Abstract
Purpose
To investigate the role of the PER3 circadian rhythm gene, located within the commonly deleted region of chromosome 1p36, in human breast cancer development.
Patients and Methods
The frequency of genetic alterations at 1p36 and PER3 gene copy number status were analyzed in 180 lymph node–negative breast cancers from patients who had received treatment with chemotherapy and/or tamoxifen. The expression levels of PER3 were also analyzed using published microarray profiles from > 400 breast cancer samples. Finally, the effect of loss of Per3 on tumor susceptibility was tested using two mouse models of breast cancer.
Results
Deletion of PER3 is directly related to tumor recurrence in patients with estrogen receptor (ER) – positive breast cancers treated with tamoxifen. Low expression of PER3 mRNA is associated with poor prognosis, particularly in a subset of tumors that are ER positive, and either luminal A or ERBB2-positive tumors. Mice deficient in Per3 showed increased susceptibility to breast cancer induced by carcinogen treatment or by overexpression of Erbb2.
Conclusion
Disruption of PER3 function may serve as an indicator of probability of tumor recurrence in patients with ER-positive tumors. Further investigations of this pathway may reveal links between deregulation of sleep homeostasis and breast tumorigenesis.
INTRODUCTION
Chromosomal region 1p36 is among the most commonly deleted regions in human cancers. Deletion of 1p36 is especially frequent in breast tumors and is associated with progression and lymph node metastasis,1 poor prognosis,2 higher rate of recurrence,3 larger tumor size, and DNA aneuploidy.4 However, no direct relationship between breast carcinogenesis or prognosis and any specific tumor suppressor gene on 1p36 has been established. Recent elegant studies have identified CHD55 and more recently KIF1B6 as candidate tumor suppressor genes in this region, but no specific roles for these genes in breast cancer development have been demonstrated.
The human PER3 gene is located within 1.5 Mb of CHD5, and the mouse homolog is a member of the period gene family that controls circadian rhythms.7,8 Members of the period family of circadian rhythm genes (Per1 and Per2) have been implicated in cell cycle control, DNA damage responses, and tumor progression.9–12 Although inactivation of Per3 in the mouse germline has only subtle effects on circadian clock function,13 it has been shown that Per3 transcripts exhibit a clear circadian rhythm both in the suprachiasmatic nucleus7 and in mouse peripheral tissues.14 Similar data have been shown in human peripheral blood cells, where circadian oscillations were more robust for PER3 expression than for other clock genes including PER1 and PER2.15,16 The possible functions of PER3 in tumor development have not been explored, but links to breast cancer are supported by biochemical studies demonstrating the existence of complexes, including proteins of the PER family together with the estrogen receptor (ER),17,18 and by reports of association between a polymorphism in the human PER3 gene and breast cancer susceptibility.19
The location of the PER3 gene within a region that is commonly deleted in breast cancers suggested a possible link to epidemiologic studies20,21 showing an association between disrupted sleep cycles and higher risk of developing breast cancer. We used a combination of human breast tumor analysis and mouse models to show that disruption of PER3 may serve as a prognostic biomarker of tumor recurrence in patients with ER-positive, luminal A, and/or ERBB2-positive tumors.
PATIENTS AND METHODS
Sample Selection
We used three previously published breast cancer data sets that included clinical, gene expression, and/or array comparative genomic hybridization (CGH) data.22–24 Data on disease-free survival (DFS; defined as the time to a first event) and overall survival (OS) were available for all patients in the three data sets except for one patient in samples from a study by Chin et al.24
Copy Number Analysis of PER3
All tumor DNA samples were obtained from frozen breast tumors with > 50% tumor cells.22 The genomic sequence of PER3 (GenBank accession NM_016831.1) was used to design a set of primers and probes specific to the PER3 gene (Primer Express software version 1.0; Applied Biosystems, Foster City, CA; see Data Supplement for detailed description).
PER3 Gene Expression Analysis
We examined PER3 expression in 413 breast tumor expression arrays taken from studies by van de Vijver et al23 (n = 295) and Chin et al24 (n = 118). In each data set, a sample si in the set S was labeled as “PER3 low,” “PER3 normal,” or “PER3 high” using the rule:
If si ≤ (mean [S] − ½ × standard deviation [S]), assign LOW.
If si ≥ (mean [S] + ½ × standard deviation [S]), assign HIGH.
Otherwise, assign NORMAL.
This method allowed us to compare relative PER3 expression levels across both data sets fused as a single group of patients.
