Abstract
Purpose
To define a gene expression profile of BRCAness that correlates with chemotherapy response and outcome in epithelial ovarian cancer (EOC).
Methods
A publicly available microarray data set including 61 patients with EOC with either sporadic disease or BRCA½ germline mutations was used for development of the BRCAness profile. Correlation with platinum responsiveness was assessed in platinum-sensitive and platinum-resistant tumor biopsy specimens from six patients with BRCA germline mutations. Association with poly-ADP ribose polymerase (PARP) inhibitor responsiveness and with radiation-induced RAD51 foci formation (a surrogate of homologous recombination) was assessed in Capan-1 cell line clones. The BRCAness profile was validated in 70 patients enriched for sporadic disease to assess its association with outcome.
Results
The BRCAness profile accurately predicted platinum responsiveness and mutation status in eight of 10 patient-derived tumor specimens and between PARP-inhibitor sensitivity and resistance in four of four Capan-1 clones. When applied to the 70 patients with sporadic disease, patients with the BRCA-like (BL) profile had improved disease-free survival (34 months v 15 months; log-rank P = .013) and overall survival (72 months v 41 months; log-rank P = .006) compared with patients with a non–BRCA-like (NBL) profile, respectively. The BRCAness profile maintained independent prognostic value in multivariate analysis, which controlled for other known clinical prognostic factors.
Conclusion
The BRCAness profile correlates with responsiveness to platinum and PARP inhibitors and identifies a subset of sporadic patients with improved outcome. Additional evaluation of this profile as a predictive tool in patients with sporadic EOC is warranted.
INTRODUCTION
Both BRCA1 and BRCA2 proteins are involved in the process of homologous recombination (HR), which mediates repair of double-stranded DNA breaks.1 Patients with ovarian cancer with germline mutations in either BRCA1 or BRCA2 genes exhibit impaired ability to repair double-stranded DNA breaks via HR, which may partly explain the heightened sensitivity to platinum and the more favorable survival compared with wild-type counterparts.2–4 Furthermore, in the setting of defective HR, it has been shown that inhibition of a second DNA repair pathway, such as base excision repair (BER), is often a lethal event.5–7 On the basis of this observation, there has been great interest in developing inhibitors of the BER pathway for use as possible therapeutic agents in patients with ovarian cancer with germline BRCA½ mutations.8,9 Drugs that target BER typically inhibit poly-ADP ribose polymerase (PARP), an enzyme critical to BER, and have already shown promising activity in patients with recurrent ovarian cancer who harbor germline BRCA½ mutations.10,11
The promise of PARP inhibitors in the management of epithelial ovarian cancer (EOC) is tempered by the presence of a germline mutation in BRCA1 or BRCA2 in only approximately 10% of such patients.12–14 At first glance, this might imply that 90% of patients with this highly lethal disease would not benefit from this novel class of drugs. However, it has been hypothesized that a subset of sporadic EOCs may harbor abnormalities in the HR pathway that could be associated with improved response rate and survival after treatment with platinum compounds in the absence of germline BRCA½ mutation.4,15 This BRCAness phenotype may be due in part to defective HR related to several mechanisms, including epigenetic hypermethylation of the BRCA1 promoter,16–19 somatic mutation of BRCA½,18,20–22 or loss of function mutations in other HR pathway genes.23,24 At present, however, it has not been possible to reliably identify such patients on the basis of molecular- or protein-based biomarkers, and the concept of BRCAness for patients with the sporadic form of the disease has remained elusive.
Given the heterogeneous mechanisms by which an ovarian cancer cell might develop defective HR, we reasoned that a broad-based approach that makes few assumptions about mechanism might have the highest chance of identifying patients with a BRCAness phenotype. Microarray gene expression profiling lends itself to this goal, because it is not mechanism based and has already been demonstrated to have prognostic as well as predictive potential in EOC.25–27 In this study, we show that it is possible to define a gene expression profile of BRCAness, associated with responsiveness to platinum and PARP inhibitors, and we correlate this profile with important outcome measures in patients with the sporadic form of EOC.
METHODS
Development of a Gene Expression Profile of BRCAness
For the purpose of profile development, we used a publicly available microarray data set that included tumor expression data from 61 patients with pathologically confirmed EOC, including 34 with BRCA½ germline mutations (n= 18, BRCA1; n = 16, BRCA2), and 27 without either mutation (ie, sporadic cancers).28 We used genome-wide hierarchical clustering to define BRCA-like (BL) and non–BRCA-like (NBL) tumors (Appendix Fig A1, online only; Data Supplement).
Patient Samples
Two patient cohorts were used in this study. The first included six patients with EOC with BRCA½ germline mutations; both have been previously described.29,30 Four patients had paired samples, before and after the development of platinum resistance, and two had samples obtained only at the time of platinum-sensitive disease.
The second patient cohort included 70 patients who were treated at Beth Israel Deaconess Medical Center, Memorial Sloan-Kettering Medical Center, and Cedars-Sinai Medical Center and who underwent exploratory laparotomy for diagnosis, staging, and debulking followed by first-line platinum-based chemotherapy. Standard post-chemotherapy surveillance included serial physical examination, serum CA-125 level, and computed tomography scanning as clinically indicated.
The study protocol for collection of tissue and clinical information for all patients was approved by the institutional review boards at all three institutions, and patients provided written informed consent authorizing the collection and use of the tissue for study purposes. Additional details are provided in the Data Supplement.
Cell Lines and RNA Isolation and Affymetrix GeneChip Hybridization
Twelve cisplatin-resistant clones of the BRCA2-mutated pancreatic cancer cell line Capan-1 have been previously described.29 Total RNA isolation, microarray hybridization (U133 Plus 2.0 Array GeneChip; Affymetrix, Santa Clara, CA), and data processing were performed as previously described.26,27,31
Statistical Analysis
The association between the BRCAness profile and various clinicopathologic factors was assessed by the Fisher's exact test. Overall survival (OS) and disease-free survival (DFS) curves were generated by the Kaplan-Meier method, and differences between survival curves were assessed for statistical significance with the log-rank test. Multivariate analyses to adjust for known prognostic factors were performed by using a Cox proportional hazards regression model that included grade (1 to 2 v 3), age (< 65 years v ≥ 65 years), stage (2 or v 3 or 4), histology (clear-cell, papillary serous, endometrioid), debulking status (optimal [less than or equal to 1 cm] or suboptimal [greater than 1 cm] residual disease), and BRCAness profile (BL v NBL).
