Guidelines and specific tests for validating macromolecular crystal structures that include TLS models are introduced. Validation may used to troubleshoot problems during refinement, to confirm the internal consistency of the model as part of deposition into the Protein Data Bank or to assess the plausibility of interpretating the boundary between two TLS groups as indicating a hinge point between structural domains.
Keywords: validation, TLS models, anisotropy
Abstract
The use of TLS (translation/libration/screw) models to describe anisotropic displacement of atoms within a protein crystal structure has become increasingly common. These models may be used purely as an improved methodology for crystallographic refinement or as the basis for analyzing inter-domain and other large-scale motions implied by the crystal structure. In either case it is desirable to validate that the crystallographic model, including the TLS description of anisotropy, conforms to our best understanding of protein structures and their modes of flexibility. A set of validation tests has been implemented that can be integrated into ongoing crystallographic refinement or run afterwards to evaluate a previously refined structure. In either case validation can serve to increase confidence that the model is correct, to highlight aspects of the model that may be improved or to strengthen the evidence supporting specific modes of flexibility inferred from the refined TLS model. Automated validation checks have been added to the PARVATI and TLSMD web servers and incorporated into the CCP4i user interface.
1. Introduction
1.1. Why new validation tools for structural models are needed
When constructing and refining a new structural model, or when examining an old one, we ask two complementary things from it. On the one hand, we would like the model to be consistent with the experimental data it was derived from. In the case of crystal structures, the most common measure of agreement between the data and the model is the crystallographic R factor. On the other hand, we would like the model to explain, or at the very least to not contradict, whatever prior knowledge we have about the biology and physical properties of the molecule being modeled. Here, a single global number such as the R factor is not so helpful. Instead, we must look for how well specific aspects of the model agree with this external knowledge. The more reliable we consider any given piece of prior knowledge to be, the more skeptical we must become if the model disagrees with it.
For example, in current practice all structural models deposited in the Protein Data Bank are examined for the agreement of their constituent bond lengths and angles with standard values known from decades of structural study and outliers are flagged (Westbrook et al., 2003 ▶). Similarly, the paired backbone torsion angles ϕ and ψ for each protein residue are examined to see whether the pair lies in a region of (ϕ, ψ) space with favorable energy, an idea that originated from G. N. Ramachandran (Ramachandran et al., 1963 ▶) and that has been refined several times since in light of the empirically observed distribution of (ϕ, ψ) values for tens of thousands of previous protein structure determinations (Kleywegt & Jones, 1996 ▶; Lovell et al., 2003 ▶). Other validation criteria have been introduced more recently, notably the set of tests collected in the validation tool MolProbity (Lovell et al., 2003 ▶; Davis et al., 2007 ▶; Chen et al., 2010 ▶). These include examination of side-chain rotamer conformations for favorable energetics, assessment of whether the conformation of non-H atoms is consistent with the known presence of H atoms and deviations from empirically determined geometry about the Cβ atom of each residue (Lovell et al., 2003 ▶). All of these tests assess whether local properties of the model conform to our prior knowledge about the physical properties of molecules.
We are fortunate to have increasingly powerful tools for constructing and refining models to agree with the experimental data. Less fortunately, the adoption of appropriate validation tests often lags behind innovations in model generation and refinement, sometimes leading to serious error (Kleywegt, 2009 ▶). In general, validation of model parameters other than the (x, y, z) coordinates tends to be overlooked. Even when an appropriate validation test is known, it may not be widely appreciated or used and may not be easily automated. For example, visual inspection of ORTEP (Burnett & Johnson, 1996 ▶) plots to assess the plausibility of anisotropic atomic displacement parameters (ADPs; colloquially called ‘thermal ellipsoids’) refined in small-molecule crystallography had been in widespread use for decades before similar anisotropic models were introduced to describe very high-resolution protein structures1. However, the generation and visual inspection of every atom via ORTEP plots does not scale well to macromolecular structures and there was a lag before more automated equivalent tools were available for validating protein and other large structures refined with anisotropic ADPs (Merritt, 1999b ▶). A similar problem faces us today arising from the introduction of new classes of structural models that include descriptions of inter-domain motion and other modes of macromolecular flexibility. The new methodology offers clear advantages if all goes well, but if a poor model is chosen to describe the flexibility, existing standard protocols and validation tools fail to catch this error because they do not assess whether this component of the overall structural model makes physical sense.
1.2. The use of TLS models in crystallographic refinement
The structural model derived from a crystallographic diffraction experiment does not describe the instantaneous state of the atoms in one unit cell, but rather an average state. The description of each atom in the model is an average over the many equivalent instances of the equivalent atom in other unit cells of the crystal. It is also an average of the state of those individual atoms over the time spent in measurement. In a good model, the variation in an atom’s position from one copy to another and from one moment to another is represented by a probability distribution function centered about the atom’s mean position that accounts for all the various factors contributing to variation in the atomic position. These range from vibrational modes of that individual atom to bulk motion of larger groups containing that atom.
The tightly packed crystal lattice of the typical small-molecule crystal mostly precludes bulk motion of large groups of atoms. In this case it is sufficient to ignore bulk motion and to assign an individual atomic displacement parameter (ADP) description to each atom. The high-resolution diffraction data typical of small-molecule crystals allows one to assign anisotropic ADPs, whose conventional mathematical form is a 3 × 3 symmetric matrix Uij (Trueblood et al., 1996 ▶). Using this representation, a model for the anisotropic probability distributions of atoms in the structure requires six parameters per atom.
The situation is different for crystals of macromolecules. The intermolecular lattice packing is much looser, allowing bulk motion of loops, secondary-structural elements, domains or whole molecules. The contribution of these bulk motions to anisotropy within the crystal tends to dominate over individual atomic vibration. Furthermore, it is rare to obtain diffraction data to sufficient resolution to allow refinement of an additional six parameters per atom. For both of these reasons, in order to describe anisotropy in crystallographic models of macromolecular structure it is desirable to model bulk motions explicitly. A choice of mathematical representations for such bulk motion is available, but the best developed of these for use in crystallographic refinement is the TLS (translation/libration/screw) formalism (Schomaker & Trueblood, 1968 ▶). TLS can be used to describe bulk motion of an arbitrarily large set of atoms acting as a rigid body. Even if this group of atoms does not in fact behave as an ideal rigid body, the TLS description may nevertheless provide a very useful approximation. This is in particular true when the total amplitude of motion is small, as is the case for atoms in a well ordered protein in a crystal lattice.
