The tandem tudor domain of Sgf29 from S. cerevisiae has been overexpressed in E. coli, purified and crystallized. Diffraction data were collected to 1.92 Å resolution.
Keywords: Sgf29, Saccharomyces cerevisiae, tudor domains
Abstract
The protein Sgf29 has been identified as a subunit of the SAGA (Spt–Ada–Gcn5 acetyltransferase) histone acetyltransferase complex in Saccharomyces cerevisiae, which is conserved from yeast to humans. The tandem tudor domain at the C-terminus of Sgf29 was crystallized using the hanging-drop vapour-diffusion method and the crystals diffracted to 1.92 Å resolution. The crystals belonged to space group P212121, with unit-cell parameters a = 49.76, b = 95.10, c = 114.43 Å, and are estimated to contain one protein molecule per asymmetric unit.
1. Introduction
Nucleosomes are composed of ∼150 bp DNA which wraps around a core histone octamer. The histone core is subjected to various post-translational modifications including methylation, acetylation, ubiquitylation, sumoylation, phosphorylation and ADP-ribosylation. These modifications play an important role in regulating gene transcription (Kouzarides, 2007 ▶). Lysine methylation at different sites of the histone core provides activation or repression marks for transcription. For example, trimethylation of lysine 4 in histone H3 (H3K4) is usually linked to gene activation, while trimethylation of lysine 9 in histone H3 (H3K9) is generally connected to gene repression (Martin & Zhang, 2005 ▶). Histone-methylation marks can be recognized by many ‘reader’ protein domains such as chromodomains, PHD domains, tudor domains, MBT domains and so on and used as a mechanism to recruit various effector-protein complexes and direct downstream events (Adams-Cioaba & Min, 2009 ▶).
SAGA (Spt–Ada–Gcn5 acetyltransferase) is a 1.8 MDa multi-protein complex that exhibits multiple functions in regulating gene transcription in response to stress conditions and contains the core components Spt7, Spt20 and Ada1. In addition to the core subunits of the SAGA complex, there are many other subunits which modulate the histone acetyltransferase and deubiquitinase activity of the core (Daniel & Grant, 2007 ▶). The Gcn5 subunit of the SAGA complex has histone acetyltransferase activity and can acetylate histone H3 lysine 14 (H3K14) in vitro. However, in the SAGA complex the enzymatic activity of Gcn5 is regulated by other subunits, expanding the substrate specificity to H3K9, H3K18 and H3K23 in addition to H3K14 both in vitro and in vivo (Grant et al., 1999 ▶).
Sgf29 has recently been identified by mass-spectrometric studies of the SAGA complex from budding yeast and its function is still unclear (Sanders et al., 2002 ▶). STAGA is a mammalian homologue of SAGA and a component of the STAGA complex, CCDC101, that is homologous to Sgf29 has been found in rats. Through its N-terminal coiled-coil domain, CCDC101 interacts with another subunit of the STAGA complex, Ada3 (Kurabe et al., 2007 ▶). The C-terminus of Sgf29 contains a conserved tandem tudor domain which is predicted to recognize methylated lysine sites in the histone core and modulate the activity of Gcn5. The mechanism of this action is unknown (Lee & Workman, 2007 ▶). To address this question, the tandem tudor domain of Saccharomyces cerevisiae Sgf29 was cloned, expressed, purified and crystallized.
2. Materials and methods
2.1. Cloning
The DNA of the Sgf29 tandem tudor domain (residues 113–259) was amplified by the polymerase chain reaction (PCR) from genomic DNA of S. cerevisiae using the oligonucleotide primers 5′-GATCACTGAGGATCCTCGTACTGGACCAGCGAA (forward) and 5′-CATCTCGAGCTATTTCCTTGCTAGGTTTGCCA (reverse) (Sangon). The PCR product was digested with BamHI and XhoI and ligated into the pET-23b vector with maltose-binding protein (MBP) fused at the N-terminus. The resulting plasmid was analyzed by DNA sequencing to confirm the insertion of the Sgf29 tandem tudor domain (Amphipound Biotech).
