Abstract
We report the 1.77-Å resolution X-ray crystal structure of a dodecamer DNA duplex with the sequence 5′-CCTCTGGTCTCC-3′ that has been modified to contain a single engineered 1,2-cis-{Pt(NH)3)2}2+-d(GpG) cross-link, the major DNA adduct of cisplatin. These data represent a significant improvement in resolution over the previously published 2.6-Å structure. The ammine ligands in this structure are clearly resolved, leading to improved visualization of the cross-link geometry with respect to both the platinum center and to the nucleobases, which adopt a higher energy conformation. Also better resolved are the deoxyribose sugar puckers, which allow us to re-examine the global structure of platinum-modified DNA. Another new feature of this model is the location of four octahedral [Mg(H2O)6]2+] ions associated with bases in the DNA major groove and the identification of 124 ordered water molecules that participate in hydrogen bonding interactions with either the nucleic acid or the diammineplatinum(II) moiety.
Keywords: cisplatin, DNA structure, X-ray crystallography
1. Introduction
Platinum-based therapy remains a highly utilized and effective option in the treatment of many types of cancer [1]. After cisplatin (cis-diamminedichloroplatinum(II)) was discovered to have antitumor properties more than 40 years ago [2, 3], much research has focused on unraveling the mode of action of this compound. Nuclear DNA is an important molecular target for platinum anticancer compounds, which bind purine bases at the N7 position. The resulting Pt-DNA damage triggers downstream effects including inhibition of replication and transcription, cell cycle arrest, and attempted repair of the damaged nucleotides. If the cell cannot remove the damage then it dies by one of several pathways [4].
Revealing the structural details of Pt-DNA adducts represented a significant milestone in platinum anticancer research. This information has helped to build structure-activity relationships that underlie transcription inhibition and cell death. The major adduct of cisplatin-DNA binding is a 1,2-intrastrand adduct between adjacent guanine bases; a minor percentage of 1,3-intrastrand and interstrand cross-links also form [4]. The 1,2-d(GpG) cisplatin cross-link was first characterized crystallographically in a DNA dodecamer duplex in 1996 at 2.6-Å resolution. This structure revealed that the Pt adduct induces a global bend of 35-40° in the DNA duplex and unwinds the double helix by ~25° [5, 6]. The major groove is compacted and the minor groove widened and flattened. A-form DNA comprises the nucleic acid to the 5′ side of the Pt cross-link, and B-DNA forms to the 3′ side of the 1,2-d(GpG) adduct. The roll angle between platinated guanine bases is 26° and, as a consequence, results in considerable strain being placed on the Pt-N7 bonds, displacing the Pt atom out of the guanine ring planes by approximately 1 Å each.
In the present article we report a higher resolution, 1.77 Å structure of this complex, a dodecamer duplex having the sequence 5′-CCTCTG*G*TCTCC-3′, where the asterisks denote platination sites. This significant increase in resolution offers a much-improved view of the electron density, particularly in the Pt-DNA adduct region. With these new data in hand, we are able to evaluate the structure of the cisplatin 1,2-cis-{Pt(NH)3)2}2+-d(GpG) cross-link on DNA at a level not previously attainable. In particular, we have important new information about the overall DNA geometry, the square-planar platinum coordination environment, the conformation of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) intrastrand cross-link, and the interactions between Pt-DNA molecules and water and metal ion components of the crystallization buffer. At 1.77-Å resolution the deoxyribose ring conformations can be clearly delineated from electron density maps, providing highly useful information about the nucleic acid structure. Improved electron density also exists for the platinum ammine ligands, so that the Pt–N bond distances and angles are more accurately determined and now match closely the values expected for a square-planar Pt coordination compound. This improved model prompted us to re-visit the unexpected observation that the Pt-N7 bonds to each guanine base are highly strained, and to ascertain the origin of this property. Finally, located in the current structure are four octahedral [Mg(H2O)6]2+] ions that interact with the DNA duplex through the primary hydration sphere; we also could define ordered water molecules that hydrogen bond to both the nucleic acid and the {Pt(NH3)2}2+ unit.