Bulk vibrational motion of a macromolecule within the crystal lattice may be approximated by assigning the entire protein molecule to a single TLS group. Depending on the particular crystal lattice packing, such a single TLS-group model can significantly improve the crystallographic model by yielding more accurate values of F calc and hence lower crystallographic residuals R and R free. This in turn may lead to improved electron-density maps and ultimately to a better structural model. Such single-group TLS models are easily generated and refined in the programs REFMAC (Winn et al., 2001 ▶), phenix.refine (Afonine et al., 2005 ▶) and BUSTER (Bricogne et al., 2009 ▶).
Partitioning the protein into more than one TLS group can yield additional improvement in the crystallographic residuals R and R free. Furthermore, correct identification of such quasi-rigid groups can be of substantial biological significance. It allows the inference of dynamic behavior, e.g. inter-domain hinge motions, directly from a single crystal structure (Painter & Merritt, 2006a ▶; Flores et al., 2008 ▶). However, until recently such models were rare because the partition of the protein chain into separate groups had to be performed empirically by guessing the likely location of hinge points or other break points (Wilson & Brunger, 2000 ▶; Papiz et al., 2003 ▶; Chaudhry et al., 2004 ▶). This changed with the introduction of an automated methodology, TLSMD, for identifying TLS groups based directly on the crystallographic experiment itself (Painter & Merritt, 2006a ▶,b ▶). TLSMD analysis of a crystal structure containing previously refined ADPs will identify multi-group TLS models for the structure that optimally explain the experimentally derived distribution of ADP values in three-dimensional space. This has the effect of replacing complex ‘noisy’ models containing separate isotropic ADPs for each atom with simpler ‘smooth’ models that describe anisotropic displacements arising from the underlying bulk motion of a small number of groups.
Particularly for low-resolution structures, the introduction of multigroup TLS models rather than conventional refinement of individual ADPs often significantly improves the standard crystallographic R factors compared with conventional refinement with no description of bulk motion. However, it does not by itself ensure that the individual TLS groups in the multi-group model make physical sense nor does it guarantee that the group represents a biologically relevant mode of motion by the protein in solution (Moore, 2009 ▶). Hence, there is a need for additional validation criteria independent of the R factors.
1.3. BEER and Skittles
Fig. 1 ▶ shows the increasing use of TLS models in structures deposited in the Protein Data Bank (PDB) over the last several years. We are particularly interested in the validation of those structures that partition each chain into multiple TLS groups, currently comprising about 10% of all PDB depositions. Nevertheless, structures with only one TLS group per chain, or one TLS group per multimeric protein, are also relevant to fundamental questions about the anisotropic behavior of proteins in crystals. They are also potentially victim to a failure to conduct validation tests. These currently comprise an additional 10% of PDB depositions.
Figure 1.
Fraction of new PDB depositions containing a TLS model. This graph necessarily accounts only for depositions in which a TLS model is described in the header records of the PDB file. We estimate that roughly 300 additional depositions (<1%) used TLS refinement to generate individual anisotropic ADP records but failed to include a description of the TLS model in the header.
Errors in deposited structural models can arise from many sources, ranging from simple bookkeeping or format errors in files prepared for deposition to hopefully rare cases in which an incorrect structural model has been refined. In between these extremes lies a class of potential problems that may be detected easily if appropriate checks are made. As one step towards a notional validation suite BEER (Best Ever Evaluation of Refinement), we have implemented and evaluated a set of tests collectively called Skittles that can highlight easily correctable errors involving the choice or refinement of TLS groups or other models of anisotropy. These include checks for global properties such as the overall distribution of anisotropy within the refined structure and checks for problems with individual atoms or residues; in particular, we introduce checks on the internal consistency of multi-group TLS models. The Skittles validation checks are being integrated into several widely used crystallographic computing environments.
2. Methods
2.1. Definitions
A single set of 20 TLS parameters describes rigid-body displacement of an arbitrary set of atoms (Schomaker & Trueblood, 1968 ▶). These parameters constitute three 3 × 3 tensors: T, L and S. T is a symmetric tensor with elements given in units of Å2; it describes the anisotropic translational displacement common to all atoms in the rigid-body group. L is also a symmetric tensor, whose elements are in units of radians2; it describes the rotational component (libration) of the rigid-body displacement. The S tensor is not usually symmetric; it describes the correlation between the rotation and translation of a rigid body undergoing rotation about three orthogonal axes that do not intersect at a common point.
A segmented TLS model is one that partitions a protein or nucleic acid chain into multiple segments. Each TLS group contains one or more of these chain segments, possibly with associated ligands. All of the atoms belonging to one TLS group are described by the set of 20 TLS parameters associated with this group. A plausible partition of a protein chain into multiple segments may be constructed manually, but is more usually performed by TLSMD analysis of the distribution of B factors in a preliminary model refined using a conventional isotropic ADP description (Fig. 2 ▶).
Figure 2.
Segmented TLS model. A partition of a single protein chain into seven TLS groups, as proposed by TLSMD analysis on the basis of the three-dimensional distribution of B values in a preliminary model.
2.2. Residuals corresponding to restraints applied during refinement
As a computational convenience, current crystallographic refinement programs implement TLS model refinement by building on the same code used to refine individual per-atom anisotropic ADPs. For each atom assigned to a particular TLS group, the 20 TLS parameters that describe the group are used to approximate the displacement of that atom as a thermal ellipsoid described by the usual 3 × 3 tensor Uij. That is, each element Uij of the tensor is expressed in terms of the TLS model parameters. During each cycle of iterative refinement, parameter shifts ΔUij are calculated as usual from the normal matrix. These are propagated back by the chain rule to yield shifts ΔTij, ΔLij and ΔSij for the TLS model parameters (Winn et al., 2001 ▶). Note that the shifts ΔUij contain contributions both from the diffraction measurements F obs(hkl) and from any restraints introduced to enforce conformity to certain a priori expectations. During refinement of per-atom anisotropic ADPs, several such restraints are typically applied as described below (equations 1–4 ; Fig. 3 ▶). In principle these restraints can also be applied during TLS refinement, although current refinement programs do not typically do so.