2.2. Overexpression and purification
The plasmid containing the MBP-fused Sgf29 tandem tudor domain (MBP-Sgf29) was transformed into Escherichia coli BL21 (DE3) cells. The cells containing the target plasmid were grown in LB medium supplemented with 50 µg ml−1 ampicillin at 310 K until the OD600nm reached 0.6–0.8 and were then induced by the addition of 0.5 mM IPTG for 20 h at 293 K. The cells were harvested by centrifugation at 6000 rev min−1 for 8 min at 277 K and the resulting bacterial pellets were resuspended in lysis buffer (200 mM NaCl, 20 mM Tris–HCl pH 8.5). The cells were lysed using a French press at 287 K and the lysate was centrifuged at 15 000g for 30 min at 277 K. The supernatant was passed over an amylose resin (New England Biolabs Inc.) column pre-equilibrated with lysis buffer. The column was washed with about 20 column volumes of lysis buffer to remove contaminants and MBP-Sgf29 was eluted with five column volumes of elution buffer (10 mM maltose, 200 mM NaCl, 20 mM Tris–HCl pH 8.5). The eluate was changed to buffer A (50 mM NaCl, 50 mM Tris–HCl pH 8.5, 5% glycerol) and loaded onto a Resource Q column (1 ml, GE Healthcare) pre-equilibrated with buffer A. The column was washed with two column volumes of buffer A. The sample was eluted with a linear NaCl gradient from 100% buffer A to 50% buffer B (50 mM Tris–HCl pH 8.5, 2 M NaCl, 5% glycerol) in 60 column volumes. The eluted fractions from the Resource Q column were concentrated and applied onto a HiLoad 16/60 Superdex 200 size-exclusion column (GE Healthcare) pre-equilibrated with column buffer (150 mM NaCl, 50 mM Tris–HCl pH 8.0). The fraction containing MBP-Sgf29 was concentrated to 10 mg ml−1 by centrifugal ultrafiltration (Millipore, 5 kDa cutoff) for further use. All purification steps were performed at 277 K and the result of each step was monitored by SDS–PAGE.
2.3. Crystallization
Crystal screening experiments were carried out by hand using the sitting-drop vapour-diffusion method at 281 K in 48-well plates (XtalQuest Co.). Initial screening was performed using Crystal Screen, Crystal Screen II (Hampton Research), Wizard I and Wizard II (Emerald BioSystems). 1 µl protein solution (10 mg ml−1 in buffer containing 150 mM NaCl, 50 mM Tris–HCl pH 8.0) was mixed with 1 µl well solution and the mixture was equilibrated against 200 µl well solution. Microcrystals appeared after one week in condition No. 32 of Crystal Screen (2 M ammonium sulfate, 0.1 M sodium acetate pH 4.6). The crystallization condition was optimized using the hanging-drop vapour-diffusion method at 281 K. 1 µl protein (10 mg ml−1 in buffer consisting of 150 mM NaCl, 50 mM Tris–HCl pH 8.0) was mixed with 1 µl well solution and the mixture was equilibrated against 500 µl well solution. After several rounds of refinement, single crystals that diffracted well were obtained.
2.4. Diffraction data collection and processing
The crystals of MBP-Sgf29 were transferred to cryoprotectant solution consisting of 2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.0, 20% glycerol and soaked for several seconds. The crystals were flash-cooled in liquid nitrogen and used for X-ray diffraction data collection at 100 K on the synchrotron-radiation beamline BL17U at Shanghai Synchrotron Radiation Facility. 180 diffraction images were recorded from a single crystal with an oscillation angle of 1° per image. Diffraction data were processed with the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). Data-collection and processing statistics are listed in Table 1 ▶.
