Abstract
Background
Alzheimer's Disease (AD) is the most common neurodegenerative disease and the leading cause of dementia among senile subjects. It has been proposed that AD can be caused by defects in mitochondrial oxidative phosphorylation. Given the fundamental contribution of the mitochondrial genome (mtDNA) for the respiratory chain, there have been a number of studies investigating the association between mtDNA inherited variants and multifactorial diseases, however no general consensus has been reached yet on the correlation between mtDNA haplogroups and AD.
Methodology/Principal Findings
We applied for the first time a high resolution analysis (sequencing of displacement loop and restriction analysis of specific markers in the coding region of mtDNA) to investigate the possible association between mtDNA-inherited sequence variation and AD in 936 AD patients and 776 cognitively assessed normal controls from central and northern Italy. Among over 40 mtDNA sub-haplogroups analysed, we found that sub-haplogroup H5 is a risk factor for AD (OR = 1.85, 95% CI:1.04–3.23) in particular for females (OR = 2.19, 95% CI:1.06–4.51) and independently from the APOE genotype. Multivariate logistic regression revealed an interaction between H5 and age. When the whole sample is considered, the H5a subgroup of molecules, harboring the 4336 transition in the tRNAGln gene, already associated to AD in early studies, was about threefold more represented in AD patients than in controls (2.0% vs 0.8%; p = 0.031), and it might account for the increased frequency of H5 in AD patients (4.2% vs 2.3%). The complete re-sequencing of the 56 mtDNAs belonging to H5 revealed that AD patients showed a trend towards a higher number (p = 0.052) of sporadic mutations in tRNA and rRNA genes when compared with controls.
Conclusions
Our results indicate that high resolution analysis of inherited mtDNA sequence variation can help in identifying both ancient polymorphisms defining sub-haplogroups and the accumulation of sporadic mutations associated with complex traits such as AD.
Introduction
Alzheimer's Disease (AD) is the most common neurodegenerative disease and the leading cause of dementia among senile subjects. Few AD cases are early onset and familial, with autosomal dominant inheritance, while the majority of cases are late-onset (over 60 years old) and sporadic [1]. Sporadic AD (SAD) has a complex aetiology due to environmental and genetic factors which taken alone are not sufficient to cause the disease. Presently the major genetic risk factor in SAD is recognized in the allele e4 of apolipoprotein E (ApoE4). However, the causal factors in the majority of late-onset AD patients are still unknown and there is likely a complex interaction between genetic and environmental factors that eventually results in the pathology [2].
It has been proposed that SAD can be caused by defects in mitochondrial oxidative phosphorylation (OXPHOS) [3]. Structurally abnormal mitochondria have been observed in AD brains [4], and deficiencies in mitochondrial OXPHOS enzymes, such as cytocrome c oxidase, have been repeatedly reported in the brains and other tissues of AD patients [5], [6]. Mitochondria are deeply involved in various cellular processes such as ATP synthesis, heat production, reactive oxygen species (ROS) generation through oxidative phosphorylation, but also calcium signalling and apoptosis [7], [8]. Defects in OXPHOS inhibit ATP production, but also increase mitochondrial ROS production, which, in turn, can damage the mitochondrial genome (mtDNA). Thus, this hypothesis on AD pathogenesis claims that mitochondrial impairment may result from accumulated mtDNA damage that accompanies normal aging, amplified by disease-specific factors [9], [3]. Although there were reports of somatic mutations specifically at sites of known mtDNA regulatory elements, the available data from hybrid studies and post-mortem brain examinations did not provide conclusive proof that somatic mtDNA mutations can play a major or dominant role in AD aetiology [10], [11].
Human mtDNA codes for the 12S and 16S mitochondrial rRNAs, 22 tRNAs, and 13 polypeptides; these polypeptides are essential subunits of mitochondrial OXPHOS enzyme complexes, which generate the principal source of intracellular energy, ATP [12], [8]. It is inherited almost exclusively through the maternal lineage, does not recombine and is highly polymorphic. Mitochondrial genomes can be affiliated within haplogroups and sub-haplogroups on the basis of their specific sequence motifs, reflecting mutation events accumulated over time along diverging maternal lineages. These haplogroups tend to show a continental or regional localization, and a very large number of European, African, and Asian/Native American-specific haplogroups have been identified so far [13], [14].
Given the fundamental contribution of the mitochondrial genome for the respiratory chain, there have been a number of studies investigating the association between mtDNA lineages, aging and multifactorial diseases. It has been hypothesised that some mtDNA haplogroups are not neutral and have been selected through adaptation to climate and nutritional conditions [15], [8], [16]. Moreover, there are many reports on the association between mtDNA haplogroups and longevity [17]–[19], Leber hereditary optic neuropathy [20], as well as complex diseases like diabetes [21], ischaemic disease [22] and neurodegenerative diseases like Parkinson's Disease [23] and AD.
As for AD, contrasting data have been published. Some studies suggested that the transition at np 4336 of the mitochondrial tRNA glutamine gene is a risk factor for AD [24]–[26]. This mutation characterizes a European sub-branch of haplogroup H [13], which was termed H5a in more recent years [27], [28]. Later it has been reported that haplogroup T is under-represented whereas haplogroup J is over-represented in AD patients [29]. Haplogroup U has been reported to be under-represented in females and over-represented in male AD patients of European ancestry [30], while recently, it has been found that HV cluster is significantly associated with the risk of AD regardless of the gender and the APOE4 status [31]. On the other side, it is to note that several studies did not find any association between mtDNA haplogroups and AD by studying different European populations [32]–[34], [11], [35]. On the whole, no general consensus has been reached on the correlation between mtDNA haplogroups and AD [36].
Since AD is a common neurodegenerative disorder, affecting globally about 3% of people older than 60 years in the world [37], even a small risk associated with mtDNA haplotype could have major causative implications at a population level. It is therefore critically important to determine the role of mtDNA polymorphisms in AD by studying large cohorts of subjects at a level of molecular and phylogenetic resolution as fine as possible. Our study is then aimed to investigate the possible association of mtDNA at the sub-haplogroup level in a population of 936 AD patients and 776 controls from central-northern Italy, in order to clarify whether specific sub-haplogroup polymorphic sites are involved in AD, and to assess the possibility that associations detectable only at the sub-haplogroup resolution level might account for the discrepancies present in the literature regarding AD and European mtDNA inherited variability.
Results
In our total sample of 1712 Italian subjects (936 AD patients and 776 controls) there is a higher proportion of female (72.9%) and APOE4+ (43.5%) subjects among AD patients relative to controls (60.6% and 13.1%, respectively). A separate comparison of genders further confirmed that APOE4 carriers were significantly different between AD patients and controls ( Table 1 ).
Table 1. Characteristics of the study participants.
| AD patients (N = 936) | Controls (N = 776) | p-value | ||
| Age (years) | (mean±SD) | 76.5±7.6 | 73.7±7.2 | |
| Gender | Female | 72.9% | 60.6% | <0.0001 |
| Male | 27.1% | 39.4% | ||
| APOE4 status | APOE4+ | 43.5% | 13.1% | <0.0001 |
| APOE4- | 56.5% | 86.9% | ||
| Female | (N = 682) | (N = 470) | ||
| Age (years) | (mean±SD) | 77.1±7.7 | 73.8±7.2 | |
| APOE4 status | APOE4+ | 43.4% | 11.1% | <0.0001 |
| APOE4- | 56.6% | 88.9% | ||
| Male | (N = 254) | (N = 306) | ||
| Age (years) | (mean±SD) | 75.1±7.2 | 73.4±7.1 | |
| APOE4 status | APOE4+ | 43.7% | 16.3% | <0.0001 |
| APOE4- | 56.3% | 83.7% | ||
The hierarchical survey of diagnostic markers in the coding region allowed the classification of mtDNAs from patients and controls into more than 40 haplogroups/sub-haplogroups (Table S1). Most of these are typical of modern European populations, but a few East Asian (M) [38] and sub-Saharan African (L1b, L2a, L2c, L3d, L3e) [16], [39] mtDNAs were also detected. This latter finding is not unexpected, since low frequencies of African and East Asian haplogroups are not uncommon in populations of southern Europe. We then grouped, using a phylogenetic rationale whenever possible, all sub-haplogroups with frequencies lower than 1.5%, thus allowing a reduction of the overall number of categories from over 39 to 19.