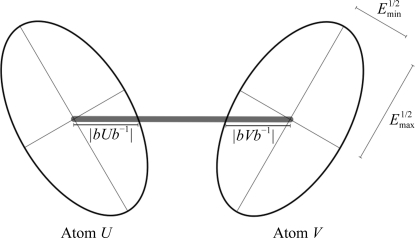
Figure 3.
A bonded pair of atoms U and V represented by anisotropic ADPs Uij and Vij. The atoms have identical eigenvalues and therefore identical anisotropy A = E min/E max even though they differ in the orientation of their principal axes (eigenvectors). The orientations shown are such that the projections of their respective bounding ellipsoids onto the connecting bond have identical length and thus the residual in (3) is zero, although this would not be true for other orientations of the principal axes. However, the residuals in (2) and (5) are nonzero owing to the difference in the orientation of their eigenvectors.
The Uij tensor can be restrained towards description of a sphere. This restraint term is called ISOR in SHELXL and SPHE in REFMAC. The degree to which an individual atom conforms to this target can also be expressed using the anisotropy
where E min and E max are the smallest and largest of the three eigenvalues for the tensor U. A perfectly spherical atom has A = 1.
The Uij terms of bonded atoms (REFMAC) or of all nearby atoms (SHELXL, phenix.refine) can be restrained to be similar to each other. This restraint is called SIMU in SHELXL and BFAC in REFMAC. The contribution from the paired atoms U and V to this restraint term is
If applied to nearby but nonbonded atoms, the restraint may be weighted by the interatomic distance. For the purpose of validation, we use here a variant of this residual that is the root-mean-square of the difference in the six unique elements of the symmetric tensors U and V,
If two atoms U and V are bonded, the projection of the two tensors Uij and Vij along the direction of the bond can be restrained to be equal. This restraint is called DELU in SHELXL and RBON in REFMAC. The residual from bonded atoms U and V contributing to this restraint term, where the along-bond direction of the bond is the vector b, is given by
In the case of refining individual anisotropic ADPs without TLS, the strength and relative weight given to these restraints can be used to guide the resulting model towards conformity with expected distributions of atomic anisotropy in much the the same way as restraints on bond lengths and angles can be used to guide the model towards conformity with expected chemical geometry (Merritt, 1999b ▶).
For a pair of atoms acting as a true rigid body, r DELU is necessarily zero as the atoms do not move relative to each other (Rosenfield et al., 1978 ▶). Similarly, adjacent atoms within a group acting as a true rigid body must necessarily have similar displacements, so r SIMU is also negligible. Thus, in the case of refining a TLS model the restraints based on the residuals r DELU and r SIMU have a negligible effect when both atoms U and V are described by the same TLS group. When atoms U and V are in two different TLS groups these restraint terms are nonzero and provide the only coupling between the parameters describing the first TLS group and the parameters describing the second TLS group. However, in practice these restraints are imposed only weakly, if at all, during TLS refinement to enforce the physical requirement for a smooth junction between adjacent TLS groups in a segmented model. Indeed, large values of the r SIMU and r DELU residuals across the bond that joins two TLS groups may remain after refinement has converged, indicating an inconsistency in the two sets of corresponding TLS parameters. As we will discuss, this provides an opportunity to use the residuals as a validation test for assessing segmented TLS models.
2.3. Other residuals
Another useful residual that quantifies the similarity of two thermal ellipsoids U and V is the correlation coefficient of the electron-density distributions described by their respective ADP tensors U and V. As with the residuals in (2) and (4), the correlation coefficient of the density for bonded atoms linking two different TLS groups can be used to check whether the two TLS descriptions are consistent at the point where they join. This value can be conveniently calculated directly from the U and V tensors (Merritt, 1999a ▶),
 |
The ccuij residual is also sensitive to disparity in the magnitudes of the isotropic components of the ellipsoids being compared, which is a disadvantage in some contexts. Variants of the residual can be constructed that first adjust the diagonal elements of the U and V tensors so that they have the same trace and hence the same equivalent isotropic B factor B eq (Trueblood et al., 1996 ▶). Another approach is to normalize the correlation calculated for the paired ellipsoids against that calculated for either ellipsoid paired with a perfect sphere (Merritt, 1999a ▶). Outliers in that normalized residual, Suij, are also reported by the PARVATI validation server. Suij was originally introduced as a validation metric for structures refined with individual anisotropic ADPs. We have found it less useful than ccuij itself for the evaluation of segmented TLS models as explored here.
2.4. Survey of PDB entries
In order to establish baseline expectations for the distribution of properties to be used as validation criteria, we surveyed all current entries in the PDB. We considered only structural models produced by X-ray crystallography and only those that contained an interpretable description of atomic anisotropy. The majority of these were either instances of full anisotropic refinement of individual ADPs or instances of TLS refinement. A small number of models containing anisotropic ADPs derived from normal-mode analysis were also included (Chen et al., 2007 ▶). Individual structural models were analyzed using the PARVATI validation tool (Merritt, 1999b ▶) to check for the overall distribution of atomic anisotropy (A) and for the presence of individual atoms with nonpositive definite ADPs. During this analysis, we also accumulated statistics on the distribution of residuals r SIMU, r DELU and ccuij so that we could set threshold levels for Skittles to flag outliers during validation.
All PDB entries as of 17 September 2009 were categorized according to the presence of REMARK records containing the TLS GROUP keyword and associated RESIDUE RANGE or SELECTION: CHAIN records. They were further grouped by the refinement method indicated in the PROGRAM or SOFTWARE records. If any chain was included in the residue-range specifications for two or more TLS groups, the entry was categorized as segmented TLS (3624 entries). Segmented TLS entries refined with REFMAC that did not already contain individual ANISOU records for the protein or DNA atoms were run through TLSANL (Howlin et al., 1993 ▶) to generate them from the TLS model. Anisotropy and correlation of anisotropy were analyzed for the entries successfully processed by TLSANL (2642 entries).
Entries refined by either REFMAC or PHENIX that already included ANISOU records were analyzed without running TLSANL (350 entries).
PDB entries that contained more than 100 ANISOU records and had been refined by SHELXL at 2.1 Å resolution or better were classified as anisotropic ADP refinement (1183 entries). To reduce any bias arising from multiple entries for isomorphous structures of the same protein, a single sample was kept whenever there were seven or more entries with similar COMPND…MOLECULE and CRYST1 records, i.e. corresponding unit-cell parameters within 5 Å and 5°. This resulted in 1070 structural models representing anisotropic ADP refinement (Fig. 4 ▶ a).
