Crystals of SAICAR synthase from S. suis serotype 2 were obtained in the presence of 40 mM aspartic acid substrate; they belonged to space group P2 and diffracted to 2.8 Å resolution.
Keywords: SAICAR synthase, Streptococcus suis
Abstract
Phosphoribosylaminoimidazole-succinocarboxamide synthase (SAICAR synthase) plays an essential role in the de novo biosynthesis of purine nucleotides. In this study, the SAICAR synthase from Streptococcus suis was cloned and overexpressed in Escherichia coli. The subsequent product was purified and crystallized using the hanging-drop vapour-diffusion method. The crystals diffracted to 2.8 Å resolution and belonged to space group P2, with unit-cell parameters a = 70.2, b = 52.2, c = 153.9 Å, β = 102.8°.
1. Introduction
Streptococcus suis serotype 2 is a Gram-positive pathogenic bacterium that causes many serious diseases, manifesting as meningitis, septicaemia, arthritis and even sudden death. As a pig-borne emerging zoonotic pathogen, S. suis also sporadically results in severe infections in humans and can pose a hazard to public health (Staats et al., 1997 ▶; Gottschalk & Segura, 2000 ▶). 35 serotypes of S. suis have been determined based on differences in their capsule antigens. S. suis serotype 2 is the most prevalent serotype isolated from both diseased piglets and patients worldwide and is often associated with a severe clinical syndrome (Staats et al., 1997 ▶). It is noteworthy that two recent large-scale outbreaks of human streptococcal toxic shock syndrome (STSS) caused by S. suis serotype 2 in China in 1998 and in 2005 have posed public health concerns worldwide (Tang et al., 2006 ▶; Yu et al., 2006 ▶). Our group has reported the whole genome sequences of two virulent S. suis serotype 2 isolates (05ZYH33 and 98HAH12; Chen et al., 2007 ▶). Here, we identified a gene encoding SAICAR synthase, which is related to purine metabolism, from S. suis strain 05ZYH33.
SAICAR synthase (phosphoribosylaminoimidazole-succinocarboxamide synthase; EC 6.3.2.6) is a central enzyme in the bacterial de novo purine nucleotide-biosynthesis pathway. Studies of purine synthesis not only increase the understanding of the mechanism and evolution of this significant pathway, but have also been used to develop effective chemotherapeutic agents. SAICAR synthase catalyses the seventh step of purine metabolism in the cells of living organisms. The enzymatic reaction involves the synthesis of 4-(N-succinylcarboxamide)-5-aminoimidazole ribonucleotide (SAICAR) from 4-carboxy-5-aminoimidazole ribonucleotide (CAIR) and aspartic acid (Asp) in the presence of adenosine triphosphate (ATP) and magnesium ions.
To date, the crystal structures of native SAICAR synthases from Saccharomyces cerevisiae (PDB code 1a48; Levdikov et al., 1996 ▶, 1998 ▶), Thermotoga maritima (PDB code 1kut; Zhang et al., 2006 ▶), Ehrlichia chaffeensis (PDB code 3kre; Seattle Structural Genomics Center for Infectious Disease, unpublished work) and Pyrococcus horikoshii OT3 (Manjunath et al., 2010 ▶) have been determined at 2.5, 2.2, 1.8 and 2.3 Å resolution, respectively. In addition, the three-dimensional structures of the enzyme in complex with ATP (PDB codes 1obd and 1obg; Antonyuk et al., 2001 ▶) and with the substrate analogues AICAR (5′-phosphoribosyl-5-aminoimidazole-4-carboxamide) and succinic acid (PDB code 2cnq; Urusova et al., 2003 ▶), as well as the structure of a complex with ADP–CAIR in the binding groove of SAICAR synthase from Escherichia coli (PDB codes 2gqs and 2gqr; Ginder et al., 2006 ▶), have also been reported. These studies enabled the definition of the ATP-binding sites, the CAIR substrate-binding sites and the residues that facilitate binding in the protein molecule at the atomic level. Nevertheless, the detailed interactions mediating the binding of the other substrate Asp to the enzyme remain elusive.
In the present study, SAICAR synthase from S. suis strain 05ZYH33 (dubbed 05SSU0026) was expressed, purified and crystallized in the presence of 0.04 M aspartic acid (Asp) using the hanging-drop vapour-diffusion method. We aim to determine the crystal structure of this enzyme and the structure of the complex of the enzyme with Asp in order to shed further light on the detailed enzyme–substrate interactions in the SAICAR synthase family.