Statistical Analysis
The association between PER3 deletion or PER3 expression and clinical-pathologic parameters was analyzed using Fisher's exact test. All reported P values were two-tailed. Significant differences in DFS and OS time were calculated using the Cox proportional hazard (log-rank) test. Multivariate Cox regression analysis was used to prove statistical independence of PER3 from other known prognostic factors. Statistical analysis was performed using SPSS version 12.0 (SPSS, Chicago, IL).
Mice and Tumor Induction
Wild-type (Per3+/+) and Per3 knockout (Per3−/−) 129/sv mice (provided by Drs. Y.H. Fu and L.J. Ptáček, University of California, San Francisco [USCF], San Francisco, CA) were bred and treated according to Laboratory Animal Resource Center regulations. Female mice 7 weeks old from the F2 intercross population (Per3+/+, Per3+/−, and Per3−/−) were treated with six doses of 1 mg of 7,12-dimethylbenz[a]anthracene (DMBA) diluted in corn oil by weekly oral gavage. A second group of mice was treated with corn oil only as a group control. In a second experiment, male Per3−/− mice were crossed with female FVB mice expressing the Neu (ErbB2) proto-oncogene under control of the mouse mammary tumor virus (MMTV) 3′ long terminal repeats promoter25 (provided by Dr. Z. Werb, UCSF) to generate F1 transgenic mice heterozygous for Per3 (Neu/Per3+/−). F1 males and females were intercrossed to produce the F2 generation consisting of Neu/Per3+/+, Neu/Per3+/−, and Neu/Per3−/− animals. Identification of animal genotypes is described in the Data Supplement.
In the DMBA gavage experiment, female mice were examined every 3 days for sickness or symptoms of tumor development for up to 19.7 months. MMTV neu/Per3 transgenic female mice were examined weekly by palpation for mammary tumor development for up to 25.8 months. Mice that showed significant weight loss, morbidity, or excessive tumor burden were killed by cervical dislocation after being anesthetized according to the UCSF Animal Care and Use protocol. Tumors and tissues were fixed in 4% neutral buffered paraformaldehyde for histologic examination. Mice found dead were censored from the study.
RESULTS
Deletion of 1p36 and Loss of PER3 Genetic Variants in Breast Cancers
We previously reported genome-wide array CGH profiles of 185 lymph node–negative breast cancers from a Spanish cohort,22 of whom 85 received anthracycline chemotherapy (chemo group) and 95 received no chemotherapy (non-chemo group). To search for genetic events related to resistance to hormonal (tamoxifen) therapy, we divided the non-chemo group into two subgroups on the basis of whether they had received hormonal treatment. Of the 95 patients in the non-chemo group, 59 patients with ER-positive and/or progesterone-positive tumors received tamoxifen, whereas 36 did not receive any treatment. Analysis of CGH profiles for these patients revealed that deletion of chromosome 1p was associated with recurrence in the subgroup of ER-positive tamoxifen-treated patients (P < .05 after multiple testing correction using method of Benjamini and Hoffberg; Fig DS1).26
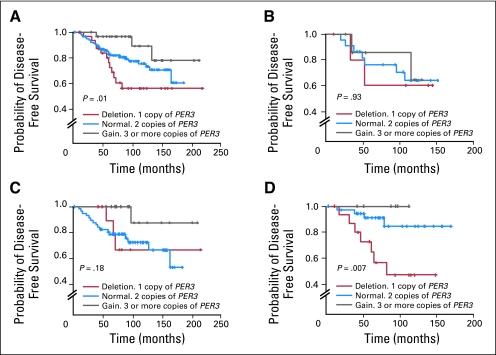
The chromosome 1p36 locus is frequently deleted in many human tumors, but the region of deletion is large. Separate, nonoverlapping chromosome fragments have been implicated,27–29 suggesting that multiple tumor suppressor genes are involved. We considered PER3 to be a good candidate for involvement in breast cancer because of its location within one of the minimal deletion regions on 1p36.2,5,6 as well as the epidemiologic19 and mechanistic17 data linking circadian rhythm genes to hormone status and breast cancer. We therefore examined the copy number status of PER3 by quantitative TaqMan analysis (Applied Biosystems) in DNA samples from 180 breast cancer patients. The relationship between the frequency of deletion or copy number gain and clinico-pathologic characteristics of the patients is shown in Table DS1. The number of copies of PER3 showed a significant gene dosage association with recurrence-free survival at 10 years (P = .01; Fig 1A). The proportion of disease-free surviving patients after 10 years was lowest in patients with single-copy PER3 deletion (56% ± 8.6; red line) compared with those having two (75% ± 4.0; blue line) or more (89% ± 5.6; gray line) copies of the PER3 gene (Fig 1A). Further analysis showed that the effect of PER3 deletion was most pronounced in the tamoxifen-treated group, with no significant association in the nontreated or chemotherapy-treated groups (Figs 1B through 1D). Among the 59 patients who received only tamoxifen treatment (Fig 1D), patients with single-copy PER3 deletions had a significantly lower DFS rate at 10 years (47% ± 12) than those with normal PER3 (84% ± 6) or copy number gains (100% DFS; P = .007). Follow-up for all patients was 82 months (range, 1.5 to 219 months). To look for potential inactivating mutations in PER3 in breast cancers, we initially sequenced the complete coding region of PER3 in a panel of 35 breast cancer cell lines. No clear pathogenic (nonsense or missense) mutation was identified. However, many known30 and some unknown polymorphisms and alternative splicing isoforms were found (see Data Supplement for full detailed description). One of the polymorphic variants identified by sequencing had been associated with breast cancer susceptibility in other studies19 and also with disruption of sleep homeostasis.31–33
Fig 1.