Figure 4.
Distribution of mean anisotropy 〈A〉 for protein atoms in PDB depositions as of September 2009. Each box-and-whisker plot element represents depositions within a single category. In (a) and (b) the categories are defined as bins, each covering a resolution range of 0.1 Å. The width of the box is proportional to the number of structures in the bin. The heavy crossbar is the median value of 〈A〉 for structural models in the bin. The vertical extent of the box represents the first and third quartiles of values in the bin, while the vertical extent of the whiskers is chosen to bound 95% of the values in the bin. Individual structural models for which the value of 〈A〉 is an outlier are shown as circles. (a) Structural models refined by SHELXL and containing individual anisotropic ADP values for each atom. (b) Structural models refined by REFMAC and containing explicit segmented TLS descriptions from which individual ADPs can be derived. (c) Mean anisotropy broken down by the program used for refinement. The SHELX category contains 1070 structures. The REFMAC and PHENIX categories represent 4446 and 290 structures, respectively, and contain both segmented and unsegmented TLS models. We also validated five models for which anisotropic ADPS were generated from normal-mode analysis by the program NMref (Poon et al., 2007 ▶).
2.5. Implementation
The PARVATI validation server, which was originally written to guide the choice of restraint weights during full anisotropic refinement of protein structures, has been extended to validate structural models in which anisotropy is described by TLS rather than by individual anisotropic ADPs. The server accepts an uploaded file in PDB or mmcif format. Alternatively, it accepts the accession code of a structural model in the Protein Data Bank for automatic retrieval. If the model is found to already contain anisotropic ADPs, i.e. ANISOU records in a PDB file, then these are validated directly. If the model contains fewer than 100 ANISOU records but does contain a recognizable TLS description, then individual anisotropic ADPs are generated for each atom from the TLS model before proceeding with validation. In either case the server generates statistical summaries and graphical output by invoking the program RASTEP, which is part of the RASTER3D molecular-graphics package (Merritt, 1999b ▶). To support this, there is a new command-line option -cn_check for the program RASTEP. This option requests tabulation of the ccuij residual for each peptide linkage in the structure being plotted or analyzed. In the case of nucleic acids the residual is calculated for each O3′—P bond linking two residues. As before, RASTEP can also be run locally rather than via the PARVATI web server to generate both graphical and tabular output.
We have similarly integrated these validation tests into the CCP4 suite of crystallographic programs (Collaborative Computational Project, Number 4, 1994 ▶). The program TLSANL (Howlin et al., 1993 ▶) has been extended to calculate the residuals ccuij, r SIMU and r DELU for each C—N bond along a protein backbone and for each O3′—P bond along a nucleic acid chain. The residuals are tabulated in the output log file in a format suitable for display by the CCP4 graphing utilities LOGGRAPH and XLOGGRAPH and optionally written to a separate output file for use by external plotting or analysis programs. The program also tabulates the distribution of anisotropy. This functionality is available via the CCP4i user interface (Potterton et al., 2004 ▶). The crystallographer can thus easily generate graphical output similar to the residual plot in Fig. 5 ▶ to check the inter-segment consistency of segmented TLS models being refined by REFMAC.
Figure 5.
Validation of ADP agreement across peptide C—N bonds linking adjacent TLS segments. Example output from analysis of a structural model drawn from the PDB. This model was refined using four TLS groups. This model is more complex than most of those deposited to date in that three of the TLS groups contained more than one segment of the protein chain. (a) The residuals corresponding to the BFAC and RBON restraints applied by REFMAC during refinement, equivalent to r SIMU and r DELU, respectively. Residues from six chains related by noncrystallographic symmetry in a single structure are shown on the same plot. (b) The density correlation ccuij (6) for the C and N atoms of each peptide linkage in the same structure. Each of the six superimposed curves corresponds to one of the six NCS-related chains in the structure being validated. The threshold values of ccuij = 0.92 and ccuij = 0.857 were set empirically on the basis of a survey of structures in the PDB. This plot was generated by the PARVATI validation server from analysis of PDB entry 3b48 (C. Chang, H. Li, S. Moy & A. Joachimiak, unpublished work).
Routines to calculate ccuij and four other residuals have also been added to the mmLib Python library (Painter & Merritt, 2004 ▶). Two variants of the code are provided, one written purely in Python and one that serves as a wrapper allowing Python code to call much faster Fortran implementations compiled into an external shared object module. The library also provides a Python script skittles.py that demonstrates the use of the mmLib routines to perform simple validation of an input structural model.
2.6. Re-refinement of structures from the PDB
We undertook re-refinement of several structures for use as examples. These structures had been flagged during validation by PARVATI as having poor ccuij residuals despite showing reasonable values for their crystallographic R factors and the overall distribution of anisotropy. Structure factors were downloaded from the PDB and converted using the CCP4 program CIF2MTZ. The refinement protocol included the addition of riding H atoms and the use of the default ‘simple’ solvent-mask treatment in REFMAC v.5.5.0106. The coordinates downloaded from the PDB were first refined using individual B iso terms, i.e. with no TLS treatment. The relative weightings of individual geometric and B-factor restraint terms were left at the program default values. However, the overall weight of geometric restraints relative to the X-ray residual was adjusted if necessary to reproduce the overall deviation of bond angles and distances from ideal values reported in the original PDB deposition. The resulting model was then used both for TLS refinement using the original TLS segmentation description from the PDB deposition and for analysis by TLSMD for possible re-assignment of TLS segment boundaries. We performed two macrocycles of TLS refinement. Each macrocycle consisted of ten rounds of TLS parameter refinement and ten rounds of conjugate-gradient refinement of the x, y, z and B iso parameters. No nonprotein atoms were included in the TLS model. At the start of the first macrocycle, the B iso values for all protein atoms were reset to 20 Å2 and any previous values for the T, L and S tensor elements were discarded. The full set of refined parameters was carried forward into the second macrocycle.