Table 1. Crystal parameters and data-collection statistics for the crystal of MBP-Sgf29.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 49.8, b = 95.1, c = 114.4, α = β = γ = 90 |
| Resolution (Å) | 50–1.92 (1.95–1.92) |
| Unique reflections | 42425 |
| Redundancy | 5.8 (4.0) |
| Average I/σ(I) | 16.5 (4.8) |
| Completeness (%) | 95.7 (79.8) |
| Rmerge† (%) | 9.6 (23.4) |
R
merge = 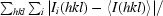
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
3. Results and discussion
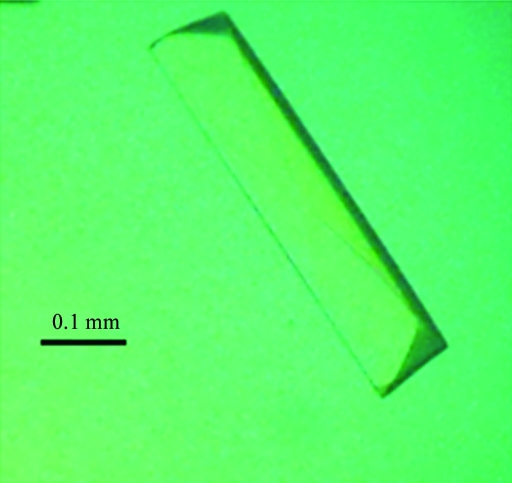
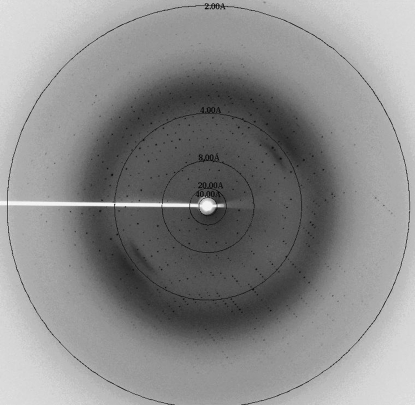
The purified MBP-Sgf29 was concentrated to about 10 mg ml−1 for initial crystallization trials and microcrystals were obtained using the condition 2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.6 at 281 K. To optimize the crystallization condition, several parameters were changed including the concentration of the protein and of ammonium sulfate, the buffer pH and the temperature. It was found that lowering the buffer pH resulted in better crystals. Crystals suitable for X-ray diffraction with a maximum dimension of 0.4 mm were obtained from 2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.0 at 281 K (Fig. 1 ▶) and these crystals diffracted to 1.92 Å resolution (Fig. 2 ▶). The crystals belonged to space group P212121, with unit-cell parameters a = 49.76, b = 95.10, c = 114.43 Å. The calculated Matthews coefficient (V M) of 2.35 Å3 Da−1, with a corresponding solvent content of 47.7%, suggests the presence of one molecule per asymmetric unit (Matthews, 1968 ▶). We are attempting to solve the structure of MBP-Sgf29 by the molecular-replacement method using MBP as a search model.
Figure 1.
Crystal of MBP-Sgf29.
Figure 2.
X-ray diffraction image collected from the crystal of MBP-Sgf29.
Acknowledgments
We thank Drs Ling Tang and Jianhua He for assistance with data collection on beamline BL17U of the Shanghai Synchrotron Radiation Facility. This work was supported by a grant from the National Natural Science Foundation of China (No. 30970576), the ‘100 Talents Program’ of the Chinese Academy of Sciences and grants from the Chinese Ministry of Science and Technology (Nos. 2009CB825502 and 2006CB806501) to JZ.
References
- Adams-Cioaba, M. A. & Min, J. R. (2009). Biochem. Cell Biol.87, 93–105. [DOI] [PubMed]
- Daniel, J. A. & Grant, P. A. (2007). Mutat. Res.618, 135–148. [DOI] [PMC free article] [PubMed]
- Grant, P. A., Eberharter, A., John, S., Cook, R. G., Turner, B. M. & Workman, J. L. (1999). J. Biol. Chem.274, 5895–5900. [DOI] [PubMed]
- Kouzarides, T. (2007). Cell, 128, 693–705. [DOI] [PubMed]
- Kurabe, N., Katagiri, K., Komiya, Y., Ito, R., Sugiyama, A., Kawasaki, Y. & Tashiro, F. (2007). Oncogene, 26, 5626–5634. [DOI] [PubMed]
- Lee, K. K. & Workman, J. L. (2007). Nature Rev. Mol. Cell Biol.8, 284–295. [DOI] [PubMed]
- Martin, C. & Zhang, Y. (2005). Nature Rev. Mol. Cell Biol.6, 838–849. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sanders, S. L., Jennings, J., Canutescu, A., Link, A. J. & Weil, P. A. (2002). Mol. Cell. Biol.22, 4723–4738. [DOI] [PMC free article] [PubMed]