No difference was found between AD patients and controls in the distribution of mtDNA groupings ( Table 2 ). We further proceeded by comparing the 19 sub-haplogroup frequencies in AD patients and controls by taking into account the multiple comparison adjustments (p-value cut-off = 0.003); again no statistical difference was observed.
Table 2. Frequencies of mtDNA sub-haplogroups in AD patients and controls from Italy.
| Sub-haplogroupa | AD patients | Controls | ||||
| (N = 936) | (N = 776) | |||||
| N | % | SEb | N | % | SEb | |
| H* | 182 | 19,4 | 0,0129 | 156 | 20,1 | 0,0144 |
| H1 | 115 | 12,3 | 0,0107 | 97 | 12,5 | 0,0119 |
| H3 | 28 | 3,0 | 0,0056 | 18 | 2,3 | 0,0054 |
| H5c | 39 | 4,2 | 0,0065 | 18 | 2,3 | 0,0054 |
| H6 | 33 | 3,5 | 0,0060 | 21 | 2,7 | 0,0058 |
| J1 | 64 | 6,8 | 0,0082 | 49 | 6,3 | 0,0087 |
| J2 | 14 | 1,5 | 0,0040 | 13 | 1,7 | 0,0046 |
| R0 | 46 | 4,9 | 0,0071 | 46 | 5,9 | 0,0085 |
| T1 | 21 | 2,2 | 0,0048 | 17 | 2,2 | 0,0053 |
| T2 | 84 | 9,0 | 0,0093 | 70 | 9,0 | 0,0103 |
| U* | 38 | 4,1 | 0,0065 | 32 | 4,1 | 0,0071 |
| K | 67 | 7,2 | 0,0084 | 68 | 8,8 | 0,0102 |
| U4 | 23 | 2,5 | 0,0051 | 16 | 2,1 | 0,0051 |
| U5a | 46 | 4,9 | 0,0071 | 38 | 4,9 | 0,0077 |
| U5b | 18 | 1,9 | 0,0045 | 22 | 2,8 | 0,0060 |
| V | 31 | 3,3 | 0,0058 | 29 | 3,7 | 0,0068 |
| W | 20 | 2,1 | 0,0047 | 16 | 2,1 | 0,0051 |
| X | 26 | 2,8 | 0,0054 | 15 | 1,9 | 0,0049 |
| Other | 41 | 4,4 | 0,0067 | 35 | 4,5 | 0,0074 |
Sub-haplogroups with frequencies lower than 1.5% were grouped. Thus R0 includes R0a, R1, HV*, HV0*, HV0a, HV1 and HV2; H* includes all mtDNAs belonging to haplogroup H, except those further classified (H1, H3, H5 and H6); the same rationale has been used for U*; whereas “Other” contains the heterogeneous haplogroups I, L1b, L2a, L2c, L3d, L3e, M, N and N1.
Standard Error.
Only for H5 there is a difference between AD patients and controls with a χ2 p-value (not adjusted for multiple comparisons) of 0.034.
Also when frequencies were compared separately for gender, we found no difference between AD patients and controls for the mtDNA, and overall no significant differences among the sub-haplogroup frequencies (Tables S2 and S3).
We then performed a univariate logistic regression analysis with the mtDNA variable to identify the potential sub-haplogroup associated with AD. Using as reference the haplogroup H, the only sub-haplogroup that resulted statistically associated with AD was H5 (OR = 1.89, 95%CI = 1.03–3.42). Hence we created a dichotomous variable (H5 versus all other sub-haplogroups) and performed again the logistic regression analysis including sex, age and APOE4.Overall sub-haplogroup H5 has a slightly higher AD risk (OR = 2.19, 95%CI = 1.06–4.51) in the female group than in the total sample (OR = 1.83, 95%CI = 1.04–3.23). As expected, in our population APOE4 carriers have a higher AD risk than non-APOE4 carriers (OR = 5.08, 95%CI = 3.98–6.50) and, among them, women have a higher risk than men (OR = 6.16, 95%CI = 4.45–8.53, OR = 3.97, 95%CI = 2.69–5.88, respectively).
Subsequently, mitochondrial sub-haplogroup H5 and the major acknowledged risk factors for AD (gender, age, APOE4) were compared in multivariate regression analysis. As shown in Table 3 , gender (female), APOE4 allele, age and sub-haplogroup H5 are confirmed to be risk factor for AD. Moreover sub-haplogroup H5 interacts with age in modifying AD risk. Subjects younger than 75 years (corresponding to the median age in the total sample) and carrying sub-haplogroup H5 harbor a four-fold increased AD risk than subjects belonging to other sub-haplogroups, while among subjects who do not carry sub-haplogroup H5, those older than 75 years have a three-fold increased risk than younger subjects.
Table 3. Wald χ2 statistic (p-value), Odds ratio and 95% Confidence Intervals for factors associated with Alzheimer's disease in the entire sample.
| AD patients (N = 936) vs Controls (N = 776)a | ||||
| p-value | OR | 95%C.I. | ||
| Gender (Female) | <.0001 | 1.72 | (1.37–2.16) | |
| APOE4 status (APOE4+) | <.0001 | 5.54 | (4.28–7.16) | |
| AGE (years)b | Haplogroup | |||
| ≤75 | Other | 1.00 | ||
| H5 | 0.0019 | 4.02 | (1.67–9.66) | |
| >75 | Other | 1.00 | ||
| H5 | 0.5326 | 0.77 | (0.33–1.77) | |
| Haplogroup | AGE (years) b | |||
| Other | ≤75 | 1.00 | ||
| >75 | <.0001 | 3.12 | (2.51–3.87) | |
| H5 | ≤75 | 1.00 | ||
| >75 | 0.3918 | 0.59 | (0.18–1.96) | |
Variables included in the model are age, gender, APOE4 and mtDNA haplogroup.
Corresponds to median value.
We then performed the multivariate logistic regression by gender. In the female group APOE4 allele, age and sub-haplogroup H5 are confirmed to be risk factor for AD, and the APOE4 allele interacts with age in modulating AD risk. In female subjects aged 76 years (corresponding to the median age in the female sample) or younger, those carrying the APOE4 allele have a ten-fold increased AD risk than non APOE4 carriers, while within subjects older than 76 years, those carrying the APOE4 allele have a nearly four-fold increased AD risk than non-APOE4 carriers. Among female non-APOE4 carriers, those older than 76 years have a four-fold increased AD risk than younger subjects ( Table 4 ). In the male group only APOE4 allele (OR = 3.87, 95%CI = 2.60–5.77) and age >74 years (OR = 2.15, 95%CI = 1.51–3.07) are confirmed to be risk factors for AD (data not shown).