3. Results and discussion
3.1. Survey of anisotropy in the PDB
3.1.1. Agreement between fully anisotropic models and segmented TLS models
Ten years ago we asked the question ‘How anisotropic are typical atoms in a protein crystal?’. At that time there were a total of 28 protein structural models in the PDB containing anisotropic models for the individual atoms, with resolution spanning the range 0.8–1.6 Å. All of them had been generated by full anisotropic refinement of individual ADPs, most of them using the program SHELXL (Sheldrick, 2008 ▶). Our preliminary conclusion was that protein atoms in well refined near-atomic resolution structures had a roughly Gaussian distribution of anisotropy, with 〈A〉 = 0.45 and σ(A) = 0.15 (Merritt, 1999b ▶). These values were maintained when we repeated the survey two years later, by which time the number of structures had more than doubled. The question was revisited again in 2007 by Kondrashov and coworkers, who surveyed Cα atoms in 83 structures with resolution better than 1 Å (Kondrashov et al., 2007 ▶). The models refined using SHELXL had 〈A〉 = 0.51, while those refined with REFMAC had 〈A〉 = 0.64. These values are more isotropic than the earlier estimate, which probably reflects both the higher average resolution and the choice to consider only Cα atoms.
Fig. 4 ▶ shows the distribution of anisotropy in all PDB entries as of September 2009, broken down by the resolution of the structure refinement and by the refinement program used. The median value of 〈A〉 for structures in the resolution range 0.7–1.2 Å refined with individual anisotropic ADPs was close to 0.45, which is consistent with the earlier surveys. Outliers were mostly in the direction of being nearly isotropic, suggesting that the atoms in these refinements were strongly restrained towards being spherical (1). It is notable that the median value of 〈A〉 is independent of resolution between 0.8 and 1.6 Å, although there is an increase in the number of outliers as the resolution worsens.
The median value of 〈A〉 in structures refined by REFMAC using TLS models is slightly higher, at roughly 0.55. There is a slight trend towards greater anisotropy (lower value of 〈A〉) at lower resolution. We found no significant difference in the distribution of anisotropy resulting from segmented TLS models and that from nonsegmented TLS models.
We were able to identify relatively few PDB entries for which TLS refinement had been performed by PHENIX and these structural models were more isotropic than models refined by other programs (Fig. 4 ▶ c). This was unexpected, as the easiest explanation for larger values of 〈A〉 would be stronger restraints toward isotropy, but PHENIX does not apply ADP restraints during refinement of TLS parameters.
3.1.2. Use of 〈A〉 as a validation criterion
The results of this comprehensive survey reinforce the idea that the atoms in the great majority of crystalline proteins exhibit a mean anisotropy of approximately 0.5, which is largely independent of the resolution of the diffraction observed. If the distribution of A for atoms in a particular structural model deviates strongly from this value, there is reason to believe that the model could be improved. This was the rationale for using the distribution of anisotropy to guide the choice of restraint weights in fully anisotropic refinements carried out at other than true atomic resolution (Merritt, 1999b ▶). On the basis of the current survey, we now suggest that the same expectation holds for structural models that use TLS to describe atomic anisotropy. Of course, as 〈A〉 approaches 1.0 the model is close to that which would have arisen from a conventional purely isotropic refinement of B factors; structural models that are outliers in this direction may be considered as being at worst no different from an isotropic model. Outliers in the direction of 〈A〉 ≪ 1 are more suspect. Many of these outliers found in the survey showed other evidence of unstable or poorly restrained refinement, e.g. the presence of nonpositive-definite ADPs.
3.2. Types of problem that were identified
3.2.1. Local discrepancies: bad joins between TLS segments
A primary motivation for this work was concern that the TLS descriptions of individual segments within a segmented TLS model might be inconsistent with each other. The ADPs of neighboring atoms whose covalent bond connects two separately refined TLS groups are by default restrained only weakly, if at all, in existing refinement programs. It seemed plausible that if the TLS groups within a segmented model were chosen poorly then discrepancies between the true atomic displacements and the modeled atomic displacements would tend to pile up at these junctions between adjacent TLS groups (Figs. 5 ▶ and 6 ▶). These are the problem cases that calculation of the residuals ccuij (5), r SIMU (3) and, to a lesser extent, r DELU (4) were intended to catch. Because residuals equivalent to r SIMU and r DELU are themselves used as restraints in some refinement protocols, we further expected that ccuij might be a more sensitive diagnostic in the general case. These expectations were borne out during validation trials (Fig. 5 ▶).
Figure 6.
Bad junction between two adjacent TLS groups. The C—N bond between residues AlaF126 and AlaF127, spanning two TLS groups, was highlighted in the validation test shown in Fig. 5 ▶. The atoms of these two residues are depicted here as thermal ellipsoids drawn at the 33% probability level. The TLS model for the group containing residue 126 describes a relatively isotropic displacement for atoms in this region of space. The TLS model for the group containing residue 127 describes a more anisotropic displacement for atoms in this same region. This discrepancy results in incompatible models for the vibrational motion of the two bonded atoms that bridge the two TLS groups. One measure of this discrepancy is the quantity ccuij (5). A small value of ccuij may indicate a poorly chosen boundary between the two groups. Alternatively, it may indicate that the description refined for one or both of the TLS groups is dominated by inclusion of other residues whose true displacements are different from those of atoms in either of the residues shown here and thus would better be split off into a TLS group of their own. Both scenarios suggest that the assignment of TLS-group boundaries within the protein chain should be reconsidered.
3.2.2. Global discrepancies: unreasonable TLS descriptions
Between 2 and 3% of the PDB entries that were surveyed contained TLS records which when applied generate nonpositive-definite ADPs for an unreasonable fraction of the atoms in the structure. These are easily identified in curves showing the distribution of anisotropy (Fig. 7 ▶). In most cases it is not possible to determine exactly what has gone wrong. The possible causes range from numerical instability during refinement to formatting problems while preparing files for deposition to a mismatch between the deposition TLS parameters and the deposited model coordinates and individual ADPs. Whatever their precise cause, the presence of such errors is easily caught. We hope that widespread adoption of Skittles or equivalent validation checks at the time of structure deposition will obviate this class of errors in the future.
Figure 7.
Distribution of anisotropy within individual PDB entries. The overall distribution of anisotropy for individual atoms in 209 structures with segmented TLS models. The 209 structures were chosen semi-randomly (the second character of the PDB code was either ‘a’ or ‘b’), but 21 structures containing ten or more nonpositive-definite atoms were discarded from the set. Structures containing fewer than ten nonpositive-definite ADPs were retained. The distribution curves for these 16 structures run off the left edge of the plot.