2. Materials and Methods
Phosphoramidites, columns, and other reagents for solid-phase oligonucleotide synthesis were purchased from Glen Research. Potassium tetrachloroplatinate(II), which was used to synthesize cisplatin according to published procedures [7], was a gift from Engelhard (Iselin, NJ, now BASF). Crystallization reagents and supplies were obtained from Hampton Research. All other reagents were purchased from commercial suppliers and used without further purification. Oligonucleotides were prepared in-house using an Applied Biosystems Model 392 DNA/RNA Synthesizer. Liquid chromatography was performed with an Agilent 1200 series HPLC equipped with a temperature-controlled autosampler and automated fraction collector. Atomic absorption spectroscopy was performed with a Perkin Elmer AAnalyst 300 system. UV-Vis spectra were collected on a Hewlett-Packard 8453 spectrophotometer.
2.1. Preparation of platinated DNA duplex
The oligonucleotide 5′-d(CCTCTGGTCTCC)-3′ (1) and its complement (2) were synthesized on a 2 × 1.0 μmol scale with dimethoxytrityl (DMT) groups on by standard solid-phase synthetic methods. The strands were purified by reversed-phase high-pressure liquid chromatography (HPLC) on an Agilent SB-300, 9.4 × 250 mm column, with a linear gradient of 100 mM triethylammonium acetate pH 7.0 and acetonitrile. After lyophilization, DMT groups were removed in 80% acetic acid for 30 min at room temperature, and the oligonucleotides were precipitated with isopropanol and desalted with Waters Sep-Pak C18 cartridges. Reaction of 1 with 1.2 equiv of cisplatin was carried out in 10 mM HEPES pH 6.8 buffer for 14 h at 37 °C. The platinated product was purified by ion-exchange HPLC on a Dionex DNA-Pac PA-100, 9.4 × 250 mm column at 35 °C, with a mobile phase of 90:10 25 mM Na acetate pH 5.2:acetonitrile and a linear NaCl gradient. Pure fractions were dialyzed against water overnight, then lyophilized and reconstituted in water, yielding 5′-d(CCTCTG*G*TCTCC)-3′ (3). The site-specifically platinated duplex was prepared by combining equimolar amounts of 2 and 3 in 200 μL of 200 mM LiCl, 100 mM HEPES pH 7.0, and 50 mM MgCl2, heating to 70 °C for 10 min, and cooling to 4 °C over 2.5 h. The duplex was purified by ion exchange HPLC as described above but with a LiCl gradient. The final product was dialyzed, lyophilized, and desalted as described above.
2.2 Crystallization and X-ray diffraction data collection
Diffraction-quality crystals were grown by the hanging-drop vapor diffusion method at 4 °C. Crystallization solutions contained 120 mM magnesium acetate, 50 mM sodium cacodylate pH 6.5, 1 mM spermine, and 28% w/v polyethylene glycol (PEG) 4000; hanging drops contained 2 μL of 0.4 mM Pt-DNA in water and 2 μL of crystallization solution, equilibrated against 1 mL of crystallization solution. All solutions were prepared and sterile filtered immediately before use. Crystals with dimensions of ~1.0 mm × 0.2 mm × 0.1 mm grew in clusters in approximately two weeks.
Single crystals were isolated from the clusters and transferred to a cryoprotectant solution of 120 mM magnesium acetate, 50 mM sodium cacodylate pH 6.5, 1 mM spermine, 28% w/v polyethylene glycol (PEG) 4000, and 15% v/v glycerol, mounted on loops, and flash frozen directly in liquid nitrogen. Diffraction data were collected at 100 K (λ = 0.979 Å) at beamline 9-2 of the Stanford Synchrotron Radiation Laboratory (SSRL), and processed in HKL2000 [8]. Data were collected over 360° with 1° oscillation per frame. Data collection statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics.a
| Beamline | SSRL BL9-2 |
| Wavelength | 0.979 Å |
| Collection temperature | 100 K |
| Space group | P1 |
| Unit cell parameters | |
| a | 31.30 Å |
| b | 35.43 Å |
| c | 45.13 Å |
| α | 80.06° |
| β | 84.09° |
| γ | 81.77° |
| Z | 2 |
| Unit cell volume (Å3) | 48,729 |
| Resolution limit (Å) | 1.77 (1.83-1.77) |
| Unique reflections | 16734 |
| Completeness (%) | 96.0 (85.2) |
| Redundancy | 2.9 (2.5) |
| Rmerge (%)b | 9.2 (38.9) |
| I/σ(I) | 8.0 (2.5) |
| Resolution range (Å) | 50 - 1.77 |
| R (%)c | 17.2 |
| Rfree (%)d | 19.8 |
| B-factors (Å2) | |
| DNA | 37.8 |
| Water | 47.2 |
| Pt | 28.7 |
| Mg | 46.5 |
| RMSD bond lengths | 0.011 Å |
| RMSD bond angles | 1.88° |
| DNA atoms | 972 |
| Water atoms | 148 |
| Mg atoms | 4 |
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|I-□I〉|/ΣI.