Association between PER3 deletion and disease-free survival in breast cancer patients. (A) TaqMan copy number analysis of PER3 in all patients, (B) in patients who received no treatment or (C) were treated with anthracycline chemotherapy, and (D) in a subset of 59 patients who were estrogen receptor–positive and/or progesterone receptor–positive and were treated only with tamoxifen.
Low Expression of PER3 Is Associated With Reduced Survival
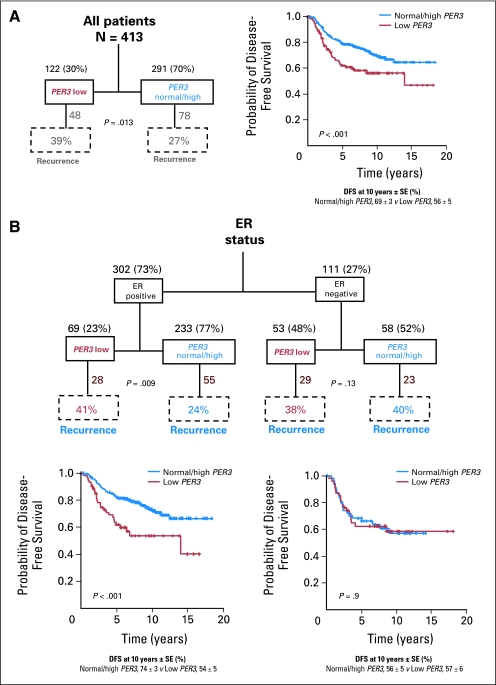
We next examined PER3 gene expression in 413 breast tumor expression arrays taken from two publicly available data sets (van de Vijver et al23 [n = 295] and Chin et al24 [n = 118]). A full description of the stratification of the patients into different subgroups according to PER3 expression together with DFS curves for all patients in each subgroup is shown in Figures 2 and 3. Patients with lower PER3 expression (“PER3 low” [n = 122]) were significantly more likely to recur than those with normal or higher expression (“PER3 normal/high” [n = 291]; P = .013; Fig 2A). DFS analysis showed that PER3 low patients had significantly worse survival rates than PER3 normal/high patients (P < .001). ER status is an important predictor of recurrence and greatly influences treatment regimens.34,35 If low expression of PER3 segregates with ER status, any effect of low PER3 expression could be confounded with the effect of ER status. We therefore performed a subset analysis of PER3 in ER-positive and ER-negative tumors. Low PER3 levels were significantly associated with recurrence (P = .01) and shorter DFS times (P < .001) in patients with ER-positive, but not ER-negative tumors (Fig 2B). We conclude that the association between low PER3 expression and recurrence in the complete patient sample set was driven by the ER-positive tumors, with no effect being detected in the ER-negative tumors. These data are in agreement with the independent association between deletion of PER3 and recurrence specifically in the tamoxifen-treated (ER-positive tumors) patients in Figure 1D.
Fig 2.
Association between PER3 gene expression and survival of breast cancer patients. (A) PER3 low expression (red) versus PER3 normal/high expression (blue) in all patients. (B) Comparison of PER3 expression with estrogen receptor (ER) status. DFS, disease-free survival; SE, standard error.