3.2.3. Errors in TLS-group assignments for individual residues
We found an additional set of PDB entries (approximately 16% of those surveyed) in which a small number of residues are clearly not described properly by the TLS-group definitions in the header records of the PDB file. In many instances the nature of the problem is evident upon inspection. For example, a suspect TLS group may contain both protein residues and an associated ligand, but the group specification incorrectly names a symmetry-related ligand belonging to a different protein chain. In other cases the TLS group describes residues that are not present in the PDB file at all. One common cause is likely to be that the chain identifiers used for ligands or other nonprotein residues have been changed during the deposition process but the references to these same residues in the TLS records of the header were not changed to match. This is particularly problematic if water molecules are included in TLS refinement but are then later renamed or moved to a symmetry-equivalent position as part of deposition processing.
We were unable to generate ANISOU records with confidence for 76 entries with duplicated residue-range records, nor were we able to automate analysis of 546 entries that had overlapping ranges, non-existent atoms or other problems in the TLS-model description. No analysis was performed on ten files produced by phenix.refine but lacking ANISOU records. The Skittles validation tools can issue warning messages in these cases but do not attempt to reconstruct the original names or TLS-group assignments of the problematic residues.
3.3. Choice of validation criteria for TLS segmentation boundaries
At the outset, we did not know the expected magnitude of the various residuals across segment breaks in well behaved refinements. To determine expectations for use in validation, we selected 2282 PDB entries containing segmented TLS models refined by REFMAC and containing no nonpositive-definite ADPs after application of the TLS description. We calculated residuals for all 16 594 TLS-segment junctions in this set of well behaved models and selected target values for validation corresponding to 95 and 99% compliance. That is, we found that the similarity of the C—N peptide linkage in 99% of the segment junctions in these well behaved models had ccuij ≥ 0.86 and 95% of this same set had ccuij ≥ 0.92. These values were chosen as validation targets (Fig. 8 ▶).
Figure 8.
Superposition of ccuij calculated for peptide linkages in 209 PDB structures with segmented TLS models. This plot shows the ccuij residual (6) calculated for every linked-residue C—N bond (the plot is truncated at 550 residues for clarity). The 209 structures were chosen semi-randomly (the second character of the PDB code was either ‘a’ or ‘b’), but structures containing more than ten nonpositive-definite atoms were discarded from the set. For the vast majority of the C—N bonds ccuij ≅ 1, either because the bond lies entirely within one TLS group or because the two adjacent TLS groups that it spans are consistent at the junction point. The eight most extreme deviations from ccuij ≅ 1 in this figure resulted from the inclusion of 16 structures that were retained in the set even though they contained 1–10 nonpositive-definite ADPs. The color and symbol encoding allowed us to identify the specific PDB entries corresponding to the relatively small number of outliers (key not shown). If ccuij = 0.92 is chosen as a threshold, then the test highlights specific junctions that may be worth reexamination in <5% of the PDB files.
We similarly identified the compliance values for the residual r SIMU (3), which directly compares the Uij terms belonging to adjacent atoms. We found that 95% of the segment junctions satisfied r SIMU < 0.43, while 99% satisfied r SIMU < 0.92. The 95 and 99% compliance values for the residual r DELU (4) were similar: 0.43 and 1.01, respectively. However, the overall distribution of these residuals within the selected set of segment junctions was less Gaussian (much more uniform) than the distribution of ccuij values, making them less useful as validation criteria (Fig. 5 ▶ a). Furthermore, as noted already, the residual r DELU can be near zero even if the ADPs for the two atoms disagree as to the direction of displacement (Fig. 4 ▶).
The overall correlation between the residual r SIMU and ccuij was only moderate (correlation coefficient = 0.66). If both of these tests are applied to the set of structures, then 392 of the 16 594 segment junctions are outliers at the 95% level for both ccuij and r SIMU; 437 are outliers according to ccuij only and 437 are outliers according to r SIMU only. All three residuals are calculated and may be plotted from the output of TLSANL and RASTEP, but we suggest that ccuij provides the most useful criteria for automated validation.
3.4. Interpretation of poor correlation across TLS-segment boundaries
A low value of ccuij for the two atoms on either side of a TLS-segment boundary indicates that the TLS descriptions of the two adjacent TLS groups are not consistent. That is, they make very different predictions for the displacement of an atom located near this shared boundary. One common case that can cause inconsistency across a segment boundary arises when a short stretch of residues has been modeled into weak density. If it is modeled using individual B iso values, these values will be larger than those of neighboring well ordered residues. In a segmented TLS model, the poor ordering of this same stretch of residues can be described by assigning them to a single TLS group, whose parameters will again describe larger displacements than those of the neighboring well ordered residues. This can lead to discontinuity at the segment boundaries at either end of the poorly ordered segment. Assigning this set of residues a shared set of TLS parameters may be a valid description of poor ordering in the structure; shifting the segment boundaries is unlikely to improve the model or to lower the crystallographic R factors. However, the low value of ccuij across this pair of segment boundaries tells us that they should not be interpreted as hinge points belonging to a well ordered intervening segment undergoing rigid-body motion.
A more interesting case of inconsistency arises when the residues making up the segments to either side of the boundary are well ordered. If both segments are in reality part of the same relatively rigid larger group, perhaps a domain or the entire protein, we expect their respective TLS descriptions to be essentially the same. Both TLS descriptions should yield equivalent predictions for all atoms in the larger group. If both segments are individually well described as approximating a rigid group but their junction acts as a hinge point, then their respective TLS descriptions may make different predictions for the displacement of atoms at arbitrary positions. Nevertheless, we still expect that the two descriptions will agree in predicting displacement at the hinge point itself. In either case, a low value of ccuij for the pair of atoms at the segment boundary, whether or not it is a hinge point, is an anomaly. It indicates disagreement between the predictions of the two TLS groups at a point where they are expected to agree.
3.5. What exactly are we validating?
Structure-validation tests are intended to assess whether a structural model is physically plausible. The tests specifically considered here evaluate the plausibility of the portion of the model that describes atomic displacements. Examination of the distribution of net anisotropy for all individual atoms of the structure evaluates a global property. In this sense, it is similar to examination of the crystallographic R factors. An unusual distribution of anisotropy may indicate that there is a problem with the model or with the refinement, but as in the case of a high R factor it does not immediately highlight specific regions within the structural model that are problematic.