R = Σ||FO|-|FC||/Σ|FO|.
Rfree = R obtained for a test set of reflections (5% of diffraction data).
2.3. Structure determination and evaluation
Phasing of the diffraction data was achieved by molecular replacement with the program Phaser [9] using the original structure (1AIO) with all water molecules removed as a search model. After one round of restrained refinement with Refmac5 [10], the Rfree value was below 24%. Platinum-nitrogen bond distances and angles were adjusted to fit the improved electron density around the ammine ligands, and [Mg(H2O)6]2+] sites were inserted manually into the 2Fo-Fc and Fo-Fc electron density maps with the program Coot [11]. Water molecules were placed automatically with Coot into areas of Fo-Fc density above 3.5σ and then adjusted manually. Subsequent rounds of refinement and model adjustment were performed to afford the final model with an Rfree value of 19.8%. Final refinement statistics are given in Table 1.
Nucleic acid geometric parameters were calculated with the program 3DNA [12]. Single point energy calculations of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) moiety were performed with Gaussian 03 suite of programs [13]. From the experimental structures, the atomic valences were completed with hydrogen atoms. Thus, the final model carries a molecular charge of 2+. In this approach, the models are treated by Density Functional Theory (DFT), with the B3LYP functional [14, 15] and the 6-31g(d,p) basis set [16] on all light atoms (H, C, N, O and P). Platinum was represented by the Los Alamos LANL2DZ basis [17, 18], which includes relativistic effective core potentials. All crystallographic images in this paper were created using Pymol [19]. Coordinates have been deposited into the Protein Data Bank (PDB) with accession code 3LPV.
3. Results and discussion
3.1 Unit cell and crystal packing
Crystals of the Pt-DNA construct were grown under different conditions than those reported from the original investigation; however, the two structures are nearly identical. The present conditions were used to obtain diffraction quality crystals of DNA duplexes containing an oxaliplatin-DNA cross-link [20] as well as a monofunctional Pt-DNA adduct of cis-diammine(pyridine)chloroplatinum(II) [21]. Despite differences in the present and previous conditions used to grow crystals, the cisplatin-modified duplex DNA crystallizes in the same space group, P1, and with nearly identical until cell dimensions as the previous structure. Unit cell dimensions for the original crystal structure are a = 31.27 Å, b = 35.46 Å, c = 47.01 Å, α = 79.81°, β = 84.75°, and γ = 82.79°, whereas the dimensions for this crystal are a = 31.30 Å, b = 35.43 Å, c = 45.13 Å, α = 80.06°, β = 84.09°, and γ = 81.77°, giving unit cell volumes of 50,770 Å3 and 48,729 Å3, respectively. The 4% decrease in volume can be attributed to differences in temperature; data for the first cisplatin-DNA structure were collected at 277 K, whereas we collected the current data at 100 K. Because the unit cells are nearly equivalent, packing interactions between molecules are also conserved between structures. Two unique DNA duplexes comprise the asymmetric unit that are related by non-crystallographic symmetry and interact via hydrogen bonding, end-to-end, and end-to-groove contacts in the crystal lattice. The combination of the latter two interactions in the same crystal is unusual, the former being typically observed in B-form DNA structures and the latter in crystals of A-form nucleic acids.
3.2. Global DNA geometry
Notable characteristics of DNA containing the cisplatin 1,2-{Pt(NH)3)2}2+-d(GpG) cross-link, determined from the original crystal structure, include bending of the double helix by 35-40° toward the major groove and local duplex unwinding of ~25°. The roll angle between Pt-bound guanine bases is 26° (see Figs. 1a and S1). DNAs from the previous and current models align nearly identically, with a root-mean-square deviation (rmsd) over all atoms of 0.472 Å (see Fig. 2). Although there are two crystallographically unique molecules in the unit cell, the DNA structures of each are also equivalent, with a rmsd over all atoms of 0.18 Å. Analysis of the DNA structure beyond what can be found in the original publication [6] will therefore be restricted to that which is clarified or changed after collection of the present high-resolution data.
Figure 1.