Table 4. Wald χ2 statistic (p-value), Odds ratio and 95% Confidence Intervals for factors associated with Alzheimer's disease in females.
| AD patients (N = 682) vs Controls (N = 470)a | ||||
| p-value | OR | 95%C.I. | ||
| mtDNA haplogroup (H5) | 0.0327 | 2.38 | (1.07–5.27) | |
| AGE (years)b | APOE4 status | |||
| ≤76 | APOE4- | 1.00 | ||
| APOE4+ | <.0001 | 9.56 | (6.13–14.89) | |
| >76 | APOE4- | 1.00 | ||
| APOE4+ | <.0001 | 3.70 | (2.26–6.08) | |
| APOE4 status | AGE (years)b | |||
| APOE4- | ≤76 | 1.00 | ||
| >76 | <.0001 | 3.90 | (2.91–5.24) | |
| APOE4+ | ≤76 | 1.00 | ||
| >76 | 0.1744 | 1.51 | (0.83–2.74) | |
Variables included in the model are age, gender, APOE4 and mtDNA haplogroup.
Corresponds to median value.
To determine whether the increased risk associated with sub-haplogroup H5 could be attributed to specific mutations or mutational motifs, the complete sequence of virtually all H5 mtDNAs (56 out of 57; 38 AD patients and 18 controls) in our collection was determined and the phylogenetic distribution among the samples was analyzed. The results of the phylogenetic distribution and of the sequence analysis are summarized in Table 5 and Figure 1 . A total of 159 mutated positions relative to the reference sequence [40] were detected and analyzed in the network, including 104 nucleotide changes in the coding regions: the count of each kind of mutation is important in order to understand the mutational spectrum of the molecule and identify mutational hotspots. Overall 17 of the mutations (underlined and in italics in Table 5 ) were not previously reported in either MITOMAP (www.mitomap.org) or mtDB (www.genpat.uu.se/mtDB) and each mutation was observed only in a single H5 mtDNA.
Table 5. Mutations relative to the revised Cambridge reference sequences (rCRS) found in 56 mtDNAs belonging to sub-haplogroup H5.
| Nucleotide Positiona | Locus | Nucleotide Change | Aminoacid Changeb | AD Patients: N = 38 (%) | Controls N = 18 (%) |
| 73 | D-loop (HVSII) | A>G | non coding | 2 (5,3) | 0 (0) |
| 146 | D-loop (HVSII) | T>C | non coding | 3 (7,9) | 1 (5,5) |
| 150 | D-loop (HVSII) | C>T | non coding | 1 (2,6) | 0 (0) |
| 151 | D-loop (HVSII) | C>T | non coding | 0 (0) | 1 (5,5) |
| 152 | D-loop (HVSII) | T>C | non coding | 7 (18,4) | 5 (27,78) |
| 189 | D-loop (HVSII) | A>G | non coding | 1 (2,6) | 0 (0) |
| 195 | D-loop (HVSII) | T>C | non coding | 1 (2,6) | 0 (0) |
| 195A | D-loop (HVSII) | T>A | non coding | 1 (2,6) | 0 (0) |
| 199 | D-loop (HVSII) | T>C | non coding | 0 (0) | 1 (5,5) |
| 204 | D-loop (HVSII) | T>C | non coding | 1 (2,6) | 0 (0) |
| 207 | D-loop (HVSII) | G>A | non coding | 5 (13,2) | 4 (22,2) |
| 252 | D-loop (HVSII) | T>C | non coding | 1 (2,6) | 0 (0) |
| 259 | D-loop (HVSII) | A>G | non coding | 0 (0) | 1 (5,5) |
| 263 | D-loop (HVSII) | A>G | non coding | 38 (100) | 18 (100) |
| 309+C | D-loop (HVSII) | ins | non coding | 7 (18,4) | 7 (38,9) |
| 309+CC | D-loop (HVSII) | ins | non coding | 5 (13,2) | 1 (5,5) |
| 315+C | D-loop (HVSII) | ins | non coding | 38 (100) | 18 (100) |
| 316 | D-loop (HVSII) | G>A | non coding | 1 (2,6) | 0 (0) |
| 334 | D-loop (HVSII) | T>C | non coding | 1 (2,6) | 0 (0) |
| 365 | D-loop (HVSII) | A>G | non coding | 1 (2,6) | 0 (0) |
| 373 | D-loop | A>G | non coding | 1 (2,6) | 1 (5,5) |
| 385 | D-loop | A>G | non coding | 1 (2,6) | 0 (0) |
| 456 | D-loop (HVSIII) | C>T | non coding | 37 (97,4) | 18 (100) |
| 456+T | D-loop (HVSIII) | ins | non coding | 1 (2,6) | 0 (0) |
| 460 | D-loop (HVSIII) | T>C | non coding | 0 (0) | 1 (5,5) |
| 489 | D-loop (HVSIII) | T>C | non coding | 1 (2,6) | 0 (0) |
| 513 | D-loop (HVSIII) | G>A | non coding | 2 (5,3) | 1 (5,5) |
| 523_524 delCA | D-loop (HVSIII) | del | non coding | 8 (21,1) | 3 (16,7) |
| 523_524+CA | D-loop (HVSIII) | ins | non coding | 2 (5,3) | 0 (0) |
| 593 | tRNA phenylalanine | T>C | - | 2 (5,3) | 0 (0) |
| 721 | rRNA 12S | T>C | - | 1 (2,6) | 0 (0) |
| 750 | rRNA 12S | A>G | - | 38 (100) | 18 (100) |
| 769 | rRNA 12S | G>A | - | 1 (2,6) | 0 (0) |
| 801 | rRNA 12S | A>G | - | 1 (2,6) | 0 (0) |
| 1120 | rRNA 12S | C>T | - | 1 (2,6) | 0 (0) |
| 1438 | rRNA 12S | A>G | - | 38 (100) | 18 (100) |
| 3368 | NADH dehydrogenase subunit 1 | T>C | Met NP > Thr P | 1 (2,6) | 0 (0) |
| 3744 | NADH dehydrogenase subunit 1 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 3753 | NADH dehydrogenase subunit 1 | T>C | synonymous | 2 (5,3) | 0 (0) |
| 3992 | NADH dehydrogenase subunit 1 | C>T | Thr P > Met NP | 0 (0) | 1 (5,5) |
| 4024 | NADH dehydrogenase subunit 1 | A>G | Thr P > Ala NP | 0 (0) | 1 (5,5) |
| 4336 | tRNA glutamine | T>C | - | 19 (50) | 6 (33,3) |
| 4659 | NADH dehydrogenase subunit 2 | G>A | Ala NP > Thr P | 1 (2,6) | 0 (0) |
| 4769 | NADH dehydrogenase subunit 2 | A>G | synonymous | 38 (100) | 18 (100) |
| 4820 | NADH dehydrogenase subunit 2 | G>A | synonymous | 0 (0) | 1 (5,5) |
| 4832 | NADH dehydrogenase subunit 2 | C>T | synonymous | 1 (2,6) | 0 (0) |
| 5004 | NADH dehydrogenase subunit 2 | T>C | synonymous | 0 (0) | 1 (5,5) |
| 5082 | NADH dehydrogenase subunit 2 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 5268 | NADH dehydrogenase subunit 2 | A>G | Ile NP > Val NP | 0 (0) | 1 (5,5) |
| 5302 | NADH dehydrogenase subunit 2 | T>C | Ile NP > Thr P | 0 (0) | 1 (5,5) |
| 5319 | NADH dehydrogenase subunit 2 | A>G | Thr P > Ala NP | 1 (2,6) | 2 (11,1) |