Other tests considered here evaluate agreement between the structural model and our expectations for certain local properties, notably the overall displacement modeled for an individual atom and the compatibility of the displacements modeled for a pair of bonded atoms. Deviations from expectation indicate physical implausibility of that specific region of the structural model, e.g. an atom whose overall ADP tensor is nonpositive-definite or a pair of bonded atoms described as having radically different displacements. Note, however, that while the problem is flagged by a local violation of expectations, the ultimate cause of the problem may lie in a more global aspect of the model: perhaps the restraints used in refinement were too weak or perhaps the choice of TLS groups was not physically realistic.
These validation tests based on atomic displacements, when considered jointly with better known validation tests based on geometry, conformation and inferred energetics, can be considered to establish the plausibility of the overall structural model as a representation of the averaged state of the protein as it exists in the crystal. However, this is not the end of the story.
3.6. Validating the choice of TLS groups in a segmented model
Given the importance of flexibility to protein function, it would be of great interest if we could reliably interpret a well refined segmented TLS model as identifying biologically relevant hinge points and flexional modes. However, even if global measures such as the crystallographic R factors and the distribution of anisotropy look entirely reasonable, this does not constitute evidence that the assignment of TLS groups corresponds to correct identification of specific sets of residues that move in concert within the actual protein. That is, the breakpoints between TLS groups in a segmented model may not correspond to points of hinging or torsional motion in the actual protein. This reservation was raised recently by Moore (2009 ▶), who proposed that the best approach to validating the interpretation of a TLS model as a description of actual flexional groups within the macromolecule would be to use the TLS model to predict additional observable properties such as thermal diffuse scattering. Analysis of separately measured thermal diffuse scatter is beyond the current scope of Skittles, but we suggest that there is an alternative approach to validating the physical plausibility of the set of groups making up a segmented TLS model.
The alternative view focuses not on the bulk motion of the body of each TLS group, but rather on the implied hinge point or flexional junction where one TLS group adjoins the next. Flores et al. (2008 ▶) recently evaluated the ability of several methods to predict hinge points based on a single structure determination. The predictions were scored by comparing them with the actual hinge points implied by the existence of multiple experimentally determined structural homologs whose conformation differed by hinge or torsional motion about this point. Predictions based on TLSMD segmentation fared well in this comparison. The true hinge points were found to lie very near junctions between TLS groups chosen by TLSMD. However, there were additional TLS junctions chosen by TLSMD that did not correspond to a previously characterized hinge point in the set of homologous structures. Some of these may in fact have been correct predictions of a hinge that by chance was not evident from the limited set of known structures, but it is likely that many of them were false positives in exactly the sense that Moore has raised concerns about.
Can we filter out these false positives by applying a validation test that evaluates the plausibility of each junction individually? This is the rationale for the Skittles test that calculates ccuij for the C—N bonds connecting adjoining TLS groups. In order for both of the TLS groups adjoining a given junction to be plausible models for actual protein motion, they must agree with each other on the implied motion, and hence displacement, of the atoms at the point where they join. This is well illustrated in Fig. 6 ▶, which shows an actual case where adjoining groups do not agree. A low value of ccuij indicates that the implied torsional or hinge motion about this junction is not physically plausible.
3.7. An example of using validation to guide model improvement
Fig. 9 ▶ shows an example of re-refinement following validation of a segmented TLS model. The structural model as retrieved from the PDB scored well overall according to the validation tests performed by the PARVATI server, but the ccuij residuals across peptide bonds were somewhat anomalous. The ccuij residual at one of the three TLS segment boundaries was an outlier at the 1% level. Furthermore, many peptide linkages internal to the individual TLS segments were flagged (Fig. 9 ▶ a), suggesting that the individual isotropic B components for successive atoms along the backbone varied more than is typical for well refined structures. Re-refinement using the default set of isotropic B restraints in REFMAC, which are stronger than those used for the deposited model, reduced this variation to more typical levels and yielded a slightly better R free. Even after re-refinement, however, the TLS-segment boundaries were flagged as having poor ccuij residuals (Fig. 9 ▶ b). We then replaced the original four-segment TLS model with a five-segment model suggested by TLSMD. This had the effect of replacing one of the original segment boundaries, which had been flagged as anomalous, with two new segment boundaries whose ccuij residuals were unremarkable after refinement (Fig. 9 ▶ c). However, this local change did not yield a further improvement in R free. The ccuij residual across the segmentation boundary nearest the C-terminus is still poor in this revised model, but is no longer an outlier at the 1% level. Increasingly fine-grained TLS models proposed by TLSMD on the basis of the original refinement, i.e. partition of the chain into more and more segments, do not shift this problematic boundary until one reaches the point of partitioning the chain into nine segments. The nine-segment model does indeed further improve the ccuij residuals and yields marginally better R and R free values. A decision as to whether this degree of improvement in model statistics would justify doubling the complexity of the TLS model would depend on more detailed consideration of the individual structure being refined.
Figure 9.
Validation plots before and after re-refinement. PDB entry 3gp0, a 1.9 Å resolution structure of human mitogen-activated protein kinase 11 (P. Filippakopoulos et al., unpublished work), is used as an illustrative example. (a) The ccuij residual plot produced by the PARVATI validation server for entry 3gp0 as retrieved from the Protein Data Bank. (b) The equivalent residual plot after re-refinement following the protocol described in §2, using the same four TLS segments as the original entry. (c) Re-refinement instead using a five-segment TLS model with segment boundaries suggested by TLSMD analysis. Crystallographic R factors and the mean anisotropy 〈A〉 after refinement are listed in each panel. TLS-segment boundaries are indicated by arrows above each plot. Re-refinement using a nine-segment TLS model with segment boundaries suggested by TLSMD further improved the ccuij residuals and improved R and R free to 0.179 and 0.221, respectively.
3.8. After validation
When validation tests indicate that a structural model deviates significantly from expectations for a global property such as the distribution of anisotropy, it is probably best to revise or replace this component of the current model. For segmented TLS models this would mean retreating to conventional refinement of B iso values only, i.e. without TLS, followed by reanalysis of the result using TLSMD to generate a new segmented TLS model. Improvement in the regenerated model should be evident both in validation tests and by a drop in the crystallographic R free. Depending on exactly what went wrong in the original refinement, the TLS boundaries in the regenerated model may or may not lie in the same places as before.