RESULTS
Characteristics of the BRCAness Profile
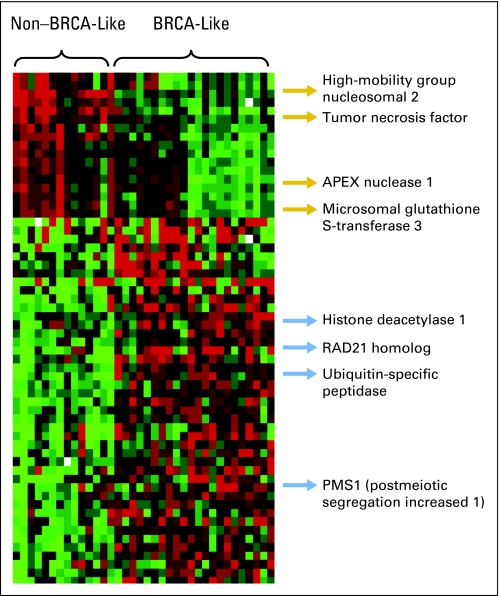
The strategy for developing the BRCAness profile is described in detail in Appendix Figure A1. The optimal classifier was a 60-gene, diagonal linear discriminant predictor that distinguished BL from NBL tumors with 94% accuracy, as assessed by leave-one-out cross-validation and a 1,000 random permutations test (Fig 1; P < .001). Other predictive algorithms performed similarly, such as compound covariate predictor (92%), nearest centroid (92%), and support vector machines (92%).32–35 For the analyses described in the Results section, the gene expression signature that correlates with BL tumors is defined as the BL profile, and the signature that correlates with NBL tumors is defined as the NBL profile. The identities of all BRCAness profile genes are provided in Appendix Table A1 (online only).
Fig 1.
Expression plot of the 60 genes that comprise the BRCAness profile. Columns represent set samples; rows, gene expression levels (normalized). Complete information regarding gene identity is provided in Appendix Table A1. Red indicates overexpressed genes; green, underexpressed genes. The gene expression signature that correlates with BRCA-like tumors is defined as the BL profile, and the signature that correlates with non–BRCA-like tumors is defined as the NBL profile.
BRCAness Profile Distinguishes Between Platinum-Sensitive and Platinum-Resistant Tumor Biopsy Samples
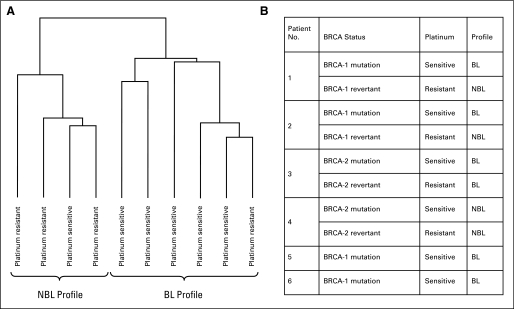
We first investigated whether the BRCAness profile could correlate with platinum responsiveness in patients with known BRCA germline mutation. For this purpose, we used 10 tumor biopsy specimens from six patients with either BRCA1 or BRCA2 germline mutation, four of whom were initially platinum sensitive but eventually developed platinum resistance (with pre- and post-biopsy pairs). These patients formed the basis of two previous reports, in which reversion of the BRCA genotype occurred (with re-establishment of BRCA function) on the development of platinum resistance.29,30 A separate report that used the CAPAN-1 cell line demonstrated similar findings.36 Thus, these samples afforded us with an opportunity to determine how the BRCAness profile correlated with both platinum responsiveness and BRCA functional status (eg, mutant v revertant BRCA gene).
As shown in Figure 2, the BRCAness profile could accurately distinguish between platinum sensitivity and platinum resistance in eight of 10 tumor specimens, which in turn correlated with the presence of mutated versus revertant (ie, functional) BRCA gene status, respectively. Specifically, five of six tumors with the BL signature were platinum sensitive (and were BRCA1 or BRCA2 mutated), whereas three of four tumors with the NBL signature were platinum resistant (and had reverted to functional BRCA1 or BRCA2).29,30 Furthermore, we observed two patients in whom the BRCAness profile dynamically tracked the development of platinum resistance during the course of therapy (ie, the profile changed from BL to NBL after the development of platinum resistance, associated with reversion to functional BRCA1; Fig 2B).
Fig 2.
BRCAness profile distinguishes between platinum-sensitive and platinum-resistant tumor biopsy specimens in patients with known BRCA germline mutation status. (A) Hierarchical clustering that is based on the expression pattern of the 60 genes of the BRCAness profile distinguished between platinum-resistant and platinum-sensitive tumor biopsy samples. Platinum sensitivity was defined as a complete response to treatment maintained without progression for at least 6 months after platinum therapy. Platinum resistance was defined as progressive disease on platinum therapy, or less than a complete response to platinum therapy, or progression within 6 months of completing platinum therapy. (B) Correlation of BRCAness profile with platinum sensitivity and BRCA germline mutation status. The BRCAness profile accurately distinguished between platinum sensitivity and platinum resistance in eight of 10 tumor specimens, which in turn correlated with the presence of mutated versus functional BRCA gene status, respectively. NBL, non–BRCA-like; BL, BRCA-like.
BRCAness Profile Correlates With PARP Inhibitor Responsiveness and RAD51 Foci Formation
As another surrogate of BRCAness, we next evaluated whether the profile could correlate with the ability to form RAD51 foci after ionizing radiation, which is a surrogate of intact HR, as well as with responsiveness to PARP inhibitors. For this purpose, we used 12 clones of the BRCA2-mutated pancreatic cancer cell line Capan-1, previously characterized (by T.T.).29 These clones were generated by exposing the parent Capan-1 cell line to platinum-selection pressure, eventually isolating 12 platinum-resistant clones. Seven of these clones formed intact RAD51 foci after ionizing radiation (six of these had reverted to functional BRCA2 because of secondary BRCA2 mutations, which canceled the effect of the inherited BRCA2 mutation), and the remaining five exhibited deficient RAD51 foci formation (all of which contained the inherited, nonfunctional BRCA2 mutation). PARP inhibitor sensitivity had been determined for four of these clones; two were PARP inhibitor sensitive, and two were PARP inhibitor resistant.29 When applied to these cell lines, the BRCAness profile correlated with RAD51 foci formation in nine of 12 Capan-1 clones and between presence of mutated versus revertant BRCA2 gene status in 10 of 12 Capan-1 clones (Table 1). Importantly, the BRCAness profile accurately distinguished between two PARP inhibitor–resistant clones (NBL signature) and two PARP inhibitor–sensitive clones (BL signature).
Table 1.
Association of BRCAness Profile With BRCA2 Mutation Status, RAD51 Foci Formation After Radiation and PARP Inhibitor Responsiveness
| Capan-1 Clone | BRCA2 Status | RAD51 Foci Formation* | PARP Inhibitor Responsiveness† | Profile |
|---|---|---|---|---|
| C2-1 | Revertant‡ | Yes | ND | NBL |
| C2-2 | Revertant | Yes | ND | NBL |
| C2-4 | Revertant | Yes | ND | NBL |
| C2-14 | Revertant | Yes | Resistant | NBL |
| C2-12 | Revertant | Yes | Resistant | NBL |
| C2-5 | Revertant | Yes | ND | BL |
| C2-16 | Mutated§ | Yes | ND | BL |
| C2-10 | Mutated | No | Sensitive | BL |
| C2-13 | Mutated | No | Sensitive | BL |
| C2-15 | Mutated | No | ND | BL |
| C2-18 | Mutated | No | ND | BL |
| C2-8 | Mutated | No | ND | NBL |
Abbreviations: PARP, poly-ADP ribose polymerase; ND, not determined; NBL, non–BRCA-like; BL, BRCA-like.