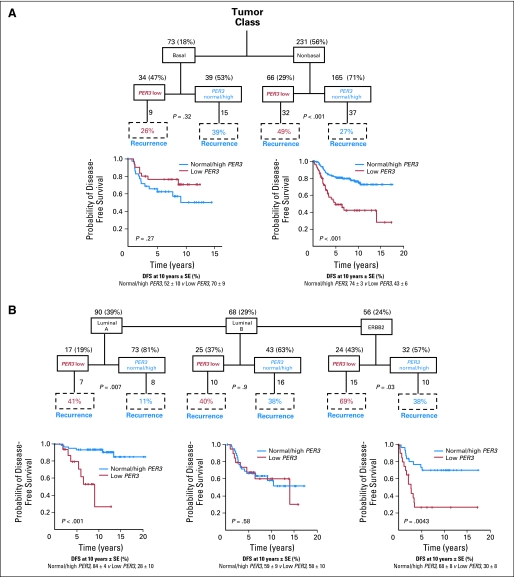
Fig 3.
Effect of PER3 expression levels on survival according to molecular subtypes. Kaplan-Meier estimates of disease-free survival (DFS) among the 413 patients, according to the PER3 expression. (A) Basal versus non-basal tumors; (B) luminal A versus luminal B and ERBB2-positive subgroups of tumors. SE, standard error.
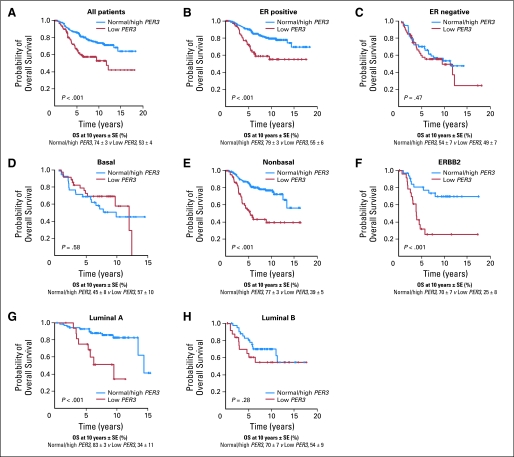
We next asked whether stratifying tumors according to their molecular subtype36,37 could reveal additional information. The tumors were labeled using a nearest centroid classifier, and a label was assigned only if correlation with a target class was above 0.1.31,32 This resulted in samples labeled luminal A (n = 90), luminal B (n = 68), ERBB2 (n = 56), normal-like (n = 17), basal (n = 73), or unclassified (n = 109; Fig 3 and Fig DS4). Of these groups, low PER3 expression had significant association with recurrence only in luminal A–type (P = .007) or ERBB2-type tumors (P = .03; Fig 3B). None of the patients with ERBB2-positive tumors received anti–ERBB2-targeted therapy. DFS analysis for luminal A–type and ERBB2-type tumors indicated that PER3 low patients had lower DFS rates at 10 years than patients with PER3 normal/high (luminal A: 28% ± 10 v 84% ± 4; P < .001 and ERBB2: 30% ± 8 v 68% ± 8; P = .004). There was also a striking effect on OS rate at 10 years in all the patients and in the subgroups of ER-positive, luminal A, and ERBB2 patients (Fig 4): The 10-year OS rate for patients with ER-positive tumors and with low PER3 was 55% ± 6 versus 79% ± 3 for normal/high patients (P < .001; Fig 4B). The OS rate was 25% ± 8 for patients with ERBB2 and low PER3 versus 70% ± 7 for patients with ERBB2 and normal/high PER3 (P < .001; Fig 4F). The OS rate at 10 years in luminal A patients with low PER3 was 34% ± 11 versus 83% ± 3 for patients with normal/high PER3 (P < .001; Fig 4G). Importantly, multivariate analysis showed that PER3 expression is independently significant from all the prognostic factors tested both for DFS (P < .001) and OS (P = .001; Table 1).
Fig 4.
Kaplan-Meier estimates of overall survival (OS). The different expression levels of PER3 were evaluated in all the patients (A) and the different subgroups of patients based on (B) ER-positive, (C) ER-negative, (D) basal, (E) nonbasal, (F) ERBB2-positive, (G) luminal A, and (H) luminal B tumors. SE, standard error.
Table 1.