| 5471 | NADH dehydrogenase subunit 2 | G>A | synonymous | 2 (5,3) | 0 (0) |
| 5821 | tRNA cysteine | G>A | - | 1 (2,6) | 0 (0) |
| 5839 | tRNA tyrosine | C>T | - | 1 (2,6) | 0 (0) |
| 5899+C | - | ins | - | 0 (0) | 1 (5,5) |
| 6425 | Cytochrome c oxidase subunit 1 | T>C | synonymous | 2 (5,3) | 2 (11,1) |
| 7364 | Cytochrome c oxidase subunit 1 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 7444 | Cytochrome c oxidase subunit 1 | G>A | STOP > Lys | 0 (0) | 1 (5,5) |
| 7474 | tRNA serine | A>G | - | 1 (2,6) | 0 (0) |
| 7645 | Cytochrome c oxidase subunit 2 | C>T | synonymous | 1 (2,6) | 0 (0) |
| 7853 | Cytochrome c oxidase subinit 2 | G>A | Val NP > Ile NP | 0 (0) | 1 (5,5) |
| 7861 | Cytochrome oxidase subunit 2 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 7961 | Cytochrome c oxidase subunit 2 | T>C | synonymous | 0 (0) | 1 (5,5) |
| 7999 | Cytochrome oxidase subunit 2 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 8011 | Cytochrome c oxidase subunit 2 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 8053 | Cytochrome c oxidase subinit 2 | A>G | synonymous | 0 (0) | 1 (5,5) |
| 8188 | Cytochrome c oxidase subunit 2 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 8277 | - | T>C | - | 1 (2,6) | 0 (0) |
| 8343 | tRNA lysine | A>G | - | 1 (2,6) | 0 (0) |
| 8348 | tRNA lysine | A>G | - | 2 (5,3) | 0 (0) |
| 8434G | ATP synthase F0 subunit 8 | C>G | Ile NP > Met NP | 1 (2,6) | 0 (0) |
| 8473 | ATP synthase F0 subunit 8 | T>C | synonymous | 0 (0) | 1 (5,5) |
| 8557 | ATP synthase F0 subunit 6 | G>A | Ala NP > Thr P | 0 (0) | 1 (5,5) |
| 8563 | ATP synthase F0 subunit 6 | A>G | Thr P > Ala NP | 1 (2,6) | 2 (11,1) |
| 8593 | ATP synthase F0 subunit 6 | A>G | Ile NP >Val NP | 0 (0) | 1 (5,5) |
| 8803T | ATP synthase F0 subunit 6 | A>T | Thr P > Ser P | 1 (2,6) | 0 (0) |
| 8857 | ATP synthase F0 subunit 6 | G>A | Gly NP > Ser P | 1 (2,6) | 0 (0) |
| 8860 | ATP synthase F0 subunit 6 | A>G | Thr P > Ala NP | 38 (100) | 18 (100) |
| 9010 | ATP synthase F0 subunit 6 | G>A | Ala NP > Thr P | 0 (0) | 1 (5,5) |
| 9123 | ATP synthase F0 subunit 6 | G>A | synonymous | 0 (0) | 1 (5,5) |
| 9515 | Cytochrome c oxidase subunit 3 | C>T | synonymous | 1 (2,6) | 0 (0) |
| 10172 | NADH dehydrogenase subunit 3 | G>A | synonymous | 2 (5,3) | 0 (0) |
| 10211 | NADH dehydrogenase subunit 3 | C>T | synonymous | 0 (0) | 1 (5,5) |
| 10217 | NADH dehydrogenase subunit 3 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 10256 | NADH dehydrogenase subunit 3 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 10275 | NADH dehydrogenase subunit 3 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 10289 | NADH dehydrogenase subunit 3 | A>G | synonymous | 0 (0) | 1 (5,5) |
| 10310 | NADH dehydrogenase subunit 3 | G>A | synonymous | 1 (2,6) | 0 (0) |
| 10410 | tRNA arginine | T>C | - | 5 (13,2) | 4 (22,2) |
| 10454 | tRNA arginine | T>C | - | 0 (0) | 1 (5,5) |
| 10646 | NADH dehydrogenase subunit 4L | G>A | synonymous | 1 (2,6) | 0 (0) |
| 10664A | NADH dehydrogenase subunit 4L | C>A | synonymous | 0 (0) | 1 (5,5) |
| 10907 | NADH dehydrogenase subunit 4 | T>C | Phe NP > Leu NP | 1 (2,6) | 0 (0) |
| 10915 | NADH dehydrogenase subunit 4 | T>C | synonymous | 2 (5,3) | 0 (0) |
| 11084 | NADH dehydrogenase subunit 4 | A>G | Thr P > Ala NP | 1 (2,6) | 0 (0) |
| 11087 | NADH dehydrogenase subunit 4 | T>C | Phe NP > Leu NP | 0 (0) | 1 (5,5) |
| 11089 | NADH dehydrogenase subunit 4 | C>T | synonymous | 0 (0) | 1 (5,5) |
| 11147 | NADH dehydrogenase subunit 4 | T>C | synonymous | 0 (0) | 1 (5,5) |
| 11329 | NADH dehydrogenase subunit 4 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 11686 | NADH dehydrogenase subunit 4 | C>T | synonymous | 0 (0) | 1 (5,5) |
| 11914 | NADH dehydrogenase subunit 4 | G>A | synonymous | 1 (2,6) | 0 (0) |
| 11959 | NADH dehydrogenase subunit 4 | A>G | synonymous | 0 (0) | 1 (5,5) |
| 11965 | NADH dehydrogenase subunit 4 | C>T | synonymous | 1 (2,6) | 0 (0) |
| 12088 | NADH dehydrogenase subunit 4 | C>T | synonymous | 0 (0) | 1 (5,5) |
| 12246 | tRNA serine2 | C>T | - | 0 (0) | 1 (5,5) |
| 12471 | NADH dehydrogenase subunit 5 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 12501 | NADH dehydrogenase subunit 5 | G>A | synonymous | 1 (2,6) | 0 (0) |
| 12723 | NADH dehydrogenase subunit 5 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 12771 | NADH dehydrogenase subunit 5 | G>A | synonymous | 1 (2,6) | 0 (0) |
| 12864 | NADH dehydrogenase subunit 5 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 13194 | NADH dehydrogenase subunit 5 | G>A | synonymous | 2 (5,3) | 0 (0) |
| 13434 | NADH dehydrogenase subunit 5 | A>G | synonymous | 1 (2,6) | 1 (5,5) |
| 13473 | NADH dehydrogenase subunit 5 | A>G | synonymous | 1 (2,6) | 0 (0) |
| 13651 | NADH dehydrogenase subunit 5 | A>G | Thr P > Ala NP | 1 (2,6) | 0 (0) |
| 13708 | NADH dehydrogenase subunit 5 | G>A | Ala NP > Thr P | 1 (2,6) | 0 (0) |
| 13725 | NADH dehydrogenase subunit 5 | C>T | synonymous | 5 (13,2) | 4 (22,2) |
| 14200 | NADH dehydrogenase subunit 6 | T>C | synonymous | 1 (2,6) | 0 (0) |
| 14311G | NADH dehydrogenase subunit 6 | T>G | Tyr NP > Asp A | 1 (2,6) | 0 (0) |
| 14319 | NADH dehydrogenase subunit 6 | T>C | Asn P > Asp A | 1 (2,6) | 0 (0) |
| 14365 | NADH dehydrogenase subunit 6 | C>T | synonymous | 0 (0) | 1 (5,5) |
| 14582 | NADH dehydrogenase subunit 6 | A>G | Val NP > Ala NP | 0 (0) | 1 (5,5) |