Ability to form RAD51 formation after ionizing radiation, as previously described.29
PARP inhibitor responsiveness in vitro, as previously described.29
Revertant refers to clones harboring secondary BRCA2 mutations that cancel the effect of the inherited 6174delT BRCA2 mutation and lead to functional BRCA2 isoforms.
Mutated refers to clones harboring only the original 6174delT BRCA2 mutation without acquiring a secondary mutation that restored BRCA2 function.
Relationship Between BRCAness Profile and Clinical Outcome in Patients With Sporadic EOC
These data suggest that the BRCAness profile may correlate with platinum and PARP-inhibitor responsiveness in the context of a known BRCA germline mutation, but they do not address whether the profile correlates with outcome in patients with sporadic disease. To test this, we applied the profile to tumor samples from 35 patients with invasive EOC who underwent sequencing for germline mutation (by using DNA obtained from peripheral-blood leukocytes) and did not harbor germline BRCA1 or BRCA2 mutations. We also studied an additional 35 patients who did not undergo genetic testing but who were enriched for sporadic disease on the basis of the following characteristics: no family history of ovarian cancer, no family history of breast cancer younger than 50 years of age, no family history of more than one breast cancer at any age, and not of Ashkenazi Jewish ethnicity.37,38 The clinical and pathologic characteristics of all 70 patients are listed in Table 2. Overall, 20 (29%) of the 70-patient cohort demonstrated the BL profile (eight of 35 in the sequenced group, and 12 of 35 in the nonsequenced group; P = .43). Compared with the nonsequenced cohort, the sequenced cohort was enriched for patients with optimally debulked disease, although this did not reach statistical significance (two-sided Fisher's exact P = .19). As listed in Table 3, there were no differences in age, stage, grade, histology, or debulking status between the BL and the NBL signature groups. The ability to achieve a clinical remission for the BL and NBL groups was 90% compared with 74%, although this did not reach statistical significance (two-sided Fisher's exact P = .2).
Table 2.
Clinical and Pathologic Characteristics
| Characteristic | Sequenced Cohort (n = 35)* |
Nonsequenced Cohort (n = 35)† |
Combined Cohort (n = 70)‡ |
|||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Age, years§ | ||||||
| Median | 62.3 | 58.7 | 60.5 | |||
| Range | 44-89 | 39-80 | 39-89 | |||
| Grade | ||||||
| 1-2 | 4 | 11.4 | 6 | 17.1 | 10 | 14.3 |
| 3 | 31 | 88.6 | 29 | 82.9 | 60 | 85.7 |
| Histology | ||||||
| Serous | 34 | 97.1 | 31 | 88.6 | 65 | 92.9 |
| Clear cell | 0 | 0 | 2 | 5.7 | 2 | 2.9 |
| Endometrioid | 1 | 2.9 | 2 | 5.7 | 3 | 4.3 |
| Stage | ||||||
| 2 | 2 | 5.7 | 1 | 2.9 | 3 | 4.3 |
| 3 | 28 | 80 | 29 | 82.9 | 57 | 81.4 |
| 4 | 5 | 14.3 | 5 | 14.3 | 10 | 14.3 |
| Debulking status‖ | ||||||
| Optimal | 27 | 79.4 | 22 | 62.9 | 49 | 71 |
| Suboptimal | 7 | 20.6 | 13 | 37.1 | 20 | 29 |
All 35 patients underwent germline DNA sequencing and had wild-type BRCA1 and BRCA2.
Not sequenced but enriched for sporadic disease on the basis of the following: no family history of ovarian cancer, no family history of breast cancer younger than 50 years of age, no family history of more than one breast cancer at any age, and not of Ashkenazi Jewish ethnicity.
All patients received first-line platinum-based chemotherapy.
There was no statistically significant difference in age, grade, histology, stage, or debulking status between sequenced and nonsequenced cohorts.
Debulking status was unknown for one patient. Optimal was defined as less than or equal to 1 cm; suboptimal, greater than 1 cm diameter residual disease.
Table 3.
Association of BRCAness Profile With Clinical Characteristics and Remission Status After First-Line Therapy
| Characteristic | NBL Profile (n = 50) |
BL Profile (n = 20) |
P | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Age, years | |||||
| Median | 61 | 59.25 | |||
| Range | 39-89 | 44-80 | .55 | ||
| Grade | |||||
| 1-2 | 9 | 18 | 1 | 5 | .477 |
| 3 | 41 | 82 | 19 | 95 | |
| Histology | |||||
| Serous | 46 | 92 | 19 | 95 | .412 |
| Clear cell | 1 | 2 | 1 | 5 | |
| Endometrioid | 3 | 6 | 0 | 0 | |
| Stage | |||||
| 2 | 3 | 6 | 0 | 0 | .479 |
| 3 | 41 | 82 | 16 | 80 | |
| 4 | 6 | 12 | 4 | 20 | |
| Debulking status* | |||||
| Optimal | 33 | 67.3 | 16 | 80 | .386 |
| Suboptimal | 16 | 32.7 | 4 | 20 | |
| Achievement of CR after first-line therapy | 37 | 74 | 18 | 90 | .2 |
Abbreviations: NBL, non–BRCA-like; BL, BRCA-like; CR, complete response.
Debulking status was unknown for one patient. Optimal was defined as less than or equal to 1 cm; suboptimal, greater than 1 cm.
