Abstract
Knowledge of protein structures and protein-protein interactions is essential for understanding of biological processes. Recent advances in protein crosslinking and mass spectrometry (MS) have shown significant potential to contribute to this area. Here we report a novel method to rapidly and accurately identify crosslinked peptides based on their unique isotope signature when digested in the presence of H218O. This method overcomes the need for specially synthesized crosslinkers and/or multiple MS runs required by other techniques. We validated our method by performing a ‘blind’ analysis of 5 proteins/complexes of known structure. Side chain repacking calculations using Rosetta show that 17 of our 20 positively identified crosslinks fit the published atomic structures. The remaining 3 crosslinks are likely due to protein aggregation. The accuracy and rapid throughput of our workflow will advance the use of protein crosslinking in structural biology.
Keywords: Protein Crosslinking, Isotope signature, Structure, Protein-protein interaction, Mass spectrometry, Rosetta, Hardklör, PepLynx
Introduction
In an era where the genomes and proteomes of many organisms are well known at the sequence level, our knowledge of the three-dimensional structure of proteins and protein complexes lags frustratingly behind. An understanding of protein structure and protein-protein interactions is essential to comprehend the molecular mechanisms underlying biological processes.
To date, high-resolution structural information on proteins has been obtained using methods such as NMR spectroscopy or X-ray crystallography. However, the structures of many proteins and complexes cannot yet be solved by these methods either because of lack of protein crystals or due to their large size.
Elucidation of protein-protein interactions has relied mainly upon two methods: Yeast two-hybrid assays1–3 and affinity purification followed by mass spectrometry.4, 5 Chemical crosslinking in combination with analysis of the reaction products by mass spectrometry (MS) not only yields information about protein-protein interactions, but also allows discrimination between direct versus indirect interactions, along with revealing which residues within protein complexes lie next to one another.6 Such methods require little sample, and purity is not of paramount importance. One of the main challenges obstructing the use of this method is the confident identification of low abundance crosslinked peptides, from the complex mixtures generated by chemical crosslinking.
Until now, two strategies have commonly been used to facilitate the detection of crosslinked peptides by MS. The first strategy makes use of isotope-coded crosslinkers.7–10 Crosslinking reactions are performed using light and heavy (often deuteriated) crosslinkers. Peptide fragments containing a crosslinker are present in two populations mass shifted by the weight difference between the heavy and light forms of the crosslinker. The second method uses trypsin facilitated incorporation of 18O.11–13 Crosslinked proteins are digested by trypsin in the presence of either H216O or H218O. Oxygen atoms from H218O specifically exchange with the two oxygen atoms of the C-termini of tryptic peptides.14 Crosslinked peptides have two C-termini, and will thus be present in +0 or +8 Da forms in the 16O and 18O samples, respectively. Non-crosslinked peptides will be present in +0 or +4 Da forms. Recently, a third method using high-charge-state driven data acquisition was utilized to enrich data sets for cross-linked peptides.15
This paper describes a new method designed to identify crosslinked peptides based on their unique isotope signature when digested in the presence of H218O. Rather than looking for a mass shift between two populations of peptides generated under different crosslinking or digestion conditions, a single digestion is performed under partial 18O enrichment conditions. This has a profound effect on the isotope signature of crosslinked peptides. This method, in conjunction with the software we have written to analyze the data generated, has advantages over previously published methods: (1) it overcomes the need for isotope-coded crosslinkers; (2) it does not rely on detecting a mass shift between two populations of peptides; (3) in a single MS run one can confidently identify the parent masses and retention times of crosslinked peptides at the same time as acquiring MS/MS spectra by data dependent acquisition (DDA) on many of those peptides. A second MS run is only required in the case that the DDA did not generate sufficient MS/MS spectra to confidently assign the detected crosslinks; (4) no user intervention is required during the analysis of the MS data.
One factor currently preventing the general adoption of MS based crosslinking methods has been their difficult and time-consuming nature. We made our initial analysis very rapid in order to test whether our method was versatile enough to yield results under non-optimized conditions and also to determine its potential in higher-throughput applications where time cannot be spent optimizing reaction and analysis conditions for each individual sample. This work aims not just to provide a novel means to do MS analysis of crosslinked proteins, but also to offer a rapid method that requires minimal manual intervention for the confident identification of crosslinks while keeping the number of false positive identifications close to zero. To test how quickly our method could generate data, we validated our method with five proteins. Crosslinking reactions were performed once and took ~ 6 hours. Data acquisition and analysis for all samples took just over one week.