| 14668 | NADH dehydrogenase subunit 6 | C>T | synonymous | 1 (2,6) | 0 (0) |
| 14831 | Cytochrome b | G>A | Ala NP > Thr P | 1 (2,6) | 0 (0) |
| 14893 | Cytochrome b | A>G | synonymous | 0 (0) | 1 (5,5) |
| 15001 | Cytochrome b | T>C | synonymous | 0 (0) | 1 (5,5) |
| 15139 | Cytochrome b | T>C | synonymous | 2 (5,3) | 0 (0) |
| 15172 | Cytochrome b | G>A | synonymous | 1 (2,6) | 0 (0) |
| 15175 | Cytochrome b | C>T | synonymous | 0 (0) | 1 (5,5) |
| 15213 | Cytochrome b | T>C | Ile NP > Thr P | 0 (0) | 1 (5,5) |
| 15326 | Cytochrome b | A>G | Tyr NP > Asp A | 38 (100) | 18 (100) |
| 15511 | Cytochrome b | T>C | synonymous | 0 (0) | 1 (5,5) |
| 15833 | Cytochrome b | C>T | synonymous | 8 (21,1) | 1 (5,5) |
| 15884 | Cytochrome b | G>A | Ala NP >Thr P | 2 (5,3) | 1 (5,5) |
| 15889 | tRNA threonine | T>C | - | 1 (2,6) | 0 (0) |
| 15910 | tRNA threonine | C>T | - | 1 (2,6) | 0 (0) |
| 15930 | D-loop | G>A | non coding | 2 (5,3) | 1 (5,5) |
| 15940 | D-loop | T>C | non coding | 1 (2,6) | 0 (0) |
| 16092 | D-loop (HVSI) | T>C | non coding | 1 (2,6) | 0 (0) |
| 16093 | D-loop (HVSI) | T>C | non coding | 2 (5,3) | 0 (0) |
| 16124 | D-loop (HVSI) | T>C | non coding | 0 (0) | 1 (5,5) |
| 16129 | D-loop (HVSI) | G>A | non coding | 1 (2,6) | 0 (0) |
| 16145 | D-loop (HVSI) | G>A | non coding | 1 (2,6) | 1 (5,5) |
| 16148 | D-loop (HVSI) | C>T | non coding | 1 (2,6) | 0 (0) |
| 16201 | D-loop (HVSI) | C>T | non coding | 1 (2,6) | 0 (0) |
| 16243 | D-loop (HVSI) | T>C | non coding | 1 (2,6) | 0 (0) |
| 16256 | D-loop (HVSI) | C>T | non coding | 1 (2,6) | 0 (0) |
| 16291 | D-loop (HVSI) | C>T | non coding | 1 (2,6) | 1 (5,5) |
| 16294 | D-loop (HVSI) | C>T | non coding | 3 (7,9) | 0 (0) |
| 16304 | D-loop (HVSI) | T>C | non coding | 37 (97,4) | 18 (100) |
| 16311 | D-loop (HVSI) | T>C | non coding | 6 (15,8) | 6 (33,3) |
| 16312 | D-loop (HVSI) | A>G | non coding | 0 (0) | 1 (5,5) |
| 16320 | D-loop (HVSI) | C>T | non coding | 2 (5,3) | 0 (0) |
| 16335 | D-loop (HVSI) | A>G | non coding | 1 (2,6) | 0 (0) |
| 16352 | D-loop (HVSI) | T>C | non coding | 1 (2,6) | 0 (0) |
| 16357 | D-loop (HVSI) | T>C | non coding | 0 (0) | 1 (5,5) |
| 16362 | D-loop (HVSI) | T>C | non coding | 2 (5,3) | 0 (0) |
| 16391 | D-loop | G>A | non coding | 0 (0) | 1 (5,5) |
| 16497 | D-loop | A>G | non coding | 0 (0) | 1 (5,5) |
| 16519 | D-loop | T>C | non coding | 3 (7,9) | 2 (11,1) |
Novel mutations are underlined and in italics.
Mutations resulting in an aminoacid change are in bold. NP (Not Polar), P (Polar), A (Acid).
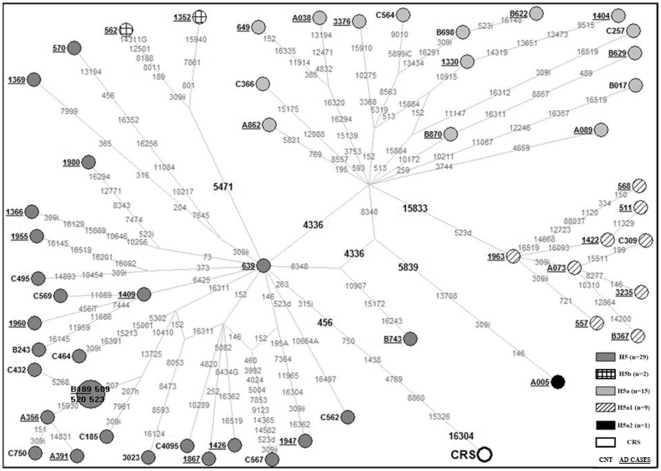
Figure 1. Phylogenetic network of the sub-haplogroup H5 complete sequences.
Phylogenetic network based on the variation of 56 mtDNA complete sequences from Italian subjects (GenBank accession numbers: GQ983055–GQ983110) relative to the revised Cambridge Reference Sequence (rCRS, GenBank accession number: NC_012920). Numbers beside the nodes denote samples (Cases and Control subjects). Unless marked otherwise, the polymorphic variants, shown on the lines connecting the nodes, are transitions. Letters following the polymorphic variants indicate transversions or insertions (i) or deletions (d). Positions in bold are H5 specific mutations. We assigned a weight = 10 to shared positions among all the samples and a weight = 30 to the rare mutations in the D-loop region, while we assigned a weight = 70 to shared positions and a weight = 90 to the rare variants in the coding region.
The topology of our mtDNA network matches published European trees in most respects [41].The network data set included 38 AD cases and 18 healthy controls, belonging to H5. A reduced-median network was constructed based on the sequence variations in the entire mtDNA molecules and by placing the rCRS as reference ( Figure 1 ).
The H5 network turned out to be star-like and constituted by a number of sub-clusters. The major dichotomy is due to the presence/absence of the 4336 mutation. The presence of this mutation characterizes H5a (N = 15) the cluster over represented in AD patients (1.1% in patients vs. 0.6% in controls). Moreover, entire mitochondrial sequence data show that H5a can be further subdivided; in particular there is one sub-branch termed H5a1 and defined by the mutation 15833 which encompasses many of mtDNAs within H5a (9 out of 15).
Twenty-nine of the mutations (in bold) listed in Table 5 result in amino acid changes, but excluding the mutations 8860 and 15326 (rCRS private mutations), none was particularly common. The highest frequencies were indeed reached by the mutations 5319 and 8563, each observed in one AD patient (2.6%) and two controls (11.1%).
Among all mutations observed in the 56 H5 mtDNAs, the mutated position at 15833, defining the H5a1 subgroup, was observed in the 21.1% of the H5 AD patients and in the 5.5% of controls, when compared with the entire sample this difference was statistical significant (0.9% vs 0.1%; p = 0.039). The nucleotide change at np 4336 in the tRNA glutamine gene, already associated to AD in previous studies [24], [25], was observed in 50.0% of the H5 AD patients and 33.3% of the controls. When the whole sample is considered, the H5a subgroup of molecules, harboring the 4336 transition, was about threefold more represented in AD patients than in controls (2.0% vs 0.8%; p = 0.031), and it might account for the H5 frequency increase in AD patients (4.2% vs 2.3%).