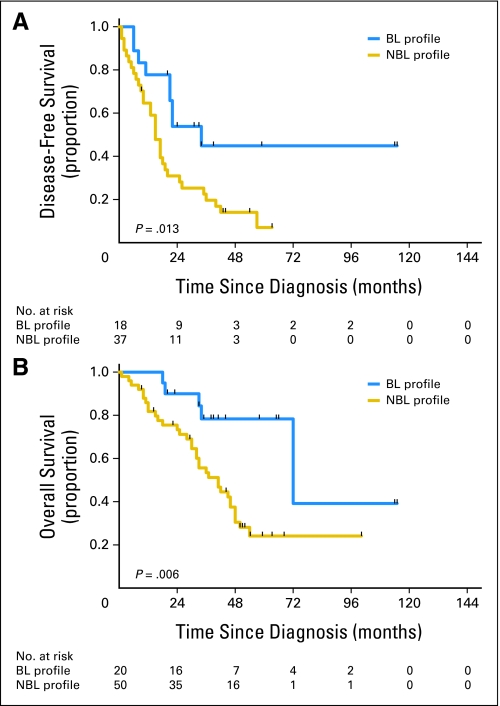
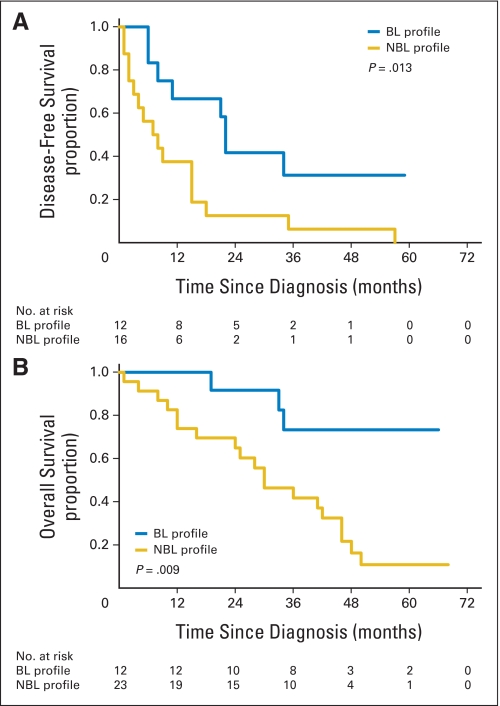
For the entire 70-patient cohort, the BRCAness profile was capable of discriminating between long and short median DFS; the patients with BL and NBL profiles had median DFS times of 34 months and 15 months, respectively (log-rank P = .013; Fig 3A). In addition, the percentages of patients who were disease free at 4 months for the BL and NBL groups were 90% and 64% (P = .04), respectively; at 6 months, percentages were 85% and 60%, respectively (P = .053); and at 18 months, percentages were 65% and 29%, respectively (P = .007). Finally, the BRCAness profile distinguished between long and short median OS, as the patients in the BL and NBL groups had median OS times of 72 and 41 months, respectively (log-rank P = .006; Fig 3B). Similar findings were observed when applying the profile separately to the group of 35 sequenced patients who had undergone germline mutation testing and who were found to have wild-type BRCA1 and BRCA2 genes or to the group of the 35 nonsequenced patients enriched for sporadic disease on the basis of clinical characteristics, as previously described (Appendix Figs A2 and A3, respectively, online only).
Fig 3.
Association of BRCAness profile with disease-free survival (DFS) and overall survival (OS) in the combined patient cohort (N = 70). (A) DFS in the combined patient cohort. The median DFS times for patients with either the BRCA-like (BL) or non–BRCA-like (NBL) profile were 34 months and 15 months, respectively (log-rank P = .013). (B) OS in the combined patient cohort. The median OS times for patients with either the BL or NBL profile were 72 months and 41 months, respectively (log-rank P = .006).
In univariate analysis, the hazard ratio for recurrence (NBL v BL group) was 2.47 (P = .018; 95% CI, 1.17 to 5.2), and the hazard ratio for death (NBL v BL group) was 3.29 (P = .009; 95% CI, 1.34 to 8.09; Table 4). Multivariate analysis, which included the BRCAness profile, age, stage, grade, histology, and debulking status, demonstrated that the profile maintained an independent association with DFS and OS. The hazard ratio for recurrence (NBL v BL group) was 2.65 (P = .016; 95% CI, 1.2 to 5.86), and the hazard ratio for death (NBL v BL group) was 3.39 (P = .009; 95% CI, 1.35 to 8.5; Table 4). The lack of correlation of characteristics such as stage, grade, and histology with outcome in either univariate or multivariate analysis is likely because the vast majority of patients in this analysis had stage III disease (81%), grade 3 tumors (86%), and serous histology (93%).
Table 4.
Predictive Value of BRCAness Profile Adjusted for Standard Clinical Prognostic Factors
| Factor | Univariate P |
Multivariate P |
||
|---|---|---|---|---|
| DFS | OS | DFS | OS | |
| Age, years | 0.44 | 0.35 | 0.96 | 0.8 |
| Grade | 0.9 | 0.34 | 0.87 | 0.32 |
| Histology | 0.97 | 0.61 | 0.9 | 0.96 |
| Stage | 0.43 | 0.54 | 0.4 | 0.61 |
| Debulking status | 0.39 | 0.05*† | 0.53 | 0.67 |
| HR‡ | 1.84† | |||
| NBL/BL profile | 0.018† | 0.009† | 0.016† | 0.009† |
| HR‡ | 2.47† | 3.29† | 2.65† | 3.39† |
Abbreviations: DFS, disease-free survival; OS, overall survival; HR, hazard ratio; NBL, non–BRCA-like; BL, BRCA-like.
Debulking status was unknown for one patient.
Statistically significant at P ≤ .05.
HR for death represented as comparison of NBL v BL groups for statistically significant associations.
DISCUSSION
PARP inhibitors have been evaluated in patients with germline BRCA½ mutations with impressive results as single agents.10,11 In addition to patients with germline BRCA½ mutations, however, it has been suggested that PARP inhibition might be a useful therapeutic strategy for the treatment of patients with sporadic cancers that have a BRCAness phenotype, characterized by defective HR.15 In this regard, a number of mechanisms have been identified in sporadic ovarian cancer that might implicate the HR pathway in pathogenesis and in chemotherapy responsiveness. Such mechanisms include somatic BRCA½ mutations in up to 20% of high-grade ovarian cancers20 as well as mutations or epigenetic silencing in Fanconi anemia genes, intrinsic HR genes, or other DNA damage-response genes.5,15,17,19,23 Amplification of genes that encode for proteins that inactivate BRCA2 function, such as EMSY, has also been described.24 BRCA1 promoter methylation, FANCF promoter methylation, and EMSY amplification have been identified in 5% to 31%, 21%, and 17% of sporadic EOCs respectively,15,17,19,23,24 which supports the notion that at least some patients with sporadic disease might harbor defects in HR, independent of the presence of a germline BRCA½ mutation.
Although it is possible to identify individual molecular mechanisms by which the HR pathway might be disrupted in some patients with sporadic EOC, only a few studies have explored the relationship between HR and response to platinum or PARP inhibitors in this setting. D'Andrea et al23 showed that inhibition of the FANCF gene in ovarian cancer cell lines through promoter methylation is associated with enhanced sensitivity to DNA damaging agents such as platinum, whereas demethylation of the FANCF promoter results in platinum resistance. Mccabe et al5 showed that cells deficient in the expression of genes involved in HR (eg, RAD51, ATR, ATM, CHK2) are sensitive to PARP inhibitors. Teodoridis et al16 used methylation-specific polymerase chain reaction and showed that BRCA1 promoter hypermethylation is associated with improved response to platinum-based chemotherapy. In addition, Quinn et al39 used siRNA knock-down to decrease the expression of the BRCA1 gene in two separate ovarian cancer cell lines, which showed that lower levels of BRCA1 mRNA correlated with enhanced in vitro sensitivity to cisplatin.