2. Materials and methods
2.1. Cloning and expression
The gene coding for SAICAR synthase from S. suis strain 05ZYH33 was amplified by PCR using the genomic DNA extract as the template. The primer sequences were 5′-ggaattccatATGAAAACAGACCTTCTC-3′ (NdeI site in lower case) and 5′-ccgctcgagTTTCACTTCCTGTAA-3′ (XhoI site in lower case). The resultant PCR product (approximately 700 bp) was first cloned into pMD18-T Simple Vector (Takara) and then digested with the restriction endonucleases NdeI and XhoI. The target gene was then linked to the prokaryotic expression vector pET21a(+) to generate the plasmid pET21a-SAICAR. The identity of the DNA construct was verified by direct DNA sequencing.
For protein expression, E. coli BL21 (DE3) cells were transformed with the above-mentioned plasmid and grown in LB medium containing 50 µg ml−1 ampicillin at 310 K until the optical density of the culture at 600 nm reached 0.6. Expression of SAICAR synthase was obtained by induction with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 289 K for 10 h.
2.2. Purification
The cells were harvested by centrifugation and resuspended in buffer containing 20 mM Tris–HCl pH 8.0, 50 mM NaCl. After sonication, the lysate was centrifuged at 16 000g for 10 min to remove cell debris. The resultant supernatant was incubated with 10 ml Ni–NTA agarose (Qiagen) for 2 h on an overhead shaker at 277 K. The resin was then recollected and washed with 10–15 bed volumes of wash buffer (20 mM Tris–HCl pH 8.0, 50 mM NaCl) to remove any contaminants. The proteins were then eluted with wash buffer containing an imidazole gradient (10, 20, 50 and 200 mM) and analyzed by SDS–PAGE with Coomassie staining. The eluant containing the protein of interest with high purity was concentrated in an Amicon Ultra-15 filter (Millipore) using a 10 kDa cutoff membrane and then loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with wash buffer prior to further purification.
2.3. Crystallization
The pooled fractions containing purified SAICAR synthase were concentrated to 12 mg ml−1 using a Millipore Amicon 10 kDa centrifugal device and stored at 277 K. Aspartic acid (Asp) was then dissolved in the protein solution to a concentration of 40 mM to yield the final protein preparation for crystallization. Initial crystal-screening trials were performed with Crystal Screens I and II and PEG/Ion Screen (Hampton Research) at 291 K using the hanging-drop vapour-diffusion method. 1.5 µl of the above-mentioned protein preparation was mixed with 1.5 µl reservoir solution and equilibrated against 200 µl reservoir solution.
2.4. X-ray data collection
X-ray diffraction data were collected using synchrotron radiation (λ = 1.0000 Å). A single piece of crystal was separated from the crystal bunch, mounted in a nylon loop and flash-cooled in an N2 gas cryostream before data collection. The cryoprotectant consisted of an additional 12%(v/v) glycerol in reservoir solution. Data were indexed, integrated and scaled using DENZO and SCALEPACK as implemented in HKL-2000 (Otwinowski & Minor, 1997 ▶). The statistics of X-ray data collection are given in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outermost resolution shell.
| Space group | P2 |
| Unit-cell parameters | |
| a (Å) | 70.2 |
| b (Å) | 52.2 |
| c (Å) | 153.9 |
| β (°) | 102.8 |
| Wavelength (Å) | 1.00000 |
| Resolution range (Å) | 50–2.8 (2.9–2.8) |
| No. of observed reflections | 138337 |
| No. of unique reflections | 26940 |
| Average redundancy | 5.1(5.2) |
| Rmerge† (%) | 0.063 (0.339) |
| Average I/σ(I) | 23.895 (4.575) |
R
merge = 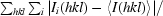
 , where 〈I(hkl)〉 is the mean intensity of reflection hkl and I
i(hkl) is the intensity of an individual measurement of reflection hkl.
, where 〈I(hkl)〉 is the mean intensity of reflection hkl and I
i(hkl) is the intensity of an individual measurement of reflection hkl.