We did not find any statistical significant result when comparing the number of mutations along the mtDNA molecule by mtDNA regions ( Table 6 ). Subsequently, we searched for groups of singleton mutations falling in specific mtDNA regions that may increase the susceptibility to AD. In particular we focused on singleton mutations falling in tRNA and rRNA genes, considering that previous studies have shown that mutations in these genes, including the transition at np 4336 (tRNAGln) and the G3196A mutation (16S rRNA), may be associated with AD [24], [42]. We found that ten sporadic mutations (single occurrences) were present in the tRNA plus rRNA genes from AD mtDNA sequences, while only two sporadic mutations were found in the same genes from controls. We verified the significance of this finding by comparing the number of the tRNA+rRNA mutations with the number of mutations falling in the remaining of coding region (43 in AD patients and 37 in controls). The χ2 test, under the hypothesis of a homogeneous distribution of mutations showed a tenuous significance (p = 0.05).
Table 6. Number of mutations found along the mtDNA molecule regions in AD patients (N = 936) vs controls (N = 776).
| mtDNA region | N. mutations in AD (N = 534) | N. mutations in CNT (N = 261) |
| D-loop | 235 | 115 |
| ND1 | 4 | 2 |
| ND2 | 44 | 24 |
| COI | 3 | 3 |
| COII | 5 | 3 |
| ATPase8 | 1 | 1 |
| ATPase6 | 41 | 24 |
| COIII | 1 | 0 |
| ND3 | 6 | 2 |
| ND4L | 1 | 1 |
| ND4 | 7 | 6 |
| ND5 | 16 | 5 |
| ND6 | 4 | 2 |
| CytB | 52 | 25 |
| Total (genes) | 185 | 98 |
| Y | 1 | 0 |
| R | 5 | 5 |
| T | 2 | 0 |
| C | 1 | 0 |
| Q | 19 | 6 |
| K | 3 | 0 |
| F | 2 | 0 |
| S | 1 | 0 |
| S2 | 0 | 1 |
| Total (tRNA) | 34 | 12 |
| 12S | 80 | 36 |
| Total (rRNA) | 80 | 36 |
Discussion
Although there is large amount of evidence about the role of mitochondria in the pathogenesis of AD, a definitive conclusion regarding the association of mtDNA haplogroup with AD has not been reached yet [36]. To date several studies reported lack of association between mtDNA haplogroups and AD [32]–[34], [11], [35], while others found positive associations with specific haplogroups as well as specific haplogroup clusters analyzing different Caucasian populations [24], [29]–[31], [43].
To our knowledge, this is the first case-control study investigating, at the sub-haplogroup level, the association of mtDNA with Alzheimer's Disease. We analyzed a large cohort of AD patients (N = 936) and controls (N = 776) comparable for age and ethnicity from the central-northern regions of Italy, where the controls were directly assessed for their cognitive status. We found that sub-haplogroup H5 appears to be associated with a higher risk of AD in both the total sample and the female group. This result was in keeping with a recent study on the Polish population which revealed that the super-haplogroup HV and haplogroup H are associated with a higher risk of AD [31].
Studies of both survival after sepsis [44] and sperm motility [45] have shown significant associations with mtDNA haplogroups, leading to the proposal that mitochondria bearing haplogroup H mtDNAs, and particularly haplogroups H3, H4, H5 and H6 [46] are associated with a more tightly coupled oxidative phosphorylation, and consequently they should lead to an increased production of ROS than those with haplogroup T. Accordingly, it was hypothesized that subjects with haplogroup H could be more prone to oxidative stress than those with other haplogroups [8], and consequently more susceptible to neurodegenerative diseases, in which oxidative stress plays a major role. This hypothesis would account for the higher AD risk we observed for H5 subjects.
We also found that sub-haplogroup H5 interacts with age in modifying AD risk. H5 subjects younger than 75 years old have a higher AD risk than non-H5 subjects, while in those older than 75 years no increased risk is observed. Age is a strong risk factor for AD [37] and this interaction suggests that age 75, corresponding to the median age in our sample, could be considered as a threshold, under which risk factors such as sub-haplogroup H5 and APOE, could independently exert their major effect on the development of the disease, while over this age value other risk factors, such as aging itself, would largely prevail.
The presence of mtDNA sub-haplogroup H5 confers a greater risk of AD to women than to the total sample of AD patients considered. In our sample the proportion of AD females was higher than in the female control group, however we can exclude that the association is merely due to a gender imbalance because the frequencies of all haplogroups in AD females were comparable to the female controls (and also to male controls), these latter being similar to the northern Italian general population [47].
In males we did not find any significant association but only a trend toward an increase in H5 frequency in AD males relative to controls. It is to note that males are about one third of our total AD sample, because they are less represented both among the elderly [48] and in AD patients [49]. Thus, the negative association between H5 sub-haplogroup and AD in men could be explained by the smaller size of the AD male group. However, a specific or more pronounced risk in women cannot at present be excluded, given the growing evidence that components of the AD phenotype differ significantly based on gender [50].
As for APOE, any association emerged between sub-haplogroup mtDNA variation and APOE4 genotype, suggesting that they likely exert an independent effect on AD. Previous results on this topic are discordant, some Authors reporting a positive association [43], [31], while others no association [30], [35] between haplogroups and APOE4. Indeed, we found a strong association of APOE4 with age, this effect being particularly evident in females younger than 76 years, i.e. the same age group where mtDNA sub-haplogroup H5 appears to exert its major influence on AD.
In order to assess if the H5 background of the patients harboured mutations responsible for the increased AD risk, we investigated the complete mtDNA sequences of our H5 subjects. We did not find any particular mutational pattern in the H5 patients (compared to controls) except for a threefold increase of the H5 subgroup (H5a) characterized by a transition at position 4336 (tRNAGln gene) that has been previously shown to be associated with AD [24], [25], and a sevenfold increase of the synonymous mutation at np 15833 which defines the H5a1 sub-haplogroup. The latter finding is difficult to explain unless that the high frequency of this C>T transition at np 15833 in AD patients is due to a mutational bias toward specific codon usage at synonymous sites [16].
In addition sporadic mutations (showing up in one sample only) are slightly more numerous in tRNA plus rRNA genes of AD samples than in controls. This finding is also in keeping with growing evidences showing that mutations in the protein synthesis machinery of mitochondria may be frequent cause of human degenerative diseases [42]. Thus our results confirm the relationship between mtDNA sequence variation and AD and suggest that both ancient polymorphisms defining sub-haplogroups and the accumulation of sporadic mutations are most likely involved.
Further study will need to better focus on the genetic variation falling in tRNA and rRNA genes, that is the mitochondrial translation machinery. In fact a better characterization of the molecular problems caused by these mutations in combination with may be of help in characterizing different subgroups of sporadic AD patients and help in their treatment.
Summarizing, the limitation of the present study is the lack of a replication study in another population. Accordingly, we cannot exclude population-specific effects. However, the large number of patients and controls in our study allows us to largely exclude possible false positive results. A replica of our study is quite demanding, owing to difficulties in enrolling another comparable large number of patients and of cognitively well assessed controls either in the same geographic area or in other European countries. The strengths are: (i) our study refers to a number of clinically well assessed AD patients (936) and controls (776) which is quite consistent; (ii) both AD patients and controls have been recruited in a specific relatively large geographic area thus avoiding possible bias related to a founder effect or population heterogeneity; (iii) the control subjects have been tested for their cognitive capability; (iv) we have identified for the first time a candidate risk sub-haplogroup for AD which extends previous data regarding H-V cluster on a more limited number of AD patients and controls in a Polish population [31]; (v) we have provided 56 new mtDNA complete sequences all referring to the H5 sub-haplogroup which not only suggest that sporadic mutations in mitochondrial tRNA and rRNA genes could be involved in AD risk but also and more in general represent a major contribution on mtDNA genetics regarding this specific haplogroup in a Caucasian population; (vi) we have pointed out the relationship between H5 mtDNA sub-haplogroup and other well known risk factors for AD, such as APOE4 genetic polymorphism and age. These data allowed us to identify the subgroup of AD patients (younger than 75 years of age; female sex) where H5 sub-haplogroup has a stronger effect.