In this article, we have broadened the concept of BRCAness by identifying a gene expression profile that is associated with platinum and PARP-inhibitor responsiveness, as well as RAD51 foci formation. The relatively small number of BRCA½-mutated tumors and Capan-1 clones used in this study precludes formal statistical analysis. Nonetheless, it is noteworthy that the BRCAness profile was capable of tracking platinum response in eight of 10 tumor specimens and PARP-inhibitor response in four of four Capan-1 clones. Moreover, when applied to a population of patients enriched for sporadic disease, the profile correlated with clinical outcome, independent of standard prognostic factors such as age, grade, histology, stage, and debulking status. It is impossible to determine from our data whether the correlation between the BL signature and improved survival is indicative of enhanced platinum responsiveness or, conversely, might identify patients with a more indolent natural history. In this regard, it is intriguing that the proportion of patients rendered into a complete clinical remission at the end of first-line chemotherapy was higher in patients with a BL signature (90%) than in those with the NBL signature (74%), although this was not statistically significant (P = .2).
It is noteworthy that BRCAness profile contained genes, such as APEX1, MGST3, and PMS1, that have been previously associated with platinum resistance or DNA repair (Fig 1).40–45 Neither BRCA1 nor BRCA2 was part of our gene expression profile, which perhaps indirectly supported the notion that, at least for some patients, genes other than BRCA1 or BRCA2 may sometimes be responsible for BRCAness in sporadic disease. However, it is possible that our profile is identifying a subset of patients with sporadic mutation in BRCA1 or BRCA2, epigenetic silencing of the promoter for BRCA1, or as yet unknown defects in the HR (or related) pathway. Our future studies will be directed at better understanding the mechanisms underlying the association between the BRCAness profile, chemotherapy response, and survival.
Although the BRCAness profile was developed in ovarian tumors, it was also capable of predicting PARP-inhibitor sensitivity and RAD51 foci formation in the pancreatic cancer cell line Capan-1 (Table 1), which suggests that the profile may be detecting a pattern of gene expression that more globally reflects the status of HR, independent of cell lineage. Furthermore, we are currently investigating the predictive value of this profile in triple-negative breast cancer, which is thought to be enriched for BRCAness and to have a high response to platinum-containing chemotherapy.46 Ultimately, we intend to apply this profile in the context of a clinical trial involving patients with sporadic ovarian cancer treated with a PARP inhibitor to gain additional insight into the predictive value of this approach. Studies are currently being performed to explore the potential value of PARP inhibitors in patients with ovarian cancer independent of BRCA mutation status. In the future, it may be possible to use gene expression profiling as an eligibility criterion in such studies, to enrich for sporadic patients who may benefit the most from this novel class of agents. Although additional study is clearly needed, the identification of a gene expression profile that seems to correlate with BRCAness may make it possible to eventually offer PARP inhibitors to a much larger number of patients with epithelial ovarian cancer, regardless of their BRCA1 or BRCA2 mutation status.
Supplementary Material
Acknowledgment
We thank Fangbing Liu, PhD, for his technical assistance with growing the Capan-1 clones, and we thank the gynecologic oncologists at Beth Israel Deaconess Medical Center, Memorial Sloan-Kettering Cancer Center, and Cedars-Sinai Medical Center for providing tissue samples used in this analysis.
Appendix
Methods
Development of a gene expression profile of BRCAness.
For the purpose of profile development, we used a publicly available microarray data set that included tumor expression data from 61 patients with pathologically confirmed epithelial ovarian cancer (EOC), including 34 with BRCA1 or BRCA2 germline mutation (n = 18, BRCA1; n = 16 BRCA2) and 27 without either mutation (ie, sporadic cancers).28 We defined BRCA-like (BL) and non–BRCA-like (NBL) tumors by using genome wide hierarchical clustering (Appendix Fig A1, online only).
The predictor that distinguished BL from NBL tumors was developed by using the diagonal linear discriminant algorithm (Dudoit S, Fridlyand J, Speed TP: J Am Stat Assoc 97:77-87, 2002). The classifier was trained by selecting genes with the highest fold-change difference between the two classes (ie, BL and NBL tumors). Classifier accuracy and statistical significance were assessed by using leave-one-out cross-validation and a 1,000 random permutation test to control for overfitting (Simon R, Radmacher MD, Dobbin K, et al: J Natl Cancer Inst 95:14-18, 2003; Molinaro AM, Simon R, Pfeiffer RM: Bioinformatics 21:3301-3307, 2005). To ascertain that classifier accuracy was not an artifact of the optimal 60-gene predictor, we assessed the performance of predictors from a range of 40 to 90 genes and found that they demonstrated good performance, with accuracy of 89% to 92%.
For sporadic samples, the prediction rule was defined by the inner sum of the weights (wi; weights listed in Appendix Table A1, online only) and the log-intensity expression (xi) of each gene. A sample was classified as BL if the sum was greater than 18.973 (ie, if ∑iwi xi > 18.973). Otherwise it was classified as NBL. The BRCAness profile was mapped across different platforms by using Affymetrix (Santa Clara, CA) annotation files before it was applied to patient and cell line samples (http://www.affymetrix.com/analysis/index.affx). Nonbiologic experimental variation (ie, batch effect) across platforms was adjusted by using empirical Bayesian methods, as previously described (Johnson WE, Li C, Rabinovic A: Biostatistics 8:118-127, 2007).
Patient samples.
Two patient cohorts were used in this study. The first was comprised of six patients with EOC with BRCA1 or BRCA2 germline mutations treated at Cedars-Sinai Medical Center, and both have been previously described.29,30 Four patients from this group had paired samples taken before and after the development of platinum resistance (n = 2 with germline BRCA1 mutation, n = 2 with germline BRCA2 mutation). In each case, the development of platinum resistance was associated with reversion to functional BRCA1 and BRCA2 protein.23,30 Tumor from the two other patients was obtained at the time of platinum-sensitive disease, without a follow-up specimen at the time of platinum resistance. In both of these patients, the tumor specimen contained a germline mutation in BRCA1.
The second patient cohort consisted of 70 patients with EOC in whom tumor was obtained at the time of diagnostic exploratory laparotomy. Twenty-eight of these patients were diagnosed between November 1994 and June 2005, and were treated at Cedars-Sinai Medical Center; they had sporadic EOC, as determined by the lack of germline mutation in BRCA1 or BRCA2 (on the basis of DNA from peripheral-blood leukocytes). The remaining 42 patients were diagnosed between January 1995 and October 2000 and were treated at Beth Israel Deaconess Medical Center and Memorial Sloan-Kettering Cancer Center, and these patients represent a subset of those previously reported.27 Seven of the 42 patients were sequenced and found to be negative for a BRCA1 or BRCA2 mutation. The remaining 35 of these 42 patients were selected on the basis of criteria that were expected to enrich for sporadic disease. Specifically, these 35 patients had no family history of ovarian cancer, had no family history of breast cancer at age younger than 50 years, had no family history of more than one breast cancer at any age, and were not of Ashkenazi Jewish ethnicity. Ovarian cancer samples were collected at the time of primary debulking surgery and were frozen at −80°C. Tumor samples were pulverized in liquid nitrogen and were homogenized in Trizol solution (Invitrogen, Carlsbad, CA), followed by RNA isolation.
