The omega-transaminase from V. fluvialis JS17 was crystallized. Crystals were found to belong to the orthorhombic space group P212121, with unit-cell parameters a = 78.43, b = 95.95, c = 122.89 Å. The crystals were obtained at 293 K and diffracted to a resolution of 2.5 Å.
Keywords: omega-transaminase, omega-amino acids
Abstract
Omega-transaminase (ω-TA) catalyzes the transfer of an amino group from a non-α-position amino acid or an amine compound with no carboxylic group to an amino acceptor. ω-TA from Vibrio fluvialis JS17 (ω-TAVf) is a novel amine:pyruvate transaminase that is capable of stereoselective transamination of aryl chiral amines. In this study, ω-TAVf was overexpressed in Escherichia coli with engineered C-terminal His tags. ω-TAVf was then purified to homogeneity and crystallized at 292 K. X-ray diffraction data were collected to a resolution of 2.5 Å from a crystal belonging to the orthorhombic space group P212121, with unit-cell parameters a = 78.43, b = 95.95, c = 122.89 Å.
1. Introduction
Transaminase (TA) plays an important role in amino-acid metabolism by removing the amino group from an amino acid, transferring it to a α-keto acid and then converting it to another amino acid (Mehta & Christen, 2000 ▶; Mehta et al., 1993 ▶). TA is ubiquitous in microorganisms and higher organisms and uses pyridoxal 5′-phosphate (PLP) as a cofactor for catalytic reaction. This enzyme is particularly important and has been studied intensively owing to its high potential for use in the production of various amino acids and chiral amines (Christen & Metzler, 1985 ▶; Chao et al., 1999 ▶; Markova et al., 2005 ▶). Despite the many advantages of the enzyme, such as its high stability, high enantioselectivity, high turnover rate and broad substrate specificity, its industrial use has been limited owing to the low equilibrium constants of the reactions that it is involved in. Efforts to overcome the limitations of the activity of this enzyme in reactions for the production of unnatural amino acids and d-amino acids have been conducted by using an engineered TA, coupling with other enzymes and whole-cell biotransformation (Cho et al., 2003 ▶, 2008 ▶; Yun, Cho et al., 2004 ▶; Yun, Lim et al., 2004 ▶; Yun et al., 2003 ▶).
TAs can be classified as alpha-tramsaminases (α-TAs) and omega-transaminases (ω-TAs) based on the relative position of the amino group to be transferred with respect to the carboxyl group of the substrate (Yonaha et al., 1987 ▶, 1983 ▶). ω-TAs such as ω-amino acid:pyruvate TA, ornithine TA and 4-aminobutyrate TA transfer amino groups at non-α positions, unlike α-TA which only acts on α-amino groups on α-amino acids.
ω-TA from Vibrio fluvialis JS17 (ω-TAVf) is a unique PLP-dependent enzyme that catalyzes amino-group transfer from an amine to a keto acid in a reversible manner (Shin et al., 2003 ▶). ω-TAVf is a novel amine:pyruvate transaminase that is capable of stereoselective transamination of aryl chiral amines and is highly suitable for the production of chiral amines owing to its strict enantioselectivity and broad amino-donor specificity for chiral amines (Shin et al., 2003 ▶). Interestingly, although ω-TAVf possesses high substrate specificity for chiral aromatic amines and enantioselectivity for the (S)-enantiomer of chiral amines, it shows narrow substrate specificity in the reaction that restricts its application to the kinetic resolution of aliphatic amines (Shin & Kim, 2002 ▶). Rational protein design by modelling and directed evolution of the enzyme has been used to improve the activity of TA (Yun et al., 2005 ▶; Yano et al., 1998 ▶). Although homology models using the reported structure of TA have successfully provided general information that is useful for the identification of mutation sites for engineering the enzyme, it is necessary to determine the actual atomic structure of ω-TAVf (Smith et al., 1989 ▶; Rossi et al., 2006 ▶; Chen et al., 2002 ▶; Watanabe et al., 1989 ▶; Yoshikane et al., 2008 ▶; Han et al., 2008 ▶).