Structural features of cisplatin-damaged DNA. (a) Overall structure of duplex DNA containing a cisplatin cross-link (shown in white/gray). (b) Stereo images of the platinum-bound base pairs in molecule A with 2Fo-Fc electron density (green around the DNA/blue around the Pt adduct) contoured at 1.5σ. (c) Stereo images of a [Mg(H2O)6]2+ octahedral site bound in the major groove of molecule A at guanine residues 23 and 24, with 2Fo-Fc electron density (shown in blue) contoured at 1.5σ.
Figure 2.

Stereo view of molecule A of the previously published structure of DNA modified with a 1,2-cis-{Pt(NH)3)2}2+-d(GpG) cross-link (PDB accession code 1AIO, shown in blue) superimposed on the current high-resolution structure (3LPV, in red). The two molecules align almost identically, with a rmsd over all atoms of 0.472 Å.
The DNA takes on A-form properties on the 5′ side of the Pt-DNA adduct and is a B-form structure on the 3′-end. In the original crystal structure the resolution limit prohibited direct visualization of deoxyribose sugar puckers, so ring conformations were indirectly determined by measuring distances between adjacent phosphate atoms and examining base-stacking patterns. However, in the current 1.77 Å structure, we can clearly differentiate sugar ring conformations as either C3′-endo (typical of A-form DNA) or C2′-endo (for B-form DNA) in the electron density maps (see Fig. S2). This unusual A/B structural feature was originally postulated to be caused by the presence of [Co(NH3)6]3+ in the crystallization solution, because this complex stabilizes A-form DNA [22]. This suggestion is clearly incorrect, however, because hexamminecobalt(III) was not present in our experiments. A-form DNA occurs in several protein complexes in vivo, including a region of the Xenopus 5S RNA gene bound by transcription factor IIIA [23] and the DNA binding site of HIV reverse transcriptase [24]. A-DNA may be preferable for protein binding because the greater rigidity of its helix makes for a more favorable recognition site. This analysis is consistent with a previous hypothesis that cisplatin acts on cells in part by forming DNA cross-links that alter DNA structure to mimic protein binding sites, possibly to hijack nuclear proteins and disrupt their function [25, 26].
In the present structure, the A-like geometry is reflected by large negative slide values for base pair steps to the 5′ side of the cisplatin damage. Displacement of base pairs in this manner causes strain between the sugar ring and phosphate group, which is relieved by flipping the 3′ carbon atom of the deoxyribose ring away from the phosphate into an endo conformation, conveying A-form properties [27]. This pressure is absent to the 3′ side of the helix, where the slide values are nearly zero at each base pair step, allowing the deoxyribose moieties to adopt a B-form structure. In other structures of duplex DNA containing the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) cross-link, including an NMR solution structure [28] and X-ray crystal structure with bound HMGB1 domain A [29], the 5′ platinated guanosine adopts the C3′-endo sugar pucker; however, the remainder of the double-stranded DNA adopts B-form structure. Thus the presence of A-like DNA in the present determination may be influenced by crystal packing interactions. Finally, although many features of the double helix to the 5′ side of the Pt cross-link resemble A-form DNA, other geometric characteristics are more like those of B-DNA. In addition to the previously mentioned C3′-endo sugar puckering and highly negative slide values, the A-like segments of the duplex have average helical twist values (30.4°, see Fig. S1) and minor groove widths (10.3 Å) similar to those of typical A-form (30.4° and 10.0 Å, respectively) compared to B-form DNA (35.6° and 6.2 Å, respectively) [30]. However, the average roll angle along the same six base-pair segment (5.1°) assumes an intermediate value between those of B-DNA (1.6°) and A-DNA (10.0°). Similarly, the mean inclination angle (9.7°) at the 5′ end of each duplex more closely resembles that of B-DNA (3.4°) than A-DNA (20.1°). Therefore, although many aspects of this portion of the structure display A-form DNA features, there are some B-DNA similarities, and nucleic acids such as the present one cannot classified as strictly A- or B-form.
