (15) |
| 1303-0500-0301 | 0.012 (8) | 0.020 (6) | 0.006 (2) | 0.013 (9) | 0.015 (7) | 0.009 (2) | 0.008 (2) | 0.014 (14) | 0.008 (5) | 0.016 (9) |
| 1401-0101-0503 | 0.041 (27) | 0.036 (11) | 0.042 (14) | 0.013 (9) | 0.015 (7) | 0.009 (2) | 0.016 (4) | 0.023 (22) | 0.028 (18) | 0.029 (16) |
| 1501-0102-0602 | 0.080 (53) | 0.046 (14) | 0.109 (36) | 0.048 (34) | 0.034 (16) | 0.085 (18) | 0.075 (19) | 0.046 (45) | 0.094 (61) | 0.147 (80) |
| 1501-0102-0603 | 0.002 (1) | 0 | 0.003 (1) | 0.004 (3) | 0.006 (3) | 0 | 0.004 (1) | 0.004 (4) | 0.002 (1) | 0 |
| 1502-0102-0602 | 0.003 (2) | 0.003 (1) | 0.003 (1) | 0.006 (4) | 0.002 (1) | 0.014 (3) | 0.004 (1) | 0.003 (3) | 0.006 (4) | 0.011 (6) |
| 1601-0102-0502 | 0.008 (5) | 0.010 (3) | 0.006 (2) | 0.008 (6) | 0.013 (6) | 0 | 0.012 (3) | 0.011 (11) | 0.005 (3) | 0.015 (8) |
Age at disease onset.
Two primary predisposing DRB1-DQA1-DQB1 haplotypes, DRB1*0801-DQA1*0400-DQB1*0402 and DRB1*1103/4-DQA1*0500-DQB1*0301, and one protective haplotype, DRB1*1501-DQA1*0102-DQB1*0602 (Table 3) are associated with disease status regardless of clinical subtype or age of onset, except in the case of later onset polyarticular disease, where no association with the DRB1*1103/4 haplotype is observed.
Table 3.
Odds ratio of developing JIA by subtype and age at onset conferred by different DRB1*DQA1*DQB1 haplotypes
| RF negative polyarticular JIA | Oligoarticular persistent JIA | Oligoarticular extended JIA | Combined JIA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | All | <6 yrs* | ≥6 yrs | All | <6 yrs | ≥6 yrs | All | <6 yrs | <6 yrs | ≥6 yrs |
| 0801-0400-0402 | 4.23 † (2.19-8.19) |
5.52 (2.73-11.12) |
3.31 (1.57-6.95) |
7.72 (4.10-14.56) |
8.70 (4.55-16.63) |
5.35 (2.53-11.30) |
5.36 (2.59-11.07) |
5.60 (2.71-11.59) |
7.14 (3.82-13.36) |
4.08 (2.10-7.92) |
| 1103/4-0500-0301 | 4.49 (2.33-8.67) |
6.90 (3.45-13.71) |
ns | 4.83 (2.52-9.26) |
5.43 (2.78-10.60) |
ns | 4.65 (2.22-9.75) |
2.26 (1.33-3.85) |
5.99 (3.19-11.24) |
3.55 (1.82-6.95) |
| 1301-0103-0603 | ns | ns | ns | 2.30 (1.50-3.52) |
2.49 (1.59-3.91) |
1.92 (1.09-3.41) |
ns | ns | 2.04 (1.35-3.09) |
ns |
| 1501-0602-0102 | 0.51 (0.35-0.73) |
0.28 (0.15-0.51) |
ns | 0.29 (0.19-0.45) |
0.21 (0.12-0.36) |
0.54 (0.32-0.93) |
0.48 (0.28-0.80) |
0.47 (0.27-0.80) |
0.28 (0.19-0.42) |
0.61 (0.43-0.87) |
| 0401-0300-0301 | ns | ns | ns | 0.35 (0.18-0.70) |
0.21 (0.08-0.54) |
ns | ns | ns | 0.34 (0.18-0.36) |
ns |
| 0701-0201-0201 | ns | ns | ns | ns | 0.20 (0.09-0.42) |
ns | ns | ns | 0.42 (0.27-0.67) |
ns |
Age at disease onset.