Several structures of TAs have been determined to date (Rossi et al., 2006 ▶; Chen et al., 2002 ▶). The most closely related structure to ω-TAVf is that of an aminotransferase from Silicibacter pomeroyi (PDB code 3hmu; R. Toro, J. B. Bonanno, U. Ramagopal, J. Freeman, K. T. Bain, S. Miller, J. M. Sauder, S. K. Burley & S. C. Almo, unpublished work), which shares 32% sequence identity. In the present study, we overexpressed, purified and crystallized ω-TAVf as the first step towards elucidating its molecular structure and catalytic mechanism. Details of the atomic structure of ω-TAVf should enable us to understand the catalytic mechanism of ω-TAVf and help in the design of a more efficient ω-TA for industrial use.
2. Materials and methods
2.1. Expression and purification
To express C-terminally His-tagged enzyme, the coding region of ω-TAVf was amplified by PCR using P1 (5′-ATATGGATCCATGAACAAACCGCAAAGCTGG-3′) and P2 (5′-GGATCCCTAAGCATATAAGCTTGGCAACCTCGGCAAAGACCT-3′) primers. The PCR product was digested with restriction enzymes (BamHI/HindIII) and then inserted into the vector pET24ma cut with the same restriction enzymes. The pET24ma vector (constructed by Dr David Sourdive, Pasteur Institute, France) contains a p15A replication origin. The plasmid was transformed into Escherichia coli BL21 (DE3) competent cells and expression was induced by treating the bacteria with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 293 K. Cells expressing ω-TAVf were pelleted by centrifugation, resuspended and lysed by sonication in 50 ml lysis buffer (20 mM Tris pH 7.9, 500 mM NaCl and 5 mM imidazole). The lysate was then centrifuged at 16 000 rev min−1 for 1 h at 277 K and the supernatant fractions were applied onto a gravity-flow column (Bio-Rad) packed with Ni–NTA affinity resin (Qiagen). Unbound bacterial proteins were removed from the column using wash buffer (20 mM Tris pH 7.9, 500 mM NaCl, 60 mM imidazole and 10% glycerol). The C-terminally His-tagged ω-TAVf was eluted from the column using elution buffer (20 mM Tris pH 7.9, 500 mM NaCl and 250 mM imidazole). The elution fractions were collected on a 1 ml scale to 10 ml. Fractions containing more than 80% homogeneous ω-TAVf, as indicated by SDS–PAGE, were selected, combined and concentrated to 20–25 mg ml−1 using a concentration kit (Millipore). The concentrated protein was then applied onto a Superdex 200 10/30 gel-filtration column (GE Healthcare) pre-equilibrated with a solution of 20 mM Tris pH 8.0 and 150 mM NaCl. ω-TAVf (molecular weight 49 000 Da) eluted at around 14.5 ml and was collected and concentrated to 10–12 mg ml−1. The peak was confirmed to contain ω-TAVf by SDS–PAGE. Purified ω-TAVf contained the extra C-terminal residues AAALEHHHHHH and the extra N-terminal residues MASMTGGQQMGRGS. The hexahistidine tag was not removed.
2.2. Crystallization
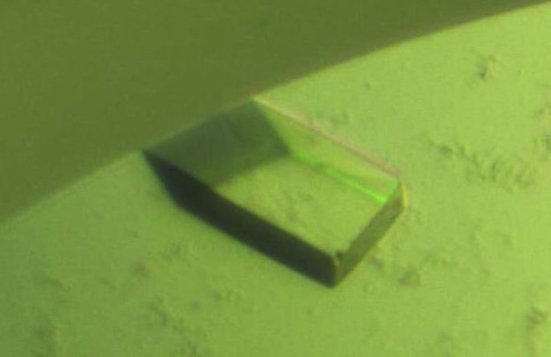
Crystallization conditions were initially screened at 293 K by the sitting-drop vapour-diffusion method using screening kits from Hampton Research (Crystal Screens I and II, Natrix, MembFac and SaltRX) and from the deCODE Biostructures Group (Wizard I, II and III). Initial crystals were grown on the plates by equilibrating a mixture containing 1 µl protein solution (10–12 mg ml−1 protein in 20 mM Tris pH 8.0, 150 mM NaCl) and 1 µl reservoir solution No. 3 from Wizard III (20% PEG 3350 and 0.2 M magnesium formate) against 0.4 ml reservoir solution. Crystallization was further optimized by searching over a range of concentrations of protein, PEG 3350 and magnesium formate. Crystals appeared within 2 d and grew to maximum dimensions of 0.2 × 0.2 × 0.1 mm (Fig. 1 ▶) in the presence of 22% PEG 3350, 0.4 M magnesium formate and 0.1 M sodium citrate pH 6.2. The crystals were rectanglar and diffracted to a resolution of 2.5 Å.