Experimental Section
Materials
BRCA1/BARD1 RING-domain heterodimer was obtained as a kind gift from Rachel Klevit and Peter Brzovic.16 Green fluorescent protein was expressed and purified from E. coli in house using standard procedures. Lysozyme (chicken egg white) was purchased from USB Corporation (Cleveland, OH). Ribonuclease A (bovine pancreas) and beta-lactoglobulin (bovine milk) were purchased from Sigma-Aldrich (St. Louis, MO). Bis(sulfosuccinimidyl) suberate (BS3) and disuccinimidyl suberate (DSS) were purchased from Pierce (Rockford, IL). H218O was from Spectra Stable Isotopes (Andover, MA). Trypsin was from Promega (Madison, WI). Other reagents were obtained from Sigma-Aldrich.
Reference Protein Structures
We used the following PDB files as reference structures for the proteins analyzed: 1jm7.pdb (model 1) for BRCA1/BARD1; 1bsq.pdb for β-lactoglobulin; 2hgd.pdb for GFP; 2lym.pdb for lysozyme and 3rsp.pdb for ribonuclase A.
Standard crosslinking reactions
Crosslinking reactions for our initial rapid analysis were done using 100 μg of protein in 234.7 μl buffer. BS3 was prepared in buffer as a 14.5 mM stock and added to the reactions to give a final concentration of 0.88 mM. All reactions were carried out in PBS (pH 8) for 5 hours at room temperature. Protein to crosslinker ratios for each protein were as follows: BRCA1/BARD1, 60:1; GFP, 60:1; RNaseA, 34:1; lysozyme, 34:1; beta-lactoglobulin 41:1. Reactions were quenched by addition of 25 μl 200 mM NH4HCO3. Quenched reactions were incubated for a further 30 minutes at room temperature. Reaction buffer was then exchanged to 50 mM NH4HCO3 using protein desalting spin columns (Pierce) according to the manufacturer’s instructions. If protein quantities are limited, 1 μg protein is sufficient to complete the MS analysis.
Additional crosslinking reactions
In addition to the standard reaction, GFP was crosslinked using DSS prepared in dry dimethylformamide as a 14.5 mM stock and added to the reaction to give a final concentration of 0.88 mM. A ‘low crosslinker concentration’ BRCA1/BARD1 reaction was performed using BS3 at low concentration (final concentration 17.6 μM) at room temperature overnight.
Digestion
Samples were suspended in 50 mM ammonium bicarbonate buffer (pH 7.8) with H218O at 25 atom percent excess (APE). At 25 APE there is 25% more 18O than in natural water, which has about 0.2% of endogenous 18O. Neutral pH conditions were maintained throughout the experiment to reduce backexchange of 18O atoms. The crosslinked proteins were reduced with 5 mM dithiothreitol (DTT), alkylated with 8 mM iodoacetamide (IAA), and digested with trypsin at a substrate to enzyme ratio of 100:1 for two hours at 37°C with shaking. The digested samples were stored at −20°C until analyzed.
Mass Spectrometry
All mass spectrometry was performed on an LTQ-FT Ultra (Thermo Fisher Scientific). 250 ng of each sample digest was loaded from the autosampler onto a fused-silica capillary column (75-μm i.d.) packed with 40 cm of Jupiter C18 300Å material (Phenomenex) mounted in an in house constructed microspray source and placed in line with an Agilent 1100 QuatPump HPLC and 1100 AS autosampler. Peptides were eluted off the column using two buffer solutions: Buffer A (94.9% water, 5% acetonitrile, 0.1% formic acid) and Buffer B (19.9% water, 80% acetonitrile, 0.1% formic acid). The gradient program consisted of five steps totaling 160 minutes: (1) A 15 minute loading phase in 5% Buffer B. (2) A 45 minute gradient of 5 to 20% Buffer B. (3) A 75 minute gradient of 20 to 68% Buffer B. (4) A 7 minute wash at 80% Buffer B. (5) Column re-equilibration with 5% Buffer B for 18 minutes.