Odds ratio (95% Confidence Interval); ns = nonsignificant
Odds ratios for haplotypes with significant disease associations are given in Table 3; these haplotypes generally show very strong effects, particularly within the oligoarticular-persistent subgroup and in those individuals with earlier age of onset. Along with stronger effects for the primary associated haplotypes, additional predisposing and protective haplotypes have been identified through RPE analysis: DRB1*1301-DQA1*0103-DQB1*0603 is predisposing and DRB1*0401-DQA1*0300-DQB1*0301 and DRB1*0701-DQA1*0201-DQB1*0201 are protective for the early-onset oligoarticular-persistent subtype only, although these associations are much weaker than the primary observations. The conditional haplotype method (CHM) applied to the DRB1 and DQB1 loci suggest that the effects of the associated haplotypes are limited to the DRB1 locus, with no evidence of a role for DQA1 or DQB1 in disease.
DPB1
Initial inspection of the data suggests several DPB1 associations with JIA (supplementary table 1). In order to ascertain whether these associations are independent or merely a product of linkage disequilibrium with associated DR-DQ haplotypes, we have studied DPB1 effects using the CHM. This analysis revealed that among polyarticular patients with later onset, no true DPB1 associations are observed. However, in the oligoarticular patients and in polyarticular patients with early age of onset, CHM confirms a predisposing effect for DPB1*0201. After adjusting for linkage disequilibrium with the associated DRB1 alleles, it is apparent that no other DPB1 allele is significantly associated with disease status.
Additive effects
Among both oligoarticular subtypes and polyarticular patients with early onset, the presence of two of the identified predisposing DRB1 alleles is associated with significantly greater predisposition to disease than a single predisposing DRB1 allele; odds ratios range from 13-23 for two predisposing DRB1 alleles relative to only a single predisposing allele, and are higher still relative to those with no predisposing DRB1 alleles, with an odds ratios ranging from 29.95 (CI 3.8-233.1) in the oligoarticular-extended subgroup to 45.68 (CI 6.11-344.0) in early polyarticular patients (supplementary table 2). Indeed, among this patient population, 13% of individuals possess two predisposing DRB1 alleles, while among the control population only one individual fits this genotype category (0.36%). In contrast, in older onset polyarticular patients the solitary predisposing DRB1*0801 allele is not enhanced by the presence of the additional DRB1 predisposing alleles, which in any event are not otherwise associated with disease in these patients.
Stratification by the predisposing DRB1 alleles reveals that in their presence, a modestly increased predisposition is also mediated by DPB1*0201 (p<0.00001; OR=2.4, CI: 1.78-3.25) relative to the predisposing DRB1 alleles alone. Most striking, disease predisposition mediated by DPB1*0201 in individuals without any predisposing DRB1 alleles is great enough to overcome even the very strong protective effect observed for DRB1*1501 (table 4). While the data show that DPB1*0201 mediates predisposition independently, an additive effect is observed in the context of disease-associated DRB1 alleles.
Table 4.
DPB1*0201 mediates predisposition independent from the strong protective effect of DRB1*1501
| DRB1-DPA1-DPB1 haplotype | Combined JIA frequency | Control frequency |
|---|---|---|
| 1501-0103-0201 | 0.232 | 0.075 |
| 1501-0201-1001 | 0.036 | 0.013 |
| 1501-0103-0301 | 0.063 | 0.125 |
| 1501 0103 0401 | 0.554 | 0.650 |
| 1501 0103 0402 | 0.036 | 0.038 |
Age of onset effects
Among polyarticular patients, there is a significant (p <0.0001) correlation between earlier age of onset and the presence of the primary predisposing DRB1*0801 or DRB1*1103/4. Survival curves for individuals with and without these predisposing DRB1 alleles are shown in Figure 2a. The median age of onset for individuals with the predisposing DRB1 alleles is 3.9 years, compared to 7.3 years for those without the predisposing alleles. Examination of the separate effects of DRB1*0801 and *1103/4 suggests that their impact on age of onset is equivalent (not shown).
Figure 2.