Cox Proportional Hazard Ratio Multivariate Analysis
| Variable | Disease-Free Survival |
Overall Survival |
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | |
| All patients | ||||||
| PER3 | 2.13 | 1.40 to 3.24 | < .001 | 2.04 | 1.34 to 3.10 | .001 |
| Tumor size | 1.72 | 1.13 to 2.63 | .012 | 2.02 | 1.31 to 3.12 | .002 |
| Age < 40 years | 0.49 | 0.32 to 0.74 | .001 | 0.54 | 0.35 to 0.83 | .005 |
| ER | 0.75 | 0.49 to 1.15 | .19 | 0.53 | 0.35 to 0.80 | .003 |
| Lymph node | 1.36 | 0.90 to 2.06 | .14 | 1.85 | 1.18 to 2.77 | .007 |
| Tumor grade | ||||||
| Good | 0.93 | 0.55 to 1.60 | .8 | 1.05 | 0.61 to 1.80 | .87 |
| Intermediate | 1.18 | 0.74 to 1.89 | .48 | 1.38 | 0.87 to 2.20 | .17 |
| ER-positive patients | ||||||
| PER3 | 2.92 | 1.71 to 4.97 | < .001 | 2.63 | 1.49 to 4.63 | .001 |
| Tumor size | 1.62 | 0.96 to 2.63 | .072 | 1.87 | 1.05 to 3.32 | .03 |
| Age < 40 years | 0.58 | 0.33 to 0.99 | .047 | 0.57 | 0.32 to 1.04 | .06 |
| ER* | ||||||
| Lymph node | 1.40 | 0.83 to 2.39 | .21 | 2.07 | 1.18 to 2.77 | .02 |
| Tumor grade | ||||||
| Good | 1.14 | 0.59 to 2.23 | .69 | 1.09 | 0.54 to 2.24 | .8 |
| Intermediate | 1.34 | 0.73 to 2.46 | .34 | 1.32 | 0.70 to 2.49 | .38 |
NOTE. Risk of distant recurrence or death among patients with breast cancer.
Abbreviation: ER, estrogen receptor.
All tumors are ER positive.
The possible links between expression levels and probability of tumor recurrence were evaluated for all 54 annotated genes in the 1p36.31 to 1p36.22 region (chromosome 1: 6,084,440 to 9,512,808 [3.5 Mb]). Gene expression was discretized as described for PER3, and log-rank P values were calculated using the survival library for R. This analysis showed that PER3 was the only gene with an uncorrected P < .05 in all data sets analyzed. Although chromosome engineering studies have previously identified CHD5 as a candidate tumor suppressor gene within the minimal deletion region on 1p36.25, no association of CHD5 expression levels with recurrence or survival was found in any of the subgroups of breast cancer patients analyzed (Figs DS5 and DS6). These data do not exclude the possibility that CHD5 plays an important role as a tumor suppressor in other tumor types.
Inactivation of Per3 Increases Breast Tumor Susceptibility in Mouse Models
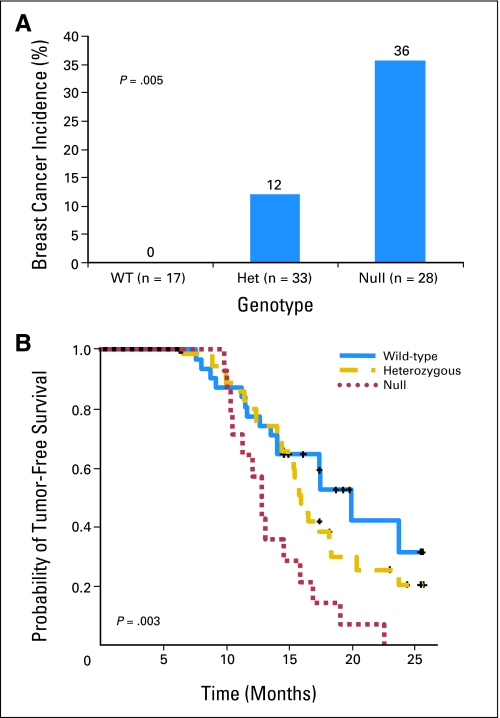
To investigate a possible causal association between loss of Per3 function and breast tumor development, we performed two studies involving mouse models of breast cancer (Fig 5 and Table 2). A total of 86 mice carrying normal or inactivated alleles of the Per3 gene (17 wild-type [Per3+/+], 35 heterozygous [Per3+/−], and 34 null [Per3−/−]) were treated by oral gavage with DMBA, a protocol known to induce breast cancer in sensitive strains of mice.38 Eight mice (two heterozygous and six null) were found dead before the end point, and no tissues were collected from them. The median follow-up of the remaining 78 mice included in the study was 8.3 months (range, 3.8 to 15.0 months). All of the mice treated with DMBA developed tumors of various kinds, including lymphoma and solid tumors of the lung, ovary, and skin (Table DS5). However, development of breast tumors was specifically associated with Per3 deficiency. Thirty-six percent of Per3−/− mice treated with DMBA developed breast tumors, while 12% of the Per3+/− mice developed breast tumors. In striking contrast, none of the control Per3+/+ mice developed a breast tumor (P = .005; Fig 3A). A group of 65 mice (19 wild-type, 25 heterozygous, and 21 null) were used as controls with no DMBA gavage treatment. Two of the Per3−/− control mice developed sporadic breast tumors, but none of the remaining mice were found sick or developed any other class of tumor during the time course of this experiment (24 months).