Figure 1.
Crystals of ω-transaminase from V. fluvialis JS17. Rectangular crystal grown in 2 d in the presence of 22% PEG 3350, 0.4 M magnesium formate and 0.1 M sodium citrate pH 6.2. The approximate dimensions of the crystal were 0.2 × 0.2 × 0.1 mm.
2.3. Crystallographic data collection
For data collection, the crystals were transiently soaked in a solution corresponding to the reservoir solution supplemented with 20%(v/v) glycerol. The soaked crystals were then frozen in liquid nitrogen. A 2.5 Å resolution native diffraction data set was collected on beamline BL-4A at Pohang Accelerator Laboratory (PAL), Republic of Korea. The data set was indexed and processed using HKL-2000 (Otwinoski & Minor, 1997 ▶). The diffraction data statistics are shown in Table 1 ▶.
Table 1. Diffraction data statistics for ω-TAVf crystals.
Values in parentheses are for the highest resolution shell.
| X-ray source | BL-4A, PAL |
| Wavelength (Å) | 1.0000 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 78.43, b = 95.95, c = 122.89 |
| Resolution limits (Å) | 50–2.5 |
| No. of observations | 237373 |
| No. of unique reflections | 32875 |
| Mean I/σ(I) | 28.36 (5.40) |
| Completeness (%) | 99.8 (98.0) |
| Rmerge (%) | 9.1 (35.6) |
3. Results and discussion
His-tag affinity chromatography followed by gel-filtration chromatography produced 99% pure ω-TAVf protein, which was analyzed by SDS–PAGE. No contaminating bands were visible on SDS–PAGE. The calculated monomeric molecular weight of ω-TAVf including the C-terminal His tag was 49 000 Da (data not shown).
The success in crystallizing ω-TAVf was the result of several factors. Firstly, many different constructs were used, most of which either led to no crystals or poorly shaped crystals that could not be optimized. Initially, a crystal shaped like a ball that diffracted poorly was obtained. Optimization of the crystallization conditions finally led to crystals of ω-TAVf that were rectangular (Fig. 1 ▶). While most of these crystals diffracted to a resolution of about 8 Å, optimization of crystal handling using a freshly frozen crystal and cryoprotection using a short soaking time by screening many crystals resulted in diffraction to a resolution of 2.5 Å. The crystals belonged to space group P212121, with unit-cell parameters a = 78.43, b = 95.95, c = 122.89 Å. Assuming the presence of one dimer in the crystallographic asymmetric unit, the Matthews coefficient (V M) was calculated to be 2.29 Å3 Da−1, which corresponds to a solvent content of 46.38% (Matthews, 1968 ▶). Most three-dimensional structures of ω-TA contain dimeric molecules and ω-TAVf has also been characterized as a dimer in solution in a previous study (Shin et al., 2003 ▶). Diffraction data statistics are given in Table 1 ▶. The data set was indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The molecular-replacement phasing method was used with the program CNS (Brünger et al., 1998 ▶) using an aminotransferase from a different class (PDB code 3hmu), which has 32% amino-acid sequence identity with that of ω-TAVf, as a search model.