3. Results and discussion
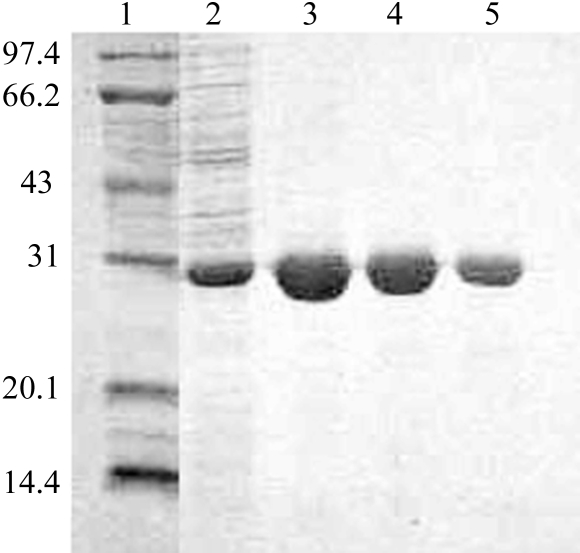
The SAICAR synthase from S. suis consists of 235 amino acids with a molecular weight of about 26.6 kDa. This protein is considerably smaller than its counterpart from yeast (about 34.5 kDa), with which it shares only 20.7% primary sequence identity. Nevertheless, in a search of the PDB several other SAICAR synthase structures that exhibited higher sequence identities with our enzyme were found, including those from E. coli (about 43% identity) and T. maritima (about 34% identity). The SAICAR synthase in this study was expressed as a C-terminally His-tagged protein in E. coli and was purified by affinity chromatography and gel filtration. The recombinant enzyme was easily obtained: a single step of purification using Ni–NTA resin yielded protein with a purity of over 99% (Fig. 1 ▶), paving the way for further enzymatic and crystallization trials in the future.
Figure 1.
One-step purification of SAICAR synthase by affinity chromatography. The cell lysate was incubated with Ni–NTA resin at 277 K for 2 h. The resin was then collected and washed with wash buffer containing an imidazole gradient (see §2 for details). The SDS–PAGE profile is shown. Lane 1, protein molecular-weight markers (kDa); lane 2, 10 mM imidazole eluant; lane 3, 20 mM imidazole eluant; lane 4, 50 mM imidazole eluant; lane 5, 200 mM imidazole eluant.
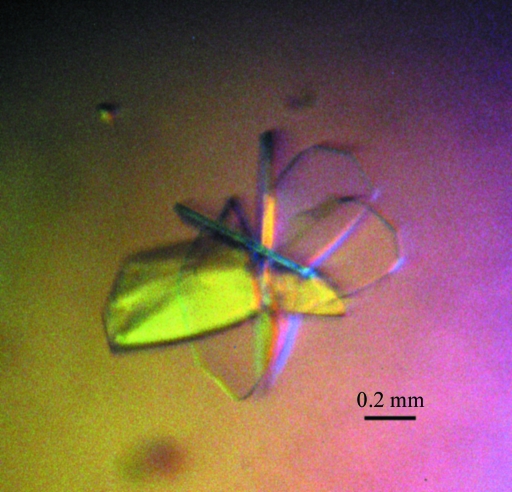
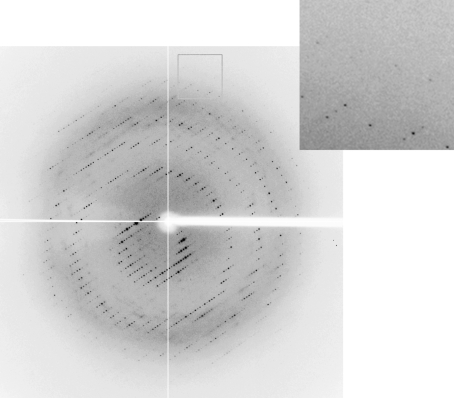
Crystallization of the SAICAR synthase using a protein preparation containing 40 mM Asp (see §2) proved to be easy under several conditions. After optimization, well diffacting crystals were finally obtained from a condition consisting of 0.2 M ammonium acetate, 16% PEG 3350 and 15% glycerol. The crystal grew to a size of 1.0–1.5 mm at a significantly rapid rate, with growth being completed within 4 d of plating at 291 K (Fig. 2 ▶). Diffraction data were collected using synchrotron radiation and the crystal diffracted to 2.8 Å resolution (Fig. 3 ▶). The final data-set statistics are summarized in Table 1 ▶. The crystal belonged to space group P2, with unit-cell parameters a = 70. 2, b = 52. 2, c = 153.9 Å, β = 102.8°. A preliminary estimation of the content of the unit cell indicated that there should be four molecules per asymmetric unit, with a Matthews coefficient of about 2.59 Å3 Da−1 and a solvent content of about 52.5%.