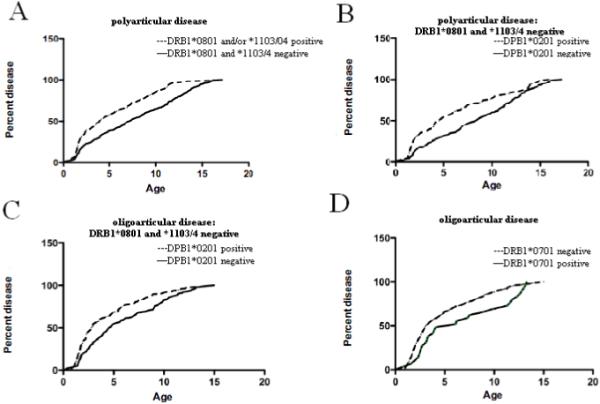
Survival curves for age of onset effects for HLA class II alleles associated with JIA. A. An earlier age of onset in polyarticular disease is mediated by DRB1*0801 and *1103/4. B-C. An earlier age of onset in oligoarticular disease (B), including both persistent and extended as well as polyarticular disease (C) is mediated by DPB1*0201 in the absence of the predisposing alleles DRB1*0801 and *1103/4. D. Later age of onset in oligoarticular disease (persistent and extended) mediated by DRB1*0701
While a similar trend exists among individuals diagnosed with oligoarticular disease, who overall tend toward earlier ages of onset, there is no significant difference in age of onset between those with and without the predisposing DRB1*0801, *1103, *1104 or *1301 alleles (not shown). However, the presence of the predisposing DPB1*0201 allele in individuals who do not bear the predisposing DRB1 alleles is associated with earlier age of onset (p <0.0002) (figure 2b). Presence of the DPB1*0201 allele is associated with a median age of onset of 2.9 years, identical to that for individuals with the predisposing DRB1 alleles; in contrast, the median age of onset for individuals without either the predisposing DRB1 or DPB1 alleles is 4.6 years. Likewise, DPB1*0201 is associated with earlier age of onset among polyarticular patients (p<0.01), where the presence of DPB1*0201 in the absence of the predisposing DRB1 alleles gives a median age of onset of 4.6 years, compared to 8.4 years in DPB1*0201 negative individuals (figure 2c).
Although not reaching statistical significance, among polyarticular patients without the predisposing DRB1 alleles, there is a strong trend toward later onset with DRB1*1501 (median age 9.1 years) compared to those without DRB1*1501 (6.8 years). The protection mediated by DRB1*0701does have significant age of onset effects (p<0.02) in oligoarticular patients, with median onset delayed from 3 years to 6 years (figure 2d). While not significant, a similar trend is observed for protection mediated by DRB1*0401, particularly among individuals with onset later than 3 years.
HLA Class I
Allele frequencies and odds ratios for significant associations for HLA-A, -B and - C in patient and controls for all disease subtypes are given in Supplementary Table 3. Although initial inspection suggests a number of class I associations, the vast majority of these can be attributed to linkage disequilibrium with the class II associated alleles. After controlling for linkage disequilibrium with the primary class II associated alleles using the CHM method, no significant associations for any of the class I loci typed are observed in the polyarticular patients. Among the oligoarticular-extended patients, however, there remains a significant association of disease predisposition with A*0201 (p<0.0001; OR=1.96 CI 1.44-2.67). This analysis also confirms that protection in the oligoarticular-extended group is mediated by A*0101 (p<0.005; OR=0.46 CI 0.27-0.77).
The class I association is different in oligoarticular-persistent patients, among whom predisposition is associated with C*0202 (p<0.05; OR=2.05 CI 1.14-3.69), which is found at frequencies of less than 6% in the patient population and less than 3% of the control population.
DISCUSSION
While several HLA associations have been documented for JIA, barriers to progress in understanding the genetics of JIA have been considerable (16, 26). The clinical phenotype in North America had hitherto used JRA criteria, and included only three subtypes (18). The more recently established JIA criteria used in this study includes seven subtypes; the two most common, oligoarticular and RF negative polyarticular, make up approximately two thirds of all JIA patients. The improved criteria allow for better homogeneity of clinical phenotype and, consequently, a likely improved ability to detect genetic and other biomarkers. The criteria are largely dependent on physical exam in the first few months of illness, relying on counts of involved joints as the basic phenotype. Other biomarkers, including IgM rheumatoid factors, are limited. Anti nuclear antibodies (ANA) are more readily available and associated with ages of onset. Recently Martini et al (4) suggested that ANA’s are less specific for oligoarticular patients and are common in both oligoarticular and polyarticular patients, while others find a split in older polyarticular cases based on gene expression profiles (27). It is anticipated that HLA typing data may provide further resolution of disease heterogeneity.