3.3. Pt adduct geometry
Unlike the overall DNA structure that is unchanged by addition of higher resolution diffraction data, the cisplatin adduct is significantly altered in the current model. Clear electron density is now visible for the ammine ligands of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) cross-link (see Fig. 1b), allowing the square-planar Pt geometry to be accurately evaluated. The Pt–N bond lengths for all four ligands average to 2.02 Å in the new model, compared to 1.90 Å in the previous structure, and 2.02 Å in the X-ray crystal structure of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) dinucleotide determined at atomic resolution [31]. The average cis N–Pt–N angles for the current and prior models and the dinucleotide structure are 90.0°, 88.0°, and 90.0° respectively, with ranges of 83.9° - 99.8°, 70.9° - 99.9°, and 86.9° - 94.3°, respectively. The trans angles are dramatically improved in the high-resolution model, with an average N–Pt–N value of 174.5°, compared to 156.9° in the 2.6 Å-resolution model and 175.1° for the isolated Pt-d(GpG) cross-link. Thus the current model depicts a platinum adduct geometry that very nearly matches that observed in the X-ray crystal structure of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) dinucleotide. A full comparison of Pt geometries is provided in Table 2.
Table 2.
Comparison of platinum adduct geometries between the current high resolution structure, the published structure 1AIO, and the X-ray crystal structure of the platinated dinucleotide 1,2-cis-{Pt(NH3)2}2+-d(GpG)
| 3LPVa | Range | 1AIOa | Range | Pt(GpG)b | Range | |
|---|---|---|---|---|---|---|
| Pt-N1 (Å) | 2.03 | 1.98 - 2.07 | 1.89 | 1.88 - 1.89 | 2.05 | 2.00 - 2.09 |
| Pt-N2 (Å) | 1.99 | 1.88 - 2.10 | 1.91 | 1.90 - 1.92 | 2.04 | 2.02 - 2.08 |
| Pt-N7(5′G) (Å) | 2.00 | 1.98 - 2.02 | 1.89 | 1.86 - 1.91 | 1.98 | 1.92 - 2.05 |
| Pt-N7(3′G) (Å) | 2.07 | 2.06 - 2.08 | 1.91 | 1.90 - 1.92 | 2.02 | 1.97 - 2.09 |
| Avg. Pt-N (Å) | 2.02 | 1.90 | 2.02 | |||
| N7(5′G)-Pt-N1 (°) | 97.1 | 92.3 - 101.3 | 85.7 | 83.2 - 88.1 | 89.0 | 88.0 - 90.3 |
| N7(5′G)-Pt-N7(3′G) (°) | 88.9 | 87.4 - 90.4 | 99.7 | 99.5 - 99.9 | 88.8 | 86.9 - 89.5 |
| N7(3′G)-Pt-N2 (°) | 89.0 | 87.7 - 90.4 | 71.4 | 70.9 - 71.9 | 90.1 | 88.1 - 92.2 |
| N1-Pt-N2 (°) | 85.1 | 83.7 - 86.5 | 95.2 | 91.8 - 98.5 | 92.0 | 90.9 - 94.3 |
| N7(5′G)-Pt-N2 (°) | 176.3 | 175.0 - 177.5 | 155.1 | 154.9 - 155.2 | 177.0 | 173.3 - 179.1 |
| N7(3′G)-Pt-N1 (°) | 173.9 | 171.4 - 176.5 | 158.8 | 157.1 - 160.5 | 175.6 | 174.2 - 178.1 |
Average of two measured values from each crystallographically unique Pt-DNA molecule.
Average of four measured values.
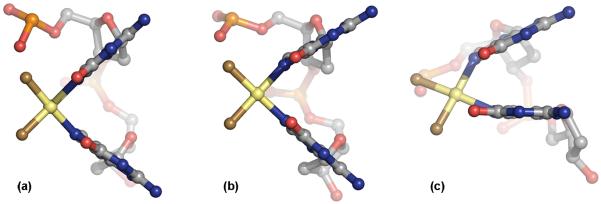
Consistent with the original model, the platinum atoms are displaced from the guanine base planes. The metal atom lies out-of-plane of the 5′ guanine by 1.2 and 1.1 Å in the two molecules, and of the 3′ nucleobase by 1.0 Å in both DNA duplexes. This feature, however, is inconsistent with data from both the X-ray crystal structure of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) dinucleotide [31] and the NMR solution structure of duplex DNA containing the lesion [28], in which the Pt atom is located within the guanine base planes and the dihedral angle between bases is closer to 90° (see Fig. 3). The shallow roll angle in the present X-ray crystal structure causes significant angular strain on the Pt–N7 bonds. Single point energy calculations indicate that the {Pt(NH)3)2}2+-d(GpG) moiety in the Pt-DNA X-ray structure is 14.6 kcal/mol higher in energy than the analogous NMR solution structure [28]. DNA end-to-end packing on the 3′ side of the Pt adduct and end-to-groove packing on the 5′ side of the cross-link appear to dictate the global curve of the molecule and prevent complete opening of the guanine-guanine base pair roll angle. However, the same crystal packing contacts are capable of stabilizing the altered higher energy conformation, since a single hydrogen bonding interaction can contribute ca. 1 - 9 kcal/mol in energy [32]. These results underscore the importance of evaluating macromolecular structures by multiple means, including NMR spectroscopy and biochemical methods in addition to X-ray crystallography, because each has its limitations.