Cell lines.
Twelve cisplatin-resistant clones of the originally cisplatin-sensitive BRCA2-mutated pancreatic cancer cell line Capan-1 have been previously described.29 The parent Capan-1 line harbors a 6174delT BRCA2 mutation, which is associated with loss of heterozygosity (Goggins M, Schutte M, Lu J, et al: Cancer Res 56:5360-5364, 1996). As a result of platinum-induced selection pressure, six of these 12 clones had acquired secondary genetic events that restored nearly full-length, functional BRCA2 protein and RAD51 foci formation in response to ionizing radiation.29 The remaining six clones showed persistent evidence of mutated BRCA2 (6174delT), lacked BRCA2 protein expression, and exhibited impaired ionizing radiation–induced RAD51 foci formation (except for one that had proficient RAD51 foci formation). Two of the clones with restored functional BRCA2 were tested for poly-ADP ribose polymerase inhibitor response and were resistant, whereas two of the clones with persistently mutated BRCA2 were tested for poly-ADP ribose polymerase inhibitor response and were sensitive.29
RNA isolation and Affymetrix GeneChip hybridization.
Total RNA was isolated from patient tumor samples using Trizol reagent (Invitrogen) and from Capan-1 cell lines by using the RNeasy Mini Kit (Qiagen Valencia, CA) according to manufacturer's instructions. cDNA synthesis and hybridization on oligonucleotide microarrays (U133 Plus 2.0 Array GeneChip, Affymetrix) that contained approximately 54,700 transcripts were carried out according to standard protocols. Samples from Beth Israel Deaconess Medical Center and Memorial Sloan-Kettering Medical Center were run on U95A2 Affymetrix Arrays, as previously described.26,27 Microarray experiments were performed at the Dana-Farber Cancer Institute Microarray Core Facility (http://chip.dfci.harvard.edu/). Raw data were processed by using Robust Multi-Array analysis. All raw microarray data are provided in GEO (Gene Expression Omnibus).
Statistical analysis.
The statistical significance of the association between the BRCAness profile and various clinicopathologic factors was assessed by the Fisher's exact test. Unsupervised hierarchical clustering was performed by using the average linkage method and the one -minus centered correlation as a distance metric in all instances (Eisen MB, Spellman PT, Brown PO, et al: Proc Natl Acad Sci U S A 95:14863-14868, 1998). The P values of all statistical tests were two sided. SPSS, version 16.0 (SPSS, Chicago, IL), and STATA, version 10.1 (College Station, TX), packages were used for statistical tests. All bioinformatic analyses were performed by using the BRB-ArrayTools, version 3.8 (developed by Richard Simon; Biometrics Research Branch, National Cancer Institute, Bethesda, MD).
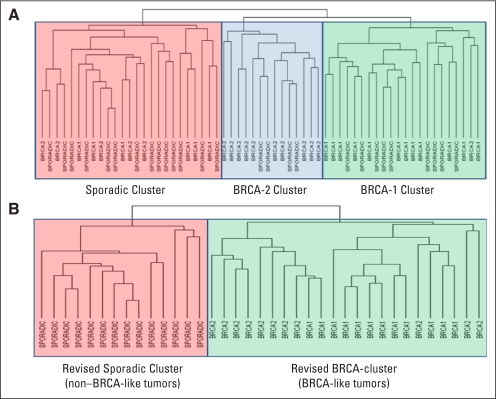
Fig A1.
Development of the BRCAness gene expression profile. Previous investigators have described gene expression differences between BRCA-mutated and sporadic cancers, although these studies have typically grouped all sporadic tumors together without taking into consideration that some of these sporadic tumors might indeed have a BRCAness phenotype (Hedenfalk IA: J Natl Cancer Inst 94:960-961, 2002; Jazaeri AA, Awtrey CS, Chandramouli GV, et al: Clin Cancer Res 11:6300-6310, 2005). Thus, the group of patients with sporadic disease in such analyses is potentially diluted by patients who may exhibit a gene expression profile more consistent with a BRCA1 or BRCA2 germline mutation carrier, and vice versa. To address this issue, we first performed genome-wide hierarchical clustering of all 61 tumors. We found that patients clustered into three groups, (A) which represeted BRCA1, BRCA2, and sporadic clusters, respectively. The BRCA1 cluster contained 22 patients, of which nine actually had sporadic (ie, nonmutated) disease. The BRCA2 cluster contained 14 patients, of which four had sporadic disease. The sporadic cluster contained 25 patients, of which six had BRCA1 and five had BRCA2 germline mutation. The clustering reproducibility index (R) was 0.934, and the three clusters did not change even if clear cell or mucinous samples were excluded from the analysis. For the purpose of defining the profile, these outliers (eg, a patient with BRCA1 contaminating the sporadic cluster, or a patient with sporadic disease contaminating the BRCA cluster) were removed from the analysis. We then proceeded to develop a 60-gene, diagonal linear discriminant predictor (B) that distinguished the BRCA clusters (ie, BRCA-like tumors) from the sporadic cluster (ie, non–BRCA-like tumors).
Fig A2.
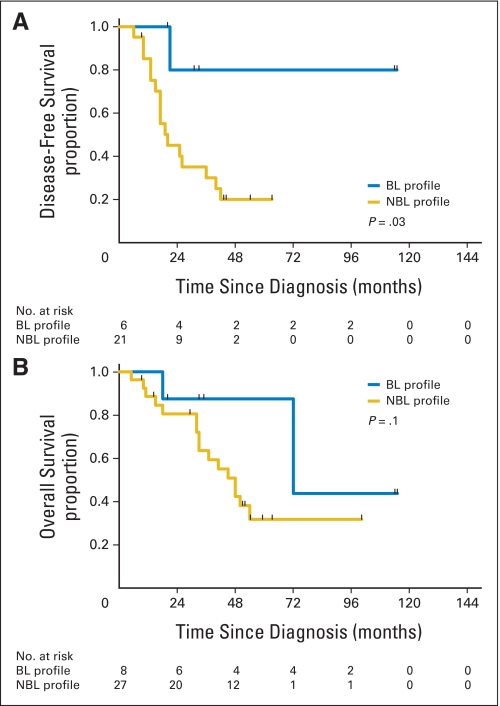
Association of BRCAness profile with disease-free survival (DFS) and overall survival (OS) in the sequenced patient cohort (n = 35). (A) DFS in the sequenced patient cohort. The median DFS times for patients with the BRCA-like (BL) and non–BRCA-like (NBL) profiles were not yet reached at median follow-up times of 53.5 months and 20 months, respectively (log-rank P = .03). (B) OS in the sequenced patient cohort. The median OS times for patients with the BL and NBL profiles were 72 months and 48 months, respectively (log-rank P = .1).