Methods
DNA samples from 1712 (936 AD patients and 776 healthy controls) unrelated central-northern Italian subjects were analyzed. The geographic origin and the number of samples for each participating group were the following: Lombardy (405 patients; 192 controls), Veneto (248 patients; 414 controls), Tuscany (138 patients; 45 controls), Marche (145 patients; 125 controls). All subjects were of Italian descent.
Patients were diagnosed by skilled evaluation units as suffering from probable AD, according to NINCDS-ADRDA criteria [51] and underwent a comprehensive geriatric assessment, including an extended neuropsychological evaluation.
The control group was comparable for age, sex and ethnicity to the patient group. A major characteristic of this control group is that they were directly assessed by MMSE in order to exclude subjects affected by cognitive deficiency. Written informed consent was obtained from all control individuals and primary caregivers on behalf of AD Patients. Written informed consent was obtained from all control individuals and primary caregivers on behalf of AD Patients. Each Institution which provided the DNA samples received the approval from their own ethical committees. In particular the Ethic committees of the: Italian National Research Center for Ageing in Ancona, Department of Neurological and Psychiatric Sciences of University of Florence, Regional Center for Cerebral Ageing in Valdagno and the S. Giovanni di Dio-Fatebenefratelli Center in Brescia have given their approval.
DNA was recovered from fresh blood by phenol-chloroform standard procedures. MtDNA profiles in patients and controls were determined by sequencing the entire mtDNA control region for each subject from nucleotide position (np) 16024 to np 576. This was followed by a hierarchical survey of haplogroup and sub-haplogroup diagnostic markers in the coding region [20], [47].
APOE genotyping was performed as previously described [52]. Patients and controls were stratified into two subgroups, according to their APOE4 status: those carrying at least one APOE4 allele (APOE4+) and non-APOE4 carriers (APOE4-).
Complete mtDNA sequences were obtained after treatment of total DNA samples with REPLI-g Mitochondrial DNA kit (Qiagen, Valencia, CA) for specific whole genome amplification of mtDNA, followed by PCR amplification with MitoALL Resequencing kit (Applera, Foster City, CA). The PCR products (5–10 ng) were purified by EXOSAPit (U.S. Biochemical, Cleveland, Ohio) and used for direct sequencing with BigDye kit version 3.1 (Applera). Electropherograms were inspected with SeqScape version 2.5 software (Applera). All the samples were aligned and compared with the revised Cambridge Reference Sequence (rCRS) [40].
Reduced median network analyses, based on nucleotide variation on the complete sequences, were carried out by Network 4.5.1.0 (http://fluxus-engineering.com) [53] with an e default value of zero.
Because of mutation rate heterogeneity in mtDNA there is not a consensus on the weights to be assigned to the mutated nucleotide positions. As reported in other studies [27], [54] and after experimental tests we choose to give different weights to nucleotide positions in the D-loop and coding regions of our samples. After an initial run, we summed the statistics (counts of each mutation) obtained for all samples and used them to weight the characters from 10 to 90 with an inverted linear relation against the number of occurrences in the statistics.
Statistical Analysis
Mitochondrial sub-haplogroups, genotype frequencies and gender were compared between AD patients and controls using the χ2 or Fisher-exact test, while differences in age were investigated through the non parametric Wilcoxon rank-sum test.
The mitochondrial sub-haplogroup variability envisages at least 40 subgroups, which have been further grouped into 19 subgroups considering their frequencies and phylogenetic relationship.
The comparisons between each of the 19 mtDNA sub-haplogroups in AD patients and controls have been computed by applying both the Bonferroni's adjustments and the Holm's step procedure. All analyses were performed separately for women and men, as well as for the total group, and multivariate logistic regression analysis using a stepwise procedure, adjusting for significant interactions (considering a threshold level of 0.15), was used to assess the relationship between independent variables (mtDNA, APOE4 status, age and gender) and the outcome. Tests for statistical significance were two-sided with = 0.05; effect size for the association was measured as odds ratio (OR) with 95% confidence intervals (CI). SAS (9.1) statistical software from SAS Institute, Cary (NC), was used for all statistical analyses.
Web Resources
Accession numbers and the URL for data presented herein are as follows: GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (complete H5 sequences [accession numbers GQ983055–GQ983110])
MITOMAP: A Human Mitochondrial Genome Database. http://www.mitomap.org, 2009
mtDB: Human Mitochondrial Genome Database. www.genpat.uu.se/mtDB
Network: Free Phylogenetic Network Software. http://www.fluxus-engineering.com
Supporting Information
Frequencies of mtDNA sub-haplogroups in 936 AD patients and 776 controls from central-northern Italy.
(0.07 MB DOC)
Frequencies of mtDNA sub-haplogroups in 682 female AD patients and 470 female controls from central-northern Italy.
(0.04 MB DOC)
Frequencies of mtDNA sub-haplogroups in 254 male AD patients and 306 male controls from central-northern Italy.