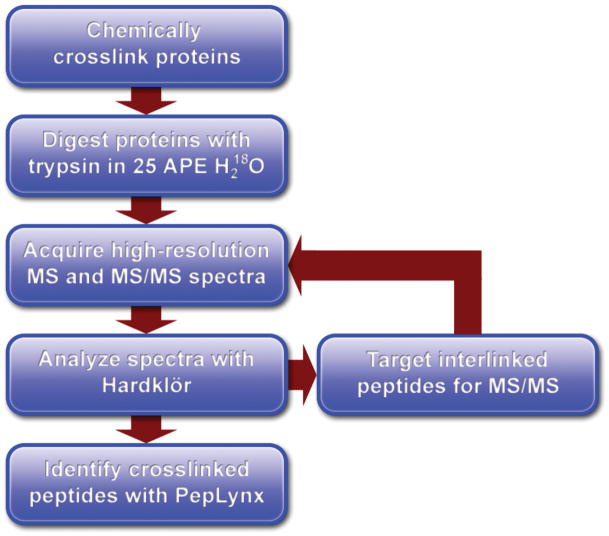
The mass spectrometer was operated using either data dependent acquisition (DDA) of MS/MS scans or targeted mass MS/MS (Figure 1). In both cases, a single high resolution mass spectrum was acquired at 50,000 resolution (at m/z 400) in the ICR. For DDA acquisition, each profile scan was followed by two high resolution MS/MS scans at 25,000 resolution in the ICR. High resolution MS/MS was performed to aid in the identification of crosslinked features that were typically of large mass and high charge state (4+, 5+, and 6+). For targeted mass scans, the analyzer method was divided into segments of 10 minutes. In each segment, each profile scan was followed by four targeted mass MS/MS scans at 25,000 resolution in the ICR. Details of targeted mass scans are found below.
Figure 1.
Flowchart describing the experimental procedure and data analysis workflow.
Data Analysis
High-resolution MS/MS spectra were analyzed by Hardklör to de-isotope the fragment distributions. Peptides were then identified from the de-isotoped fragment distribution using in house software, called PepLynx, which matched the monoisotopic fragment masses using a two-step comparison process (Figure 1). In the first step, PepLynx compared b- and y-ion fragmentation masses for single peptides generated from a protein sequence database. Single peptides that contribute to one-half of an interlinked peptide pair partially match the observed fragment pattern. The single peptides that scored the highest were passed to the second comparison step, where they were paired with peptides from the database that contribute the remaining mass of the precursor ion. Pairs of peptides are scored to the MS/MS spectrum by matching b- and y-ion fragment masses at 10 ppm accuracy. If one or both peptides contained more than one lysine, the pair was scored for every possible combination of linkages across those lysines. This two-step process rapidly searched the most likely peptide pairs, rather than all peptide combinations, which allowed for sizeable databases (up to several hundred proteins) to be used.
Results and Discussion
Method and Software Development for Isotope Labeling and Analysis of Peptides
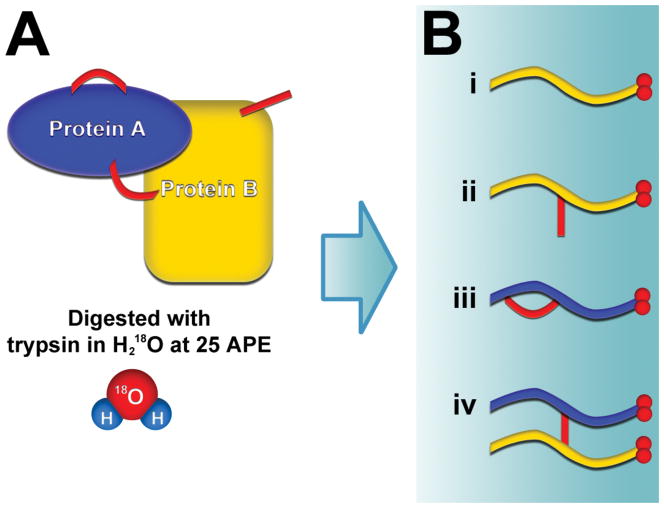
Proteins were crosslinked with BS3, as described in the experimental section. Crosslinked proteins were digested with trypsin in buffer containing H218O at 25 atom percent excess (APE) and the resulting peptides were then analyzed by LC-MS/MS. Digestion of the crosslinked samples produced four types of peptides: (i) unlinked peptides without crosslinker, (ii) dead-end peptides containing hydrolyzed or quenched crosslinker, (iii) loop-linked peptides where two lysines on a single peptide were linked to each other, and (iv) interlinked peptides where two distinct peptides were conjoined by the crosslinker (Figure 2).