In a case where validation highlights inconsistency of adjacent TLS groups in a segmented model, as in Figs. 5 ▶, 6 ▶ and 9 ▶, the appropriate action depends on what the model is to be used for. It is possible, but not inevitable, that a segment boundary flagged by Skittles can be remedied by reanalysis via TLSMD or by manual adjustment of the segment boundaries. Except for pure TLS models, however, this is unlikely to be reflected by improvement in the crystallographic residuals. This is because the residuals are calculated using the net ADP for each atom, which contains contributions from both TLS and individual B iso refinement. If shifting the segment boundary changes the TLS contribution to a particular atom, the refined value of its individual B iso contribution will tend to compensate, leaving the R factor unchanged. Nevertheless, shifting the segment boundary to a self-consistent position may be of intrinsic value if it is to be interpreted as a possible hinge point for inter-domain motion or local flexibility.
4. Availability of the validation tools
We have integrated the validation checks described here into several common crystallographic computing environments, including the mmLib library of Python routines for manipulating macromolecular structures, the PARVATI validation server and the CCP4i graphical user interface to the CCP4 suite. The source code for a Python version of the Skittles validation tool is hosted on SourceForge (http://pymmlib.sourceforge.net/), as is the underlying crystallographic toolkit mmLib. The source code is currently available under the Artistic License v.2.0, but other licensing arrangements are possible if requested. The modified version of RASTEP is part of version 2.9 of the RASTER3D graphics package, which may be downloaded from http://www.bmsc.washington.edu/raster3d/ and other places. Modifications to the program TLSANL and to the CCP4i interface have been contributed to the CCP4 project.
Acknowledgments
This work was supported by NIH award R01GM080232. We appreciate the information provided by Pavel Afonine on the implementation of TLS refinement in phenix.refine and helpful discussion with Garib Murshudov and others at the 2009 CCP4 summer school at Argonne, Illinois, USA. We are grateful that these interactions were facilitated through support of the Summer School by the US National Institutes of Health (NCI Y1-CO-1020; NIGMS Y1-GM-1104) and by a grant from the UK Science and Technology Facilities Council.
Footnotes
Even in small-molecule crystallography the need for such validation checks has not been universally appreciated, leading to avoidable errors in archived structures. This problem has been made more publicly visible by external pro bono spot-checking of published structures, for instance Richard Harlow’s ‘ORTEP of the Year’ awards (Harlow, 1996 ▶) and a series of papers in Acta Crystallographica by Dick Marsh pointing out probable space-group errors in published structures. It could be further reduced by establishing a uniform battery of validation checks to be performed by scientific journals at the time of publication (Spek, 2009 ▶), but even these measures do not assure universal coverage.
References
- Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). Acta Cryst. D61, 850–855. [DOI] [PMC free article] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Smart, O., Vonrhein, C. & Womack, T. O. (2009). BUSTER v.2.8.0. Global Phasing Ltd, Cambridge.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEP-III: Oak Ridge Thermal Ellipsoid Plot Program for Crystal Structure Illustrations. Oak Ridge National Laboratory Report ORNL-6895.
- Chaudhry, C., Horwich, A. L., Brunger, A. T. & Adams, P. D. (2004). J. Mol. Biol.342, 229–245. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chen, X., Poon, B. K., Dousis, A., Wang, Q. & Ma, J. (2007). Structure, 15, 955–962. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res.35, W375–W383. [DOI] [PMC free article] [PubMed]
- Flores, S. C., Keating, K. S., Painter, J., Morcos, F., Nguyen, K., Merritt, E. A., Kuhn, L. A. & Gerstein, M. B. (2008). Proteins, 73, 299–319. [DOI] [PubMed]
- Harlow, R. L. (1996). J. Res. Natl Inst. Stand. Technol.101, 327–339. [DOI] [PMC free article] [PubMed]
- Howlin, B., Butler, S. A., Moss, D. S., Harris, G. W. & Driessen, H. P. C. (1993). J. Appl. Cryst.26, 622–624.
- Kleywegt, G. J. (2009). Acta Cryst. D65, 134–139. [DOI] [PMC free article] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1996). Structure, 4, 1395–1400. [DOI] [PubMed]
- Kondrashov, D. A., Wynsberghe, A. W. V., Bannen, R. M., Cui, Q. & Phillips, G. N. Jr (2007). Structure, 15, 169–177. [DOI] [PMC free article] [PubMed]
- Lovell, S., Davis, I., Arendall, W. B. III, de Bakker, P., Word, J., Prisant, M., Richardson, J. & Richardson, D. (2003). Proteins, 50, 437–450. [DOI] [PubMed]
- Merritt, E. A. (1999a). Acta Cryst. D55, 1997–2004. [DOI] [PubMed]
- Merritt, E. A. (1999b). Acta Cryst. D55, 1109–1117. [DOI] [PubMed]
- Moore, P. B. (2009). Structure, 17, 1307–1315. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2004). J. Appl. Cryst.37, 174–178.
- Painter, J. & Merritt, E. A. (2006a). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006b). J. Appl. Cryst.39, 109–111.
- Papiz, M. Z., Prince, S. M., Howard, T., Cogdell, R. J. & Isaacs, N. W. (2003). J. Mol. Biol.326, 1523–1538. [DOI] [PubMed]
- Poon, B. K., Chen, X., Lu, M., Vyas, N. K., Quiocho, F. A., Wang, Q. & Ma, J. (2007). Proc. Natl Acad. Sci. USA, 104, 7869–7874. [DOI] [PMC free article] [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed]
- Ramachandran, G. N., Ramakrishnan, C. & Sasisekharan, V. (1963). J. Mol. Biol.7, 95–99. [DOI] [PubMed]
- Rosenfield, R. E., Trueblood, K. N. & Dunitz, J. D. (1978). Acta Cryst. A34, 828–829.
- Schomaker, V. & Trueblood, K. N. (1968). Acta Cryst. B24, 63–76.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Trueblood, K. N., Bürgi, H.-B., Burzlaff, H., Dunitz, J. D., Gramaccioli, C. M., Schulz, H. H., Shmueli, U. & Abrahams, S. C. (1996). Acta Cryst. A52, 770–781.
- Westbrook, J., Feng, Z., Burkhardt, K. & Berman, H. (2003). Methods Enzymol.374, 370–385. [DOI] [PubMed]
- Wilson, M. A. & Brunger, A. T. (2000). J. Mol. Biol.301, 1237–1256. [DOI] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]