Fig A3.
Association of BRCAness profile with disease-free survival (DFS) and overall survival (OS) in the nonsequenced patient cohort (n = 35). (A) DFS in the nonsequenced patient cohort. The median DFS times for patients with the BL and NBL profiles were 22 and 7 months, respectively, (log-rank P = .013). (B) OS in the nonsequenced patient cohort. The median OS times for patients with the BL and NBL profiles were not yet reached and 30 months, respectively, (log-rank P = .009).
Table A1.
Gene Symbols and Characteristics
| No. | Gene Symbol | Gene Description | Weight |
|---|---|---|---|
| 1 | DAD1 | Defender against cell death 1 | 0.0997 |
| 2 | RAD21 | RAD21 homolog (Schizosaccharomyces pombe) | 0.1743 |
| 3 | LDHA | Lactate dehydrogenase A | 0.0165 |
| 4 | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 0.1571 |
| 5 | SKP1 | S-phase kinase-associated protein 1 | 0.137 |
| 6 | PPP1CC | Protein phosphatase 1, catalytic subunit, γ isoform | 0.0249 |
| 7 | RAN | RAN, member RAS oncogene family | 0.1043 |
| 8 | USP9X | Ubiquitin specific peptidase 9, X-linked | −0.0066 |
| 9 | ECHS1 | Enoyl coenzyme A hydratase, short chain, 1, mitochondrial | −0.0495 |
| 10 | GNAI3 | Guanine nucleotide binding protein (G protein), α inhibiting activity polypeptide 3 | 0.1404 |
| 11 | HDAC1 | Histone deacetylase 1 | −0.0272 |
| 12 | LGMN | Legumain | −0.0683 |
| 13 | CYR61 | Cysteine-rich, angiogenic inducer, 61 | −0.0454 |
| 14 | SH3BGRL | SH3 domain binding glutamic acid-rich protein like | 0.0659 |
| 15 | MGST3 | Microsomal glutathione S-transferase 3 | −0.0162 |
| 16 | RCN2 | Reticulocalbin 2, EF-hand calcium binding domain | 0.2087 |
| 17 | SMC1A | Structural maintenance of chromosomes 1A | 0.0313 |
| 18 | BST2 | Bone marrow stromal cell antigen 2 | 0.2118 |
| 19 | ENG | Endoglin | 0.058 |
| 20 | SRGN | Serglycin | −0.1577 |
| 21 | GBP1 | Guanylate binding protein 1, interferon-inducible, 67 kDa | 0.0778 |
| 22 | UNC119B | Unc-119 homolog B (Caenorhabditis elegans) | 0.139 |
| 23 | RGS1 | Regulator of G-protein signaling 1 | −0.2573 |
| 24 | CDC2 | Cell division cycle 2, Gap1 to synthesis and Gap2 to mitosis | 0.1101 |
| 25 | SAT1 | Spermidine/spermine N1-acetyltransferase 1 | −0.0386 |
| 26 | GGH | Gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | −0.0169 |
| 27 | GUCY1B3 | Guanylate cyclase 1, soluble, β3 | 0.0248 |
| 28 | WFDC2 | WAP four-disulfide core domain 2 | 0.0719 |
| 29 | NMI | N-myc (and STAT) interactor | 0.0873 |
| 30 | PRAME | Preferentially expressed antigen in melanoma | 0.0267 |
| 31 | CCL4 | Chemokine (C-C motif) ligand 4 | −0.1545 |
| 32 | MGST2 | Microsomal glutathione S-transferase 2 | −0.0045 |
| 33 | MMP7 | Matrix metallopeptidase 7 (matrilysin, uterine) | 0 |
| 34 | SNCA | Synuclein, alpha (non A4 component of amyloid precursor) | −0.1605 |
| 35 | VTN | Vitronectin | −0.0399 |
| 36 | ALPP | Alkaline phosphatase, placental (Regan isozyme) | −0.0274 |
| 37 | MTAP | Methylthioadenosine phosphorylase | −0.0511 |
| 38 | IDUA | Iduronidase, α-l- | 0.097 |
| 39 | SERPINF2 | Serpin peptidase inhibitor, clade F (α-2 antiplasmin, pigment epithelium derived factor), member 2 | −0.0053 |
| 40 | WAS | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | −0.0191 |
| 41 | CD1D | CD1d molecule | −0.1431 |
| 42 | GFI1 | Growth factor independent 1 transcription repressor | −0.0318 |
| 43 | P11 | 26 serine protease | 0.0091 |
| 44 | SEMA3F | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F | 0.0805 |
| 45 | TNF | Tumor necrosis factor (TNF superfamily, member 2) | −0.024 |
| 46 | ROS1 | C-ros oncogene 1, receptor tyrosine kinase | 0.1667 |
| 47 | MADCAM1 | Mucosal vascular addressin cell adhesion molecule 1 | −0.0322 |
| 48 | PDIA4 | Protein disulfide isomerase family A, member 4 | 0.1564 |
| 49 | HMGN2 | High-mobility group nucleosomal binding domain 2 | −0.0247 |
| 50 | HLA-B | Major histocompatibility complex, class I, B | 0.0956 |
| 51 | TM9SF1 | Transmembrane 9 superfamily member 1 | 0.1125 |
| 52 | CCDC93 | Coiled-coil domain containing 93 | −0.0865 |
| 53 | APEX1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | 0.0212 |
| 54 | VEGFA | Vascular endothelial growth factor A | 0.0612 |
| 55 | POSTN | Periostin, osteoblast specific factor | 0.2142 |
| 56 | PSTPIP1 | Proline-serine-threonine phosphatase interacting protein 1 | −0.041 |
| 57 | PMS1 | PMS1 postmeiotic segregation increased 1 (Saccharomyces cerevisiae) | −0.0579 |
| 58 | HLA-A | Major histocompatibility complex, class I, A | 0.0202 |
| 59 | PCTP | Phosphatidylcholine transfer protein | −0.0536 |
| 60 | SEH1L | SEH1-like (Saccharomyces cerevisiae) | 0.1127 |
Footnotes
See accompanying article on page 3545
Supported in part through the Ovarian Cancer Specialized Program of Research Excellence (SPORE) Grant No. P50 CA105009 Career Development Award, the Bernice Shopkin Weisman Fund, the Ovarian Cancer Research Fund in memory of Amy Sachs Simon, LeAnn's Project, and the Sisters Against Ovarian Cancer.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Beth Y. Karlan, AstraZeneca Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Panagiotis A. Konstantinopoulos, Dimitrios Spentzos, Stephen A. Cannistra
Financial support: Stephen A. Cannistra
Administrative support: Stephen A. Cannistra
Provision of study materials or patients: Beth Y. Karlan, Toshiyasu Taniguchi, Douglas A. Levine, Stephen A. Cannistra
Collection and assembly of data: Panagiotis A. Konstantinopoulos, Dimitrios Spentzos, Beth Y. Karlan, Elena Fountzilas, Nancy Francoeur, Douglas A. Levine, Stephen A. Cannistra
Data analysis and interpretation: Panagiotis A. Konstantinopoulos, Dimitrios Spentzos, Beth Y. Karlan, Elena Fountzilas, Nancy Francoeur, Douglas A. Levine, Stephen A. Cannistra
Manuscript writing: Panagiotis A. Konstantinopoulos,Stephen A. Cannistra
Final approval of manuscript: Panagiotis A. Konstantinopoulos, Dimitrios Spentzos, Beth Y. Karlan, Toshiyasu Taniguchi, Elena Fountzilas, Nancy Francoeur, Douglas A. Levine, Stephen A. Cannistra
REFERENCES