Figure 2.
Labeling procedure for the detection of crosslinked peptides. (A) Crosslinked proteins are digested with trypsin in buffer containing H218O at 25 atom percent excess. All resulting peptides become 18O labeled on their C-terminus. (B) Four possible types of peptides are produced: (i) unlinked peptides without crosslinker; (ii) dead-end peptides that contain hydrolyzed or aminolyzed crosslinker; (iii) loop-linked peptides where two lysines on a single peptide are linked to each other; (iv) interlinked peptides. Interlinked peptides are distinguished from the other peptides because they have twice the amount of labeling on their C-termini.
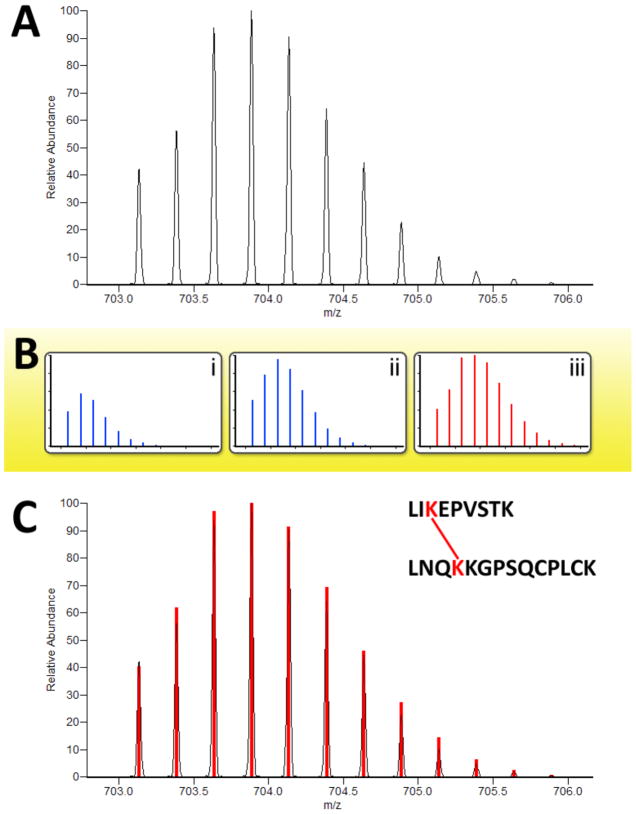
Digestion of the proteins in the presence of H218O resulted in isotopic labeling of the carboxy terminus of the digested peptides. Partial labeling at 25 APE H218O was used to create a characteristic, and predictable, isotopic peak distribution profile when observing the peptides in the mass analyzer. Because interlinked peptides have two C-termini, they incorporate twice the amount of labeling as unlinked, dead-end, and loop-linked peptides (Figure 2B). Thus, interlinked peptides could be distinguished from the other peptides in the sample by their characteristic 18O-labeled isotopic peak distribution. Precursor MS spectra were analyzed with the feature detection software Hardklör to compute monoisotopic masses for the observed peptide isotope distributions (PIDs).17 Additionally, Hardklör determined whether or not the observed PIDs contained the distinguishing 18O-labeled profile of interlinked peptides. As depicted in Figure 3, Hardklör compared each observed PID to a series of models representing the different amounts of 18O labeling at 25 APE for each type of peptide. Interlinked peptides were identified in the precursor spectra from PIDs that exhibited twice the amount of 18O-labeling as unlinked, dead-end, and loop-linked peptides.
Figure 3.
Identification of crosslinked peptide isotope distributions. A candidate peptide isotope distribution (A) is analyzed by Hardklör. Possible isotope signatures are modeled for (B-i) natural isotope abundance, (B-ii) 18O2, and (B-iii) 18O4 containing peptides. A best match to an 18O4 containing peptide (C) indicates an