Figure 3.
Views of the 1,2-cis-{Pt(NH)3)2}2+-d(GpG) adduct as depicted by (a) the X-ray crystal structure of the isolated dinucleotide [31], (b) the NMR solution structure of the Pt moiety in 8-mer duplex DNA [28], and (c) the X-ray crystal structure determination of dodecamer DNA presented here.
3.4. Magnesium sites identified
Four fully aquated magnesium(II) cations were located in the major groove on the A-form side of the nucleic acid double helix. As in other DNA crystal structures, interaction of the Mg(aq)2+ cation and the DNA major groove occurs via hydrogen bonding, the water ligands serving as hydrogen bond donors to the N7 atoms of guanine and adenine bases and to the O6 atom of guanine (see Fig. 1c) [33, 34]. In all cases the [Mg(H2O)6]2+ cations interact with adjacent purine bases, either the terminal d(GpG) or the d(ApG) dinucleotide sequence at the 3′ end of the unplatinated strand (see Fig. 4).
Figure 4.

Binding of [Mg(H2O)6]2+ cations to purine dinucleotides in the cisplatin-DNA dodecamer duplex. Hydrogen bonding interactions between water ligands and nucleobases are depicted in cyan.
Significantly better electron density exists for two Mg(aq)2+ complexes bound to the terminal d(GpG) sequence than the other two sites. Crystallographic B-factors for the former are 38.9 and 41.0 Å2, compared to 50.5 and 51.6 Å2 for the latter. The d(GpG)-bound [Mg(H2O)6]2+ complexes contribute four hydrogen bonding interactions with the DNA bases, whereas the less ordered sites participate in only three such contacts. This difference is one possible reason why the first two sites have lower B-factors. As a result, the Mg–OH2 bond distances for the former, which average 2.05 ± 0.08 Å, are close to the reported literature values of 2.06 Å [34], and the complexes have nearly ideal octahedral symmetry. The magnesium(II) complexes that bind d(ApG) sequences near the platinum adduct, bases 20-21 and 44-45 as depicted in Fig. 5, have Mg–OH2 bond distances as long as 2.46 Å if left unrestrained (data not shown), which is clearly unrealistic and an artifact of insufficiently well defined electron density around those water ligands. These complexes were therefore refined with restraints on bond distances and angles, whereas the d(GpG) coordinated Mg(aq)2+ sites were refined without restraints. No [Mg(H2O)]2+-DNA interactions were observed at the B-form end of this palindromic sequence, although they are probably present but not visible in the electron density. Average crystallographic B-factors for the four terminal base pairs on the 5′ side of the Pt cross-link, where Mg(aq)2+ coordination is resolved, are 33.8 and 36.0 Å2 for the two DNA duplexes. In contrast, these values are 45.6 and 46.0 Å2 for the last four base pairs on the 3′ ends of the two molecules, indicating that the B-form sections of each nucleic acid are less ordered than the respective A-DNA segments.
Figure 5.
Schematic depicting hydrogen-bonding interactions between solvent molecules and Pt-DNA. For clarity, many contacts between water molecules are omitted.
3.5. Ordered water molecules
Of the 148 ordered water molecules located in the model, 24 are bound to magnesium in an octahedral geometry as described above. The remaining 124 solvent molecules interact with either the platinum adduct, phosphodiester backbone, or the DNA bases. A schematic depiction of each of these solvent-DNA interactions is given in Fig. 5, and a full list of water contacts appears in Table S1. One water molecule participates in bridging hydrogen-bonding interactions with each of the platinum ammine ligands. Molecule A has four solvent molecules bound to the Pt-DNA adduct, whereas molecule B has only two ammine-bound waters. Unlike another high-resolution structure of Pt-modified DNA, no direct interaction between ordered waters and the platinum atom is observed [35, 36].