- 1.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 2.Cass I, Baldwin RL, Varkey T, et al. Improved survival in women with BRCAassociated ovarian carcinoma. Cancer. 2003;97:2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- 3.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 4.Tan DS, Rothermundt C, Thomas K, et al. “BRCAness” syndrome in ovarian cancer: A case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol. 2008;26:5530–5536. doi: 10.1200/JCO.2008.16.1703. [DOI] [PubMed] [Google Scholar]
- 5.McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Brody LC. Treating cancer by targeting a weakness. N Engl J Med. 2005;353:949–950. doi: 10.1056/NEJMcibr052331. [DOI] [PubMed] [Google Scholar]
- 8.Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 9.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 10.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 11.Audeh MW, Penson RTF M, Friedlander M, et al. Phase II trial of the oral PARP inhibitor olaparib (AZD2281) in BRCAdeficient advanced ovarian cancer. J Clin Oncol. 2009;27(suppl):277s. abstr 5500. [Google Scholar]
- 12.Pal T, Permuth-Wey J, Betts JA, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 13.Boyd J. Specific keynote: Hereditary ovarian cancer—What we know. Gynecol Oncol. 2003;88:S8–S10. doi: 10.1006/gyno.2002.6674. discussion S11-S13. [DOI] [PubMed] [Google Scholar]
- 14.Risch HA, McLaughlin JR, Cole DE, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: A kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 15.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 16.Teodoridis JM, Hall J, Marsh S, et al. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961–8967. doi: 10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- 17.Baldwin RL, Nemeth E, Tran H, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: A population-based study. Cancer Res. 2000;60:5329–5333. [PubMed] [Google Scholar]
- 18.Geisler JP, Hatterman-Zogg MA, Rathe JA, et al. Frequency of BRCA1 dysfunction in ovarian cancer. J Natl Cancer Inst. 2002;94:61–67. doi: 10.1093/jnci/94.1.61. [DOI] [PubMed] [Google Scholar]
- 19.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 20.Hennessy B, Timms K, Carey MS, et al. Somatic BRCA status in ovarian tumors. J Clin Oncol. 2009;27(suppl):284s. abstr 5528. [Google Scholar]
- 21.Foster KA, Harrington P, Kerr J, et al. Somatic and germline mutations of the BRCA2 gene in sporadic ovarian cancer. Cancer Res. 1996;56:3622–3625. [PubMed] [Google Scholar]
- 22.Hilton JL, Geisler JP, Rathe JA, et al. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002;94:1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 24.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 25.Konstantinopoulos PA, Spentzos D, Cannistra SA. Gene-expression profiling in epithelial ovarian cancer. Nat Clin Pract Oncol. 2008;5:577–587. doi: 10.1038/ncponc1178. [DOI] [PubMed] [Google Scholar]
- 26.Spentzos D, Levine DA, Kolia S, et al. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J Clin Oncol. 2005;23:7911–7918. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 27.Spentzos D, Levine DA, Ramoni MF, et al. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22:4700–4710. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 28.Jazaeri AA, Yee CJ, Sotiriou C, et al. Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J Natl Cancer Inst. 2002;94:990–1000. doi: 10.1093/jnci/94.13.990. [DOI] [PubMed] [Google Scholar]
- 29.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swisher EM, Sakai W, Karlan BY, et al. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinopoulos PA, Fountzilas E, Pillay K, et al. Carboplatin-induced gene expression changes in vitro are prognostic of survival in epithelial ovarian cancer. BMC Med Genomics. 2008;1:59. doi: 10.1186/1755-8794-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9:505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 33.Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 34.Dabney AR, Storey JD. Optimality driven nearest centroid classification from genomic data. PLoS One. 2007;2:e1002. doi: 10.1371/journal.pone.0001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CC, Lin CJ. Training nu-support vector classifiers: Theory and algorithms. Neural Comput. 2001;13:2119–2147. doi: 10.1162/089976601750399335. [DOI] [PubMed] [Google Scholar]
- 36.Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 37.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: Analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 38.Shattuck-Eidens D, Oliphant A, McClure M, et al. BRCA1 sequence analysis in women at high risk for susceptibility mutations: Risk factor analysis and implications for genetic testing. JAMA. 1997;278:1242–1250. [PubMed] [Google Scholar]
- 39.Quinn JE, James CR, Stewart GE, et al. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clin Cancer Res. 2007;13:7413–7420. doi: 10.1158/1078-0432.CCR-07-1083. [DOI] [PubMed] [Google Scholar]
- 40.Freitas S, Moore DH, Michael H, et al. Studies of apurinic/apyrimidinic endonuclease/ref-1 expression in epithelial ovarian cancer: Correlations with tumor progression and platinum resistance. Clin Cancer Res. 2003;9:4689–4694. [PubMed] [Google Scholar]
- 41.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: The 3′ ends justify the means. Mutat Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 42.Godwin AK, Meister A, O'Dwyer PJ, et al. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci U S A. 1992;89:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson SW, Ozols RF, Hamilton TC. Mechanisms of drug resistance in ovarian cancer. Cancer. 1993;71:644–649. doi: 10.1002/cncr.2820710224. [DOI] [PubMed] [Google Scholar]
- 44.Aebi S, Kurdi-Haidar B, Gordon R, et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 45.Durant ST, Morris MM, Illand M, et al. Dependence on RAD52 and RAD1 for anticancer drug resistance mediated by inactivation of mismatch repair genes. Curr Biol. 1999;9:51–54. doi: 10.1016/s0960-9822(99)80047-5. [DOI] [PubMed] [Google Scholar]
- 46.Ryan PD, Tung NM, Isakoff SJ, et al. Neoadjuvant cisplatin and bevacizumab in triple negative breast cancer (TNBC): Safety and efficacy. J Clin Oncol. 2009;27(suppl):18s. abstr 551. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.