Fig 5.
Effect of loss of Per3 on tumor susceptibility in two different mouse models. (A) Breast cancer incidence in a group of mice treated with 7,12-dimethylbenz[a]anthracene on the basis of the different genotypes (wild type+/+[WT], heterozygous+/−[Het], and null −/−). (B) Kaplan-Meier estimates of probability of tumor-free survival in the group of mouse mammary tumor virus neu-Per3 mice.
Table 2.
Tumor-Free Rate for MMTV-Neu–Positive Mice
| No. of Mice | Median Follow–Up (months) | Range(months) | Tumor-Free Rate at 15 Months ± SE (%) |
|---|---|---|---|
| 30 | 16 | 7.5-26.4 | 63 ± 6 |
| 35 | 16 | 6.3-26.5 | 63 ± 6 |
| 14 | 13 | 9.8-22.5 | 21 ± 8 |
Abbreviations: MMTV, mouse mammary tumor virus; SE, standard error.
The second mouse model was based on the observation that low levels of PER3 expression were strongly associated with recurrence in ERBB2-type human breast cancers. MMTV-Neu mice overexpress ErbB2 in the mammary gland and spontaneously develop breast tumors.26 We generated a total of 79 MMTV-Neu–positive mice, of which 30 (38%) were Per3+/+, 35 (44%) were Per3+/−, and 14 (18%) were Per3−/−. The median follow-up of all mice was 14.9 months (range, 6.3 to 25.8 months). All Per3−/− mice developed breast tumors, whereas 25 (71%) of the Per3+/− and 14 (47%) of the Per3+/+ mice developed breast tumors. The proportion of Per3−/− null mice free of tumors at 15 months (21% ± 8) was significantly lower than the proportion in the heterozygous and the wild-type mice (63% ± 6 in both Per3+/− and Per3+/+; P = .003). Histologic analysis of tumors from both models of breast cancer showed that loss of Per3 did not affect the tumor class or morphology, since both DMBA-induced and MMTV-Neu–induced tumors in Per3−/− mice resembled equivalent tumors from Per3 wild-type animals (data not shown). We also evaluated the possible loss of the wild-type Per3 allele in tumors from the Per3 heterozygous mice. No loss was observed, suggesting that homozygous loss is not essential in this mouse model.
DISCUSSION
Our data indicate that deletion and/or reduced expression of the PER3 gene on human chromosome 1p36 is associated with breast cancer recurrence, particularly in patients with ER-positive tumors treated with tamoxifen who did not receive chemotherapy. No effect of deletion was seen in patients with basal type ER-negative breast tumors. Within the ER-positive category, the effect was primarily in tumors classified as luminal A–type or ERBB2-type, but not in the luminal B–type, which shares some expression features with basal tumors.36,37 Direct evidence for a causal role for loss of PER3, rather than an alternative gene in this commonly deleted region of the genome,5,6 comes from analysis of two different mouse models of breast cancer. Both chemically induced and Neu (ErbB2) –induced breast cancers are increased in frequency and/or reduced in latency in mice carrying inactivated Per3 alleles. Although these data do not prove that Per3 is the only functional tumor suppressor gene in this chromosome interval, they indicate that Per3 is a bona fide tumor suppressor in these mouse models, with a key role in breast tissue.
While disruption of the mouse period gene family members Per1 and Per2 by gene targeting induces biologic clock phenotypes,39 loss of Per3 function induces only subtle effects on circadian rhythm.13,40 Nevertheless, evidence in favor of PER3 involvement in both sleep disruption and breast cancer comes from studies of a human structural polymorphism in the PER3 coding sequence that has been associated with delayed sleep phase syndrome, diurnal preference, and waking performance,31,41,42 but also with increased breast cancer risk,19 particularly in premenopausal women.