Early HLA studies were completed with low resolution DNA or serological typing in patients that were incompletely classified. The associations observed in this study are consistent with those reported in the literature for HLA-DR haplotypes (16), and also include a class I association with HLA-A2 that has been long recognized (11) as well as an association of HLA DPB1*0201 with oligoarticular disease, which was one of the first HLA-DP associations identified in an autoimmune disease (10). In drawing together a substantial cohort of patients classified by the JIA criteria, we have allowed consideration of HLA associations in the context of high resolution DNA based typing. Analysis of extended haplotypes now makes possible better to distinguish the roles of the class I and of class II associations, as well as pinpointing the associated loci, amino acid residues and age of onset effects, each with regard to distinct disease subtypes.
In agreement with previous studies, the DR-DQ haplotypes which are associated with disease predisposition here are DRB1*0801-DQA1*0400-DQB1*0402, DRB1*1103/4-DQA1*0500-DQB1*0301 and DRB1*1301-DQA1*0103-DQB1*0603. One protective haplotype, DRB1*1501-DQA1*0102-DQB1*0602 is observed in all subsets of individuals, as well as more limited protective associations for DRB1*0401-DQA1*0300-DQB1*0301 and DRB1*0701-DQA1*0201-DQB1*0201-DQB1*0201. While these associations have been observed previously, there is significant variation in the specificities and strength of these associations depending upon clinical subtype and age of onset which could only be revealed with a sufficiently large cohort.
Furthermore, we have shown that a DRB1 genotype with two predisposing alleles confers considerably greater risk than a single DRB1 predisposing allele. While the DRB1*0801 haplotype is associated with disease regardless of age of onset or clinical subtype, the remaining predisposing alleles show significant associations only in early-onset disease. This suggests that the very strong effect mediated by two copies of predisposing DRB1. alleles may be related to an age of onset effect of the non-DRB1*0801 allele
High-resolution DNA typing and analysis of extended haplotypes has permitted evaluation of the amino acid residues likely associated with disease predisposition by comparing closely related haplotypes that differ in disease risk. While DRB1*1101 is seen at generally equivalent frequencies and on the same DQA1-DQB1 haplotype as the predisposing DRB1*1103/4 in controls, there is no disease association with this haplotype. Likewise DRB1*1302 is not associated with disease, although it occurs at roughly equal frequencies to *1301 (disease associated) in controls. In each of these cases, the predisposing DRB1 allele differs from its related non-predisposing allele by only one amino acid; position 86 Valine in the predisposing alleles and 86 Glycine in the non predisposing alleles. In contrast, the primary predisposing DRB1*0801 allele has a Glycine residue at position 86, suggesting that this residue alone is not responsible for the predisposing effect, or that a different mechanism is responsible for DRB1*0801 predisposition.
Similarly DPB1*0201 differs from the most common DPB1 allele, *0402, at only one amino acid position, 69, with Lysine for *0402 and Glutamic Acid for *0201. As with position 86 in the DRB1 molecule, this residue, 69, plays a critical role in antigen presentation as part of the peptide binding groove in the DP molecule.
In addition to conferring disease predisposition independently, the results also suggest a modifying effect of DPB1 on the DRB1 alleles associated with JIA, whereby DPB1*0201 acts to either augment predisposition or offset protection. The additive effect mediated by DPB1 however is of a much lesser magnitude than that for two predisposing DRB1 alleles, suggesting further that these DRB1 alleles are the primary predisposing variants in JIA. The survival analysis indicates that this modification by DPB1*0201 may be mediated via a tendency to earlier onset disease.
These findings not only allow elucidation of these complex relationships but also provide new insight into the relevance of current diagnostic criteria. We observe distinct differences in the HLA associations in early vs. late onset polyarticular patients. This is further illustrated in the PCA, which suggests that the early onset polyarticular group may be more similar to the oligoarticular group, which itself shows some heterogeneity with respect to age of onset. The oligoarticular group is further distinguished between the persistent and extended subtypes. While some variation with regard to DR-DQ associations may be attributable to lack of power to detect the weaker predisposing and protective variants, it is clear that significant differences exist in class I associations. Specifically, the oligoarticular-extended subtype is characterized by HLA-A associations, while among the oligoarticular-persistent group, only HLA-C is associated with disease status. On the whole, with regard to HLA variation, it appears as though the difference between these subtypes is mediated predominately by class I (after adjusting for the DR-DQ effects). Early-onset polyarticular disease, while sharing many HLA class II associations with the oligoarticular types, does not appear to be mediated in any way by HLA class I. It is clear that individuals with later onset polyarticular disease comprise a disease subtype which is quite distinct from the others and which has very limited HLA associations. In contrast, the subgroup with an early-onset polyarticular diagnosis may be comprised of individuals whose true disease status is mixed, some of whom are more likely destined to follow a disease course similar to that of the oligoarticular patients, while others may ultimately resemble the later-onset polyarticular patients. Overall, while these predisposing DRB1 and DPB1 alleles are found in 40% of controls, they are observed among 78% of patients with age of onset prior to six years, and among 68% of oligoarticular patients with onset after six years. Among polyarticular patients with age of onset later than six years, only DRB1*0801 is seen at increased frequencies, while the remaining predisposing alleles are not associated with disease. Accordingly, it is possible that in the future, HLA typing will be utilized as a diagnostic criterion especially when viewed in the larger genomic context. It is noteworthy that the cohort described in this report coincides with one used for a SNP-based genome-wide association study, and that functional genomic studies to relate genotype, including HLA, to global gene expression levels are possible for about 200 of the subjects. Thus, this HLA data will anchor multiple integrated analyses directed at providing a molecular and biological basis for JIA classification.