Hydrogen bonding interactions between water molecules and nucleobases occur primarily at guanine N7, O6, or N3 atoms, adenine N7, N6, or N3, thymine O2 or O4, and cytosine N4 or O2 positions, consistent with previous observations of Watson-Crick base pair hydration patterns [37, 38]. Water contacts with oxygen atoms of the deoxyribose rings (O3′, O4′, or O5′) are also observed, although less regularly. There are more resolved water-nucleobase interactions at the central base pairs in the duplexes; terminal base pairs do not show as much detail in the hydration sphere, presumably due to increased thermal motion.
The phosphodiester backbone of each DNA double helix is extensively hydrated, with water molecules participating in hydrogen bonding with the phosphate oxygen atoms. Mono- and bidentate interactions, either with two oxygen atoms from a single phosphate or from adjacent nucleotides, occur. Most bridging interactions between two phosphates are found in the central four base pairs, where the DNA backbone structure is distorted by the cisplatin cross-link. Some of the atoms modeled here as oxygen from water may actually be sodium ions, given that 50 mM sodium cacodylate was present in the crystallization buffer and that DNA exists as a polyanion and must be neutralized by cations. However, these two species cannot be differentiated by X-ray crystallography since Na+ and O2− each contain ten electrons; therefore, all non-magnesium solvent molecules were modeled as water.
4. Conclusions
The structure of a dodecamer DNA duplex containing a centrally located 1,2-cis-{Pt(NH)3)2}2+-d(GpG) intrastrand cross-link of cisplatin was determined by X-ray crystallography to 1.77 Å resolution, a greater than 0.8 Å improvement over the previous structure solved in 1996. This improvement in resolution advances our understanding of the structure of cisplatin-modified duplex DNA in four primary areas: (i) we can now identify deoxyribose sugar puckers directly from the electron density and compare the resulting A/B DNA hybrid to other Pt-DNA structures and to biologically relevant non-standard DNA geometries; (ii) we more accurately determine the structure of the platinum adduct, which now closely matches idealized square-planar geometry, and can eliminate the possibility that the DNA duplex distorts the structure of the primary coordination sphere; (iii) we have sharpened the characteristic features of the {Pt(NH)3)2}2+-d(GpG) moiety, specifically with regard to the guanine plane orientation; and (iv) we identify four Mg(aq)2+ complexes and dozens of additional interacting solvent molecules.
Understanding the structural changes to DNA that occur upon binding of platinum antitumor agents is a critical step in deciphering the mechanism of these compounds. X-ray crystallography is a powerful technique for answering some of these questions, and should continue to be relied upon in the future as other platinum complexes are synthesized and studied. From these data we can better understand how Pt-DNA adducts might be repaired and inhibit cellular transcription. Understanding these two functions is key to the design of improved platinum candidates for cancer chemotherapy.
Synopsis.
The structure of a cisplatin-modified DNA dodecamer duplex containing a 1,2-cis-{Pt(NH)3)2}2+-d(GpG) cross-link has been determined at 1.77-Å resolution. Improved electron density reveals new features not seen in the originally published 2.6 Å structure.
Supplementary Material
Acknowledgements
This work was supported by the National Cancer Institute (grant CA034992 to SJL). RCT is grateful for a Koch Fund fellowship from the Koch Institute for Integrative Cancer Research at MIT. We thank Engelhard Corporation (now BASF) for the generous donation of K2PtCl4 used in this research and Juliana Fedoce Lopes for conducting single point energy calculations. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
R.C.T. designed and performed experiments and wrote the paper. S.J.L. designed experiments and wrote the paper.