Figure 2.
Representative crystals of SAICAR synthase. The size of the crystal was measured as approximately 1.0–1.5 mm.
Figure 3.
A typical X-ray diffraction image collected from a crystal of SAICAR synthase. The crystal diffracted to 2.8 Å resolution.
In order to provide further insights into catalysis by SAICAR synthase, it will be necessary to obtain high-resolution structures of the enzyme in complex with aspartic acid. In this study, we successfully crystallized the protein in the presence of 40 mM Asp. We failed to obtain any crystals under the same conditions in the absence of aspartic acid. This phenomenon strongly indicates that we have obtained complex crystals. Initial molecular-replacement calculations were performed using the program AMoRe (Navaza, 1994 ▶) with the structure of SAICAR synthase from E. coli (PDB code 2gqr; Ginder et al., 2006 ▶) as the search model, but no distinct peaks were observed. Another trial using the structure of the T. maritima synthase (PDB code 1kut) as the input probe was also unsuccessful. It should be noted that the SAICAR protein from T. maritima is a disulfide-bond-linked dimer through Cys126, which is not conserved in our enzyme; as several reports have shown that the functional unit of SAICAR synthase is a monomer (Levdikov et al., 1998 ▶; Zhang et al., 2006 ▶), only one molecule from the T. maritima SAICAR structure was selected when we performed the calculation, but no distinct peaks could be obtained. Further optimization of the crystallization conditions as well as structure-solution trials using other programs are currently under way.
Acknowledgments
This work was completed in the laboratory of Professor George F. Gao at the Institute of Microbiology, Chinese Academy of Sciences (IMCAS) and was supported by grants from the Ministry of Science and Technology (National Key Technology R&D Program 2006BAD06A04 and International Collaborative Projects 2007DFC30240 and 2006DFB32010).
References
- Antonyuk, S. V., Grebenko, A. I., Levdikov, V. M., Urusova, D. V., Melik-Adamyan, V. R., Lamzin, V. S. & Wilson, K. S. (2001). Crystallogr. Rep.46, 620–625.
- Chen, C. et al. (2007). PLoS One, 2, e315. [DOI] [PMC free article] [PubMed]
- Ginder, N. D., Binkowski, D. J., Fromm, H. J. & Honzatko, R. B. (2006). J. Biol. Chem.281, 20680–20688. [DOI] [PubMed]
- Gottschalk, M. & Segura, M. (2000). Vet. Microbiol.76, 259–272. [DOI] [PubMed]
- Levdikov, V. M., Barynin, V. V., Grebenko, A. I., Melik-Adamyan, W. R., Lamzin, V. S. & Wilson, K. S. (1998). Structure, 6, 363–376. [DOI] [PubMed]
- Levdikov, V. M., Grebenko, A. I., Barynin, V. V., Melik-Adamyan, W. R., Lamzin, V. S. & Wilson, K. S. (1996). Crystallogr. Rep.41, 275–286.
- Manjunath, K., Jeyakanthan, J., Nakagawa, N., Shinkai, A., Yoshimura, M., Kuramitsu, S., Yokoyama, S. & Sekar, K. (2010). Acta Cryst. F66, 180–183. [DOI] [PMC free article] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Staats, J. J., Feder, I., Okwumabua, O. & Chengappa, M. M. (1997). Vet. Res. Commun.21, 381–407. [DOI] [PubMed]
- Tang, J. et al. (2006). PLoS Med.3, e151. [DOI] [PMC free article] [PubMed]
- Urusova, D. V., Antonyuk, S. V., Grebenko, A. I., Lamzin, V. S. & Melik-Adamyan, V. R. (2003). Crystallogr. Rep.48, 763–767.
- Yu, H. et al. (2006). Emerg. Infect. Dis.12, 914–920. [DOI] [PMC free article] [PubMed]
- Zhang, R., Skarina, T., Evdokimova, E., Edwards, A., Savchenko, A., Laskowski, R., Cuff, M. E. & Joachimiak, A. (2006). Acta Cryst. F62, 335–339. [DOI] [PMC free article] [PubMed]