Supplementary Material
Acknowledgments
Susan D. Thompson, PhD - supported by NIH/NIAID U01-AI-1067150, NIH/NIAMS contract N01-AI-42272, NIH/NIAMS P30-AR-46373 and the Children’s Hospital Research Foundation.
Mary Ryan, BS- - supported by NIH/NIAMS contract N01-AI-42272 and the Children’s Hospital Research Foundation
Marc Sudman, BA - supported by NIH/NIAMS contract N01-AI-42272 and the Children’s Hospital Research Foundation
Miranda Marion, BS, MS supported by NIH/NIAMS contract N01-AI-42272 and Center for Public Health Genomics
Carl D. Langefeld, PhD supported by NIH/NIAMS contract N01-AI-42272 and Center for Public Health Genomics
Glenys Thomson, PhD - supported by NIH/NIAID U01-AI-1067150
David N. Glass, MD supported by supported by NIH/NIAID U01-AI-1067150, NIH/NIAMS contract N01-AI-42272, NIH/NIAMS P30-AR-46373 and the Children’s Hospital Research Foundation
References
- 1.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–77. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Inocencio J, Giannini EH, Glass DN. Can genetic markers contribute to the classification of juvenile rheumatoid arthritis. J Rheumatol. 1993;20(supplement 40):12–18. [PubMed] [Google Scholar]
- 3.Donn RP, Ollier WE. Juvenile chronic arthritis--a time for change? Eur J Immunogenet. 1996;23(3):245–60. doi: 10.1111/j.1744-313x.1996.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 4.Ravelli A, Martini A. Juvenile idiopathic arthritis. Lancet. 2007;369(9563):767–78. doi: 10.1016/S0140-6736(07)60363-8. [DOI] [PubMed] [Google Scholar]
- 5.Hall PJ, Burman SJ, Laurent MR, Briggs DC, Venning HE, Leak AM, et al. Genetic susceptibility to early onset pauciarticular juvenile chronic arthritis: a study of HLA and complement markers in 158 British patients. Ann Rheum Dis. 1986;45(6):464–74. doi: 10.1136/ard.45.6.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stastny P, Fink C. Different HLA-D associations in adult and juvenile rheumatoid arthritis. J Clin Invest. 1979;63:124–130. doi: 10.1172/JCI109265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Kerckhove C, Luyrink L, Elma MS, Maksymowych WP, Levinson JE, Larson MG, et al. HLA-DP/DR interaction in children with juvenile rheumatoid arthritis. Immunogenetics. 1990;32(5):364–8. doi: 10.1007/BF00211652. [DOI] [PubMed] [Google Scholar]
- 8.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30(11):1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 9.Lundstrom E, Kallberg H, Smolnikova M, Ding B, Ronnelid J, Alfredsson L, et al. Opposing effects of HLA-DRB1*13 alleles on the risk of developing anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60(4):924–930. doi: 10.1002/art.24410. [DOI] [PubMed] [Google Scholar]
- 10.Begovich AB, Bugawan TL, Nepom BS, Klitz W, Nepom GT, Erlich HA. A specific HLA-DP beta allele is associated with pauciarticular juvenile rheumatoid arthritis but not adult rheumatoid arthritis. Proc Natl Acad Sci U S A. 1989;86(23):9489–93. doi: 10.1073/pnas.86.23.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oen K, Petty RE, Schroeder ML. An association between HLA-A2 and juvenile rheumatoid arthritis in girls. J Rheumatol. 1982;9(6):916–20. [PubMed] [Google Scholar]
- 12.Smerdel A, Lie BA, Finholt C, Ploski R, Forre O, Undlien DE, et al. An additional susceptibility gene for juvenile idiopathic arthritis in the HLA class I region on several DR-DQ haplotypes. Tissue Antigens. 2003;61(1):80–4. doi: 10.1034/j.1399-0039.2003.610107.x. [DOI] [PubMed] [Google Scholar]