References
- [1].Kelland L. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- [2].Rosenberg B, Van Camp L, Krigas T. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- [3].Rosenberg B, Van Camp L, Trosko JE, Mansour VH. Nature. 1969;222:385–386. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- [4].Wang D, Lippard SJ. Nat. Rev. Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- [5].Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- [6].Takahara PM, Frederick CA, Lippard SJ. J. Am. Chem. Soc. 1996;118:12309–12321. [Google Scholar]
- [7].Dhara SC. Indian J. Chem. 1970;8:193–194. [Google Scholar]
- [8].Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- [9].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- [11].Emsley P, Cowtan K. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- [12].Lu X-J, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Frisch MJ, Trucks HBSGW, Scuseria GE, Robb JRCMA, Montgomery JA, Jr., Vreven T, Kudin JCBKN, Millam JM, Iyengar SS, Tomasi J, Barone BMV, Cossi M, Scalmani G, Rega N, Petersson HNGA, Hada M, Ehara M, Toyota K, Fukuda JHR, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai MKH, Li X, Knox JE, Hratchian HP, Cross JB, Bakken CAV, Jaramillo J, Gomperts R, Stratmann RE, Yazyev AJAO, Cammi R, Pomelli C, Ochterski JW, Ayala KMPY, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski SDVG, Daniels AD, Strain MC, Farkas DKMO, Rabuck AD, Raghavachari K, Foresman JVOJB, Cui Q, Baboul AG, Clifford S, Cioslowski BBSJ, Liu G, Liashenko A, Piskorz P, Komaromi RLMI, Fox DJ, Keith T, Al-Laham MA, Peng ANCY, Challacombe M, Gill PMW, Johnson WCB, Wong MW, Gonzalez C, Pople JA. Gaussian, Inc.; Wallingford CT: 2004. [Google Scholar]
- [14].Lee C, Yang W, Parr RG. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- [15].Becke AD. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- [16].Hehre WJ, Ditchfield R, Pople JA. J. Chem. Phys. 1972;56:2257–2261. [Google Scholar]
- [17].Hay PJ, Wadt WR. J. Chem. Phys. 1985;82:299–310. [Google Scholar]
- [18].Hay PJ, Wadt WR. J. Chem. Phys. 1985;82:270–283. [Google Scholar]
- [19].DeLano WL. DeLano Scientific. Palo Alto, CA, USA: 2002. [Google Scholar]
- [20].Spingler B, Whittington DA, Lippard SJ. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- [21].Lovejoy KS, Todd RC, Zhang S, McCormick MS, D’Aquino JA, Reardon JT, Sancar A, Giacomini KM, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gao Y-G, Robinson H, van Boom JH, Wang AH-J. Biophys. J. 1995;69:559–568. doi: 10.1016/S0006-3495(95)79929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fairall L, Martin S, Rhodes D. EMBO J. 1989;8:1809–1817. doi: 10.1002/j.1460-2075.1989.tb03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jacobo-Molina A, Ding J, Nanni R, Clark AD, Jr., Lu X, Tantillo C, Williams R, Kamer G, Ferris A, Clark P, Hizi A, Hughes S, Arnold E. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vichi P, Coin F, Renaud J-P, Vermeulen W, Hoeijmakers JHJ, Moras D, Egly J-M. The EMBO Journal. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cullinane C, Mazur SJ, Essigmann JM, Phillips DR, Bohr VA. Biochemistry. 1999;38:6204–6212. doi: 10.1021/bi982685+. [DOI] [PubMed] [Google Scholar]
- [27].Dickerson RE, Ng H-L. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6986–6988. doi: 10.1073/pnas.141238898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Marzilli LG, Saad JS, Kuklenyik Z, Keating KA, Xu Y. J. Am. Chem. Soc. 2001;123:2764–2770. doi: 10.1021/ja0007915. [DOI] [PubMed] [Google Scholar]
- [29].Ohndorf U-M, Rould M, He Q, Pabo C, Lippard SJ. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- [30].Ng H-L, Kopka ML, Dickerson RE. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2035–2039. doi: 10.1073/pnas.040571197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sherman SE, Gibson D, Wang AH-J, Lippard SJ. J. Am. Chem. Soc. 1988;110:7368–7381. [Google Scholar]
- [32].Bandyopadhyay D, Bhattacharyya D. Biopolymers. 2006;83:313–325. doi: 10.1002/bip.20542. [DOI] [PubMed] [Google Scholar]
- [33].Soler-López M, Malinina L, Subirana JA. J. Biol. Chem. 2000;275:23034–23044. doi: 10.1074/jbc.M002119200. [DOI] [PubMed] [Google Scholar]
- [34].Subirana JA, Soler-Lopez M. Ann. Rev. Biophys. Biomol. Struct. 2003;32:27–45. doi: 10.1146/annurev.biophys.32.110601.141726. [DOI] [PubMed] [Google Scholar]
- [35].Coste F, Malinge J-M, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Coste F, Shepard W, Zelwer C. Acta Crystallogr. 2001;D58:431–440. doi: 10.1107/s0907444901021709. [DOI] [PubMed] [Google Scholar]
- [37].Schneider B, Cohen DM, Schleifer L, Srinivasan AR, Olson WK, Berman HM. Biophys. J. 1993;65:2291–2303. doi: 10.1016/S0006-3495(93)81306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Berman HM. Biopolymers. 1997;44:23–44. doi: 10.1002/(SICI)1097-0282(1997)44:1<23::AID-BIP3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.