(0.04 MB DOC)
Acknowledgments
We are grateful to Giovanna De Benedictis for suggestions and criticisms in planning the study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the “Promozione Regionale della Ricerca Industriale, dell'Innovazione e del Trasferimento Tecnologico” (PRRIITT) program of the Emilia-Romagna Region, the University of Bologna Grant “Ricerca Fondamentale Orientata (RFO ex 60%) 2008”, the Programma Strategico 2007, conv. PS39, the Ricerca Corrente, Italian Ministry of Health Grant “An integrated approach to identify functional, biochemical and genetic markers in centenarians and Alzheimer's Disease patients” and the Roberto and Cornelia Pallotti Legacy for Cancer Research Grants (to C. F.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kamboh MI. Molecular genetics of late-onset Alzheimer's disease. Ann Hum Genet. 2004;68:381–404. doi: 10.1046/j.1529-8817.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 2.Petrozzi L, Ricci G, Giglioli NJ, Siciliano G, Mancuso M. Mitochondria and neurodegeneration. Biosci Rep. 2007;27:87–104. doi: 10.1007/s10540-007-9038-z. [DOI] [PubMed] [Google Scholar]
- 3.Nunomura A, Castellani RJ, Zhu X, Moreira PI, Perry G, et al. Involvement of oxidative stress in Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:631–641. doi: 10.1097/01.jnen.0000228136.58062.bf. [DOI] [PubMed] [Google Scholar]
- 4.Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, et al. Role of mitochondrial dysfunction in Alzheimer's disease. J Neurosci Res. 2002;70:357–360. doi: 10.1002/jnr.10389. [DOI] [PubMed] [Google Scholar]
- 5.Bosetti F, Brizzi F, Barogi S, Mancuso M, Siciliano G, et al. Cytochrome c oxidase and mitochondrial F1F0-ATPase (ATP synthase) activities in platelets and brain from patients with Alzheimer's disease. Neurobiol Aging. 2002;23:371–376. doi: 10.1016/s0197-4580(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 6.Cottrell DA, Borthwick GM, Johnson MA, Ince PG, Turnbull DM. The role of cytochrome c oxidase deficient hippocampal neurones in Alzheimer's disease. Neuropathol Appl Neurobiol. 2002;28:390–396. doi: 10.1046/j.1365-2990.2002.00414.x. [DOI] [PubMed] [Google Scholar]
- 7.Smeitink J, van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 8.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson GE, Sheu KF, Blass JP. Abnormalities of mitochondrial enzymes in Alzheimer disease. J Neural Transm. 1998;105:855–870. doi: 10.1007/s007020050099. [DOI] [PubMed] [Google Scholar]
- 10.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elson JL, Herrnstadt C, Preston G, Thal L, Morris CM. Does the mitochondrial genome play a role in the etiology of Alzheimer's disease? Hum Genet. 2006;119:241–254. doi: 10.1007/s00439-005-0123-8. [DOI] [PubMed] [Google Scholar]
- 12.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 13.Torroni A, Wallace DC. Mitochondrial DNA variation in human populations and implications for detection of mitochondrial DNA mutations of pathological significance. J Bioenerg Biomembr. 1994;26:261–271. doi: 10.1007/BF00763098. [DOI] [PubMed] [Google Scholar]
- 14.Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 16.Kivisild T, Shen P, Wall DP, Do B, Sung R, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. Faseb J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 18.Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, et al. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 19.Bilal E, Rabadan R, Alexe G, Fuku N, Ueno H, et al. Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan. PLoS One. 2008;11:e2421. doi: 10.1371/journal.pone.0002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghelli A, Porcelli AM, Zanna C, Vidoni S, Mattioli S, et al. The background of mitochondrial DNA haplogroup J increases the sensitivity of Leber's hereditary optic neuropathy cells to 2,5-hexanedione toxicity. PLoS One. 2009;19:e7922. doi: 10.1371/journal.pone.0007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007;80:407–415. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinnery PF, Elliot HR, Syed A, Rothwell PM Oxford Vasculary Study. Mitochondrial DNA haplogroups and risk of transient ischaemic attack and ischaemic stroke: a genetic association study. Lancet Neurol. 2010;9:498–503. doi: 10.1016/S1474-4422(10)70083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaweda-Walerych K, Maruszak A, Safranow K, Bialecka M, Klodowska-Duda G, et al. Mitochondrial DNA haplogroups and subhaplogroups are associated with Parkinson's disease risk in a Polish PD cohort. J Neural Transm. 2008;115:1521–1526. doi: 10.1007/s00702-008-0121-9. [DOI] [PubMed] [Google Scholar]
- 24.Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MF, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 25.Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown MD, Shoffner JM, Kim YL, Jun AS, Graham BH, et al. Mitochondrial DNA sequence analysis of four Alzheimer's and Parkinson's disease patients. Am J Med Genet. 1996;61:283–289. doi: 10.1002/(SICI)1096-8628(19960122)61:3<283::AID-AJMG15>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 27.Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roostalu U, Kutuev I, Loogväli EL, Metspalu E, Tambets K, et al. Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: the Near Eastern and Caucasian perspective. Mol Biol Evol. 2007;24:436–448. doi: 10.1093/molbev/msl173. [DOI] [PubMed] [Google Scholar]
- 29.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, et al. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am J Med Genet. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 30.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 31.Maruszak A, Canter JA, Styczynska M, Zekanowski C, Barcikowska M. Mitochondrial haplogroup H and Alzheimer's disease - is there a connection? Neurobiol Aging. 2009;30:1749–1755. doi: 10.1016/j.neurobiolaging.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Chinnery PF, Taylor GA, Howell N, Andrews RM, Morris CM, et al. Mitochondrial DNA haplogroups and susceptibility to AD and dementia with Lewy bodies. Neurology. 2000;55:302–304. doi: 10.1212/wnl.55.2.302. [DOI] [PubMed] [Google Scholar]
- 33.Pyle A, Foltynie T, Tiangyou W, Lambert C, Keers SM, et al. Mitochondrial DNA haplogroup cluster UKJT reduces the risk of PD. Ann Neurol. 2005;57:564–567. doi: 10.1002/ana.20417. [DOI] [PubMed] [Google Scholar]
- 34.van der Walt JM, Scott WK, Slifer S, Gaskell PC, Martin ER, et al. Maternal lineages and Alzheimer disease risk in the Old Order Amish. Hum Genet. 2005;118:115–122. doi: 10.1007/s00439-005-0032-x. [DOI] [PubMed] [Google Scholar]
- 35.Mancuso M, Nardini M, Micheli D, Rocchi A, Nesti C, et al. Lack of association between mtDNA haplogroups and Alzheimer's disease in Tuscany. Neurol Sci. 2007;28:142–147. doi: 10.1007/s10072-007-0807-z. [DOI] [PubMed] [Google Scholar]
- 36.Santoro A, Salvioli S, Raule N, Capri M, Sevini F, et al. Mitochondrial DNA involvement in human longevity. Biochim Biophys Acta. 2006;1757:1388–1399. doi: 10.1016/j.bbabio.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 37.Qiu C, De Ronchi D, Fratiglioni L. The epidemiology of the dementias: an update. Curr Opin Psychiatry. 2007;20:380–385. doi: 10.1097/YCO.0b013e32816ebc7b. [DOI] [PubMed] [Google Scholar]
- 38.Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 39.Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 41.Brandstätter A, Zimmermann B, Wagner J, Göbel T, Röck AW, et al. Timing and deciphering mitochondrial DNA macro-haplogroup R0 variability in Central Europe and Middle East. BMC Evol Biol. 2008;8:191. doi: 10.1186/1471-2148-8-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scaglia F, Wong LJ. Human mitochondrial transfer RNAs: role of pathogenic mutation in disease. Muscle Nerve. 2008;37:150–171. doi: 10.1002/mus.20917. [DOI] [PubMed] [Google Scholar]
- 43.Carrieri G, Bonafè M, De Luca M, Rose G, Varcasia O, et al. Mitochondrial DNA haplogroups and APOE4 allele are non-indipendent variables in sporadic Alzheimer's disease. Hum. Genet. 2001;108:194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- 44.Baudouin SV, Saunders D, Tiangyou W, Elson JL, Poynter J, et al. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. doi: 10.1016/S0140-6736(05)67890-7. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, Perez-Martos A, Montoya J, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67:682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, et al. Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;51:111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achilli A, Olivieri A, Pala M, Metspalu E, Fornarino S, et al. Mitochondrial DNA variation of modern Tuscans supports the Near Eastern origin of Etruscans. Am J Hum Genet. 2007;80:759–768. doi: 10.1086/512822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrucci L, Giallauria F, Guralnik JM. Epidemiology of aging. Radiol Clin North Am. 2008;46:643–652. doi: 10.1016/j.rcl.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azad NA, Al Bugami M, Loy-English I. Gender differences in dementia risk factors. Gend Med. 2007;4:120–129. doi: 10.1016/s1550-8579(07)80026-x. [DOI] [PubMed] [Google Scholar]
- 50.Cahill L. Why sex matters for neuroscience. Nat Rev Neurosci. 2006;7:477–484. doi: 10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- 51.McKhann G, Drachman D, Folstein M, Katzman R, Price D, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 52.Hixson JE, Vernieri DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 53.Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soares P, Ermini L, Thomson N, Mormina M, Rito T, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequencies of mtDNA sub-haplogroups in 936 AD patients and 776 controls from central-northern Italy.
(0.07 MB DOC)
Frequencies of mtDNA sub-haplogroups in 682 female AD patients and 470 female controls from central-northern Italy.
(0.04 MB DOC)
Frequencies of mtDNA sub-haplogroups in 254 male AD patients and 306 male controls from central-northern Italy.
(0.04 MB DOC)