- 13.Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, et al. Age-specific effects of juvenile rheumatoid arthritis-associated HLA alleles. Arthritis Rheum. 1999;42(9):1843–53. doi: 10.1002/1529-0131(199909)42:9<1843::AID-ANR8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Haas JP, Andreas A, Rutkowski B, Brunner H, Keller E, Hoza J, et al. A Model for the Role of HLA-DQ Molecules in the Pathogenesis of Juvenile Chronic Arthritis. Rheumatology International. 1991;11:191–197. doi: 10.1007/BF00332561. [DOI] [PubMed] [Google Scholar]
- 15.Runstadler JA, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, et al. Analysis of MHC region genetics in Finnish patients with juvenile idiopathic arthritis: evidence for different locus-specific effects in polyarticular vs pauciarticular subsets and a shared DRB1 epitope. Genes Immun. 2003;4(5):326–35. doi: 10.1038/sj.gene.6364002. [DOI] [PubMed] [Google Scholar]
- 16.Thomson W, Barrett JH, Donn R, Pepper L, Kennedy LJ, Ollier WE, et al. Juvenile idiopathic arthritis classified by the ILAR criteria: HLA associations in UK patients. Rheumatology (Oxford) 2002;41(10):1183–9. doi: 10.1093/rheumatology/41.10.1183. [DOI] [PubMed] [Google Scholar]
- 17.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31(2):390–2. [PubMed] [Google Scholar]
- 18.Brewer EJ, Jr., Bass J, Baum J, Cassidy JT, Fink C, Jacobs J, et al. Current proposed revision of JRA Criteria. JRA Criteria Subcommittee of the Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Section of The Arthritis Foundation. Arthritis Rheum. 1977;20(2 Suppl):195–9. [PubMed] [Google Scholar]
- 19.European League against Rheumatism: EULAR . National Zeitungs. AG Basel; 1977. Bulletin 4: Nomenclature and classification of Arthritis in Children. 1977. [Google Scholar]
- 20.Bugawan TL, Apple R, Erlich HA. A method for typing polymorphism at the HLA-A locus using PCR amplification and immobilized oligonucleotide probes. Tissue Antigens. 1994;44(3):137–47. doi: 10.1111/j.1399-0039.1994.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 21.Lancaster A, Nelson MP, Meyer D, Thomson G, Single RM. PyPop: a software framework for population genomics: analyzing large-scale multi-locus genotype data. Pac Symp Biocomput. 2003:514–25. [PMC free article] [PubMed] [Google Scholar]
- 22.Payami H, Joe S, Farid NR, Stenszky V, Chan SH, Yeo PP, et al. Relative predispositional effects (RPEs) of marker alleles with disease: HLA-DR alleles and Graves disease. Am J Hum Genet. 1989;45(4):541–6. [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson G, Valdes AM, Noble JA, Kockum I, Grote MN, Najman J, et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens. 2007;70(2):110–27. doi: 10.1111/j.1399-0039.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- 24.Thomson G, Valdes AM. Conditional genotype analysis: detecting secondary disease loci in linkage disequilibrium with a primary disease locus. BMC Proc. 2007;1(Suppl 1):S163. doi: 10.1186/1753-6561-1-s1-s163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peakall R, Smouse PE. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6(1):288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas JP, Nevinny-Stickel C, Schoenwald U, Truckenbrodt H, Suschke J, Albert ED. Susceptible and Protective MHC-class II Haplotypes in Early Onset Pauciarticular Juvenile Chronic Arthritis. Human Immunol. 1994;41:225–233. doi: 10.1016/0198-8859(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 27.Griffin TA, Barnes MG, Ilowite NT, Olson JC, Sherry DD, Gottlieb BS, et al. Gene expression signatures in polyarticular juvenile idiopathic arthritis demonstrate disease heterogeneity and offer a molecular classification of disease subsets. Arthritis Rheum. 2009;60(7):2113–2123. doi: 10.1002/art.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.