Acknowledgments
We are grateful to Dr Yeon Gil Kim of BL-4A at Pohang Accelerator Laboratory. We also thank Dr Soo Hyun Eom and the members of his laboratory for sharing their beam time. This research was supported by Yeungnam University research grants in 2009.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chao, Y.-P., Lai, Z. J., Chen, P. & Chern, J.-T. (1999). Biotechnol. Prog.15, 453–458. [DOI] [PubMed]
- Chen, C. C., Zhang, H., Kim, A. D., Howard, A., Sheldrick, G. M., Mariano-Dunaway, D. & Herzberg, O. (2002). Biochemistry, 41, 13162–13169. [DOI] [PubMed]
- Cho, B.-K., Cho, H. J., Park, S.-H., Yun, H. & Kim, B.-G. (2003). Biotechnol. Bioeng.81, 783–789. [DOI] [PubMed]
- Cho, B.-K., Park, H.-Y., Seo, J.-H., Kim, J., Kang, T.-J., Lee, B.-S. & Kim, B.-G. (2008). Biotechnol. Bioeng.99, 275–284. [DOI] [PubMed]
- Christen, P. & Metzler, D. E. (1985). Transaminases. New York: Wiley.
- Han, Q., Robinson, H. & Li, J. (2008). J. Biol. Chem.283, 3567–3573. [DOI] [PubMed]
- Markova, M., Peneff, C., Hewlins, M. J., Schirmer, T. & John, R. A. (2005). J. Biol. Chem.280, 36409–36416. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mehta, P. K. & Christen, P. (2000). Adv. Enzymol. Relat. Areas Mol. Biol.74, 129–184. [DOI] [PubMed]
- Mehta, P. K., Hale, T. I. & Christen, P. (1993). Eur. J. Biochem.214, 549–561. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rossi, F., Garavaglia, S., Giovenzana, G. B., Arca, B., Li, J. & Rizzi, M. (2006). Proc. Natl Acad. Sci. USA, 103, 5711–5716. [DOI] [PMC free article] [PubMed]
- Shin, J.-S. & Kim, B.-G. (2002). Biotechnol. Bioeng.77, 832–837. [DOI] [PubMed]
- Shin, J.-S., Yun, H., Jang, J.-W., Park, I. & Kim, B.-G. (2003). Appl. Microbiol. Biotechnol.61, 463–471. [DOI] [PubMed]
- Smith, D. L., Almo, S. C., Toney, M. D. & Ringe, D. (1989). Biochemistry, 28, 8161–8167. [DOI] [PubMed]
- Watanabe, N., Sakabe, K., Sakabe, N., Higashi, T., Sasaki, K., Aibara, S., Morita, Y., Yonaha, K., Toyama, S. & Fukutani, H. (1989). J. Biochem.105, 1–3. [DOI] [PubMed]
- Yano, T., Oue, S. & Kagamiyama, H. (1998). Proc. Natl Acad. Sci. USA, 95, 5511–5515. [DOI] [PMC free article] [PubMed]
- Yonaha, K., Toyama, S. & Kagamiyama, H. (1983). J. Biol. Chem.258, 2260–2265. [PubMed]
- Yonaha, K., Toyama, S. & Soda, K. (1987). Methods Enzymol.143, 500–504. [DOI] [PubMed]
- Yoshikane, Y., Yokochi, N., Yamasaki, M., Mizutani, K., Ohnishi, K., Mikami, B., Hayashi, H. & Yagi, T. (2008). J. Biol. Chem.283, 1120–1127. [DOI] [PubMed]
- Yun, H., Cho, B.-K. & Kim, B.-G. (2004). Biotechnol. Bioeng.87, 772–778. [DOI] [PubMed]
- Yun, H., Hwang, B.-Y., Lee, J.-H. & Kim, B.-G. (2005). Appl. Environ. Microbiol.71, 4220–4224. [DOI] [PMC free article] [PubMed]
- Yun, H., Lim, S., Cho, B.-K. & Kim, B.-G. (2004). Appl. Environ. Microbiol.70, 2529–2534. [DOI] [PMC free article] [PubMed]
- Yun, H., Yang, Y.-H., Cho, B.-K., Hwang, B.-Y. & Kim, B.-G. (2003). Biotechnol. Lett.25, 809–814. [DOI] [PubMed]