Although the specific molecular mechanisms remain to be elucidated, increasing evidence points to a role for circadian rhythm genes in cell cycle control and DNA damage responses11,43 as well as in hormonal control of gene expression.17,18 PER2 has been identified as an estrogen-inducible ER corepressor that forms heterodimers with PER3 to enter the nucleus. Deletion of PER3 prevents nuclear import and, instead, promotes accumulation of PER2 in the cytoplasm.44 Whether coordinated functional deregulation of all period family genes occurs in breast cancers remains to be determined.
There are several clinical implications of these observations. First, the presence of PER3 deletions in ER-positive tumors may identify patients who do not respond to tamoxifen-based hormone therapy and who may benefit from other therapeutic regimens. Second, previous data from clinical trials of chronotherapy suggest that the timing of cancer treatment during the day may affect individual patient responses.45,46 Elucidation of the relationship between control of sleep homeostasis and circadian rhythms, PER gene expression, and DNA damage responses may help in understanding the epidemiologic data linking sleep disruption to breast cancer susceptibility,17,20,21 but further detailed studies will be required to elucidate the exact mechanisms involved. Finally, small-molecule drugs that help to restore the balance of the biologic clock in individuals with frequent sleep disruption may have potential as chemopreventive agents for breast and some other cancer types.
Supplementary Material
Acknowledgment
We thank Y.H. Fu and L.J. Ptáček for providing Per3 knockout mice, Z. Werb for providing FVB-MMTV-neu mice, and R.D. Cardiff for the histologic examination of mouse tumors. We also thank M.D. To and R.D. Cardiff for helpful discussion of the manuscript.
Footnotes
Supported by Grants No. U01 CA84244 from the National Cancer Institute (A.B.), EX-2005-1059 from the Spanish Ministry of Education and Culture (J.C.), BC063443 from the Department of Defense (J.C.), PI070057 from Federación Española de Enfermedades Raras and Fondo de Investigaciones Sanitarias, PLE2009-O119 from Ministerio de Ciencia e Innóvación, SA078A09 and SAN126/SA66/09 from Junta de Castilla y Léon, 200920I137 from Consejo Superior de Investigaciones Cientificas, I5FB-0099 from the California Breast Cancer Research Program (K.-Y.J.), and by the Sandra Ibarra Foundation (J.P.-L.) and the Barbara Bass Bakar Chair of Cancer Genetics (A.B.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Joan Climent, Jesus Perez-Losada, Jian-Hua Mao, Allan Balmain
Financial support: Joan Climent, Allan Balmain
Provision of study materials or patients: Reyno Delrosario, Ana Lluch
Collection and assembly of data: Joan Climent, Jesus Perez-Losada, Il-Jin Kim, Reyno Delrosario, Ana Bosch, Ana Lluch, Allan Balmain
Data analysis and interpretation: Joan Climent, David A. Quigley, Il-Jin Kim, Kuang-Yu Jen, Jian-Hua Mao, Allan Balmain
Manuscript writing: Joan Climent, David A. Quigley, Allan Balmain
Final approval of manuscript: Joan Climent, Jesus Perez-Losada, David A. Quigley, Il-Jin Kim, Reyno Delrosario, Kuang-Yu Jen, Ana Bosch, Ana Lluch, Jian-Hua Mao, Allan Balmain
REFERENCES
- 1.Tsukamoto K, Ito N, Yoshimoto M, et al. Allelic loss on chromosome 1p is associated with progression and lymph node metastasis of primary breast carcinoma. Cancer. 1998;82:317–322. [PubMed] [Google Scholar]
- 2.Ragnarsson G, Eiriksdottir G, Johannsdottir JT, et al. Loss of heterozygosity at chromosome 1p in different solid human tumours: Association with survival. Br J Cancer. 1999;79:1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han W, Han MR, Kang JJ, et al. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer. 2006;6:92. doi: 10.1186/1471-2407-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borg A, Zhang QX, Olsson H, et al. Chromosome 1 alterations in breast cancer: Allelic loss on 1p and 1q is related to lymphogenic metastases and poor prognosis. Genes Chromosomes Cancer. 1992;5:311–320. doi: 10.1002/gcc.2870050406. [DOI] [PubMed] [Google Scholar]
- 5.Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 6.Schlisio S, Kenchappa RS, Vredeveld LC, et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–893. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takumi T, Taguchi K, Miyake S, et al. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tei H, Okamura H, Shigeyoshi Y, et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Toh KL, Jones CR, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ST, Choo KB, Hou MF, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 11.Hunt T, Sassone-Corsi P. Riding tandem: Circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Gery S, Komatsu N, Baldjyan L, et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 13.Shearman LP, Jin X, Lee C, et al. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T, Nakahata Y, Soma H, et al. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archer SN, Viola AU, Kyriakopoulou V, et al. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008;31:608–617. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hida A, Kusanagi H, Satoh K, et al. Expression profiles of PERIOD1, 2, and 3 in peripheral blood mononuclear cells from older subjects. Life Sci. 2009;84:33–37. doi: 10.1016/j.lfs.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Gery S, Koeffler HP. The role of circadian regulation in cancer. Cold Spring Harb Symp Quant Biol. 2007;72:459–464. doi: 10.1101/sqb.2007.72.004. [DOI] [PubMed] [Google Scholar]
- 18.Gery S, Virk RK, Chumakov K, et al. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y, Brown HN, Zhang Y, et al. Period3 structural variation: A circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–270. [PubMed] [Google Scholar]
- 20.Megdal SP, Kroenke CH, Laden F, et al. Night work and breast cancer risk: A systematic review and meta-analysis. Eur J Cancer. 2005;41:2023–2032. doi: 10.1016/j.ejca.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Schernhammer ES, Laden F, Speizer FE, et al. Rotating night shifts and risk of breast cancer in women participating in the Nurses' Health Study. J Natl Cancer Inst. 2001;93:1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 22.Climent J, Dimitrow P, Fridlyand J, et al. Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res. 2007;67:818–826. doi: 10.1158/0008-5472.CAN-06-3307. [DOI] [PubMed] [Google Scholar]
- 23.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 24.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Muller WJ, Sinn E, Pattengale PK, et al. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 27.Rubio-Moscardo F, Climent J, Siebert R, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 28.Bièche I, Champème MH, Matifas F, et al. Two distinct regions involved in 1p deletion in human primary breast cancer. Cancer Res. 1993;53:1990–1994. [PubMed] [Google Scholar]
- 29.Matsuzaki M, Nagase S, Abe T, et al. Detailed deletion mapping on chromosome 1p32–p36 in human colorectal cancer: Identification of three distinct regions of common allelic loss. Int J Oncol. 1998;13:1229–1233. doi: 10.3892/ijo.13.6.1229. [DOI] [PubMed] [Google Scholar]
- 30.Ebisawa T, Uchiyama M, Kajimura N, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Archer SN, Robilliard DL, Skene DJ, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 32.Arendt J. Managing jet lag: Some of the problems and possible new solutions. Sleep Med Rev. 2009;13:249–256. doi: 10.1016/j.smrv.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 33.von Schantz M. Phenotypic effects of genetic variability in human clock genes on circadian and sleep parameters. J Genet. 2008;87:513–519. doi: 10.1007/s12041-008-0074-7. [DOI] [PubMed] [Google Scholar]
- 34.Khan SA, Rogers MA, Obando JA, et al. Estrogen receptor expression of benign breast epithelium and its association with breast cancer. Cancer Res. 1994;54:993–997. [PubMed] [Google Scholar]
- 35.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 36.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medina D, Butel JS, Socher SH, et al. Mammary tumorigenesis in 7,12-dimethybenzanthracene-treated C57BL x DBA/2f F1 mice. Cancer Res. 1980;40:368–373. [PubMed] [Google Scholar]
- 39.Liu AC, Welsh DK, Ko CH, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shearman LP, Zylka MJ, Weaver DR, et al. Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 41.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–618. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 42.Groeger JA, Viola AU, Lo JC, et al. Early morning executive functioning during sleep deprivation is compromised by a PERIOD3 polymorphism. Sleep. 2008;31:1159–1167. [PMC free article] [PubMed] [Google Scholar]
- 43.Collis SJ, Boulton SJ. Emerging links between the biological clock and the DNA damage response. Chromosoma. 2007;116:331–339. doi: 10.1007/s00412-007-0108-6. [DOI] [PubMed] [Google Scholar]
- 44.Yagita K, Yamaguchi S, Tamanini F, et al. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 45.Eriguchi M, Levi F, Hisa T, et al. Chronotherapy for cancer. Biomed Pharmacother. 2003;57(suppl 1):92s–95s. doi: 10.1016/j.biopha.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Lévi F. Chronotherapeutics: The relevance of timing in cancer therapy. Cancer Causes Control. 2006;17:611–621. doi: 10.1007/s10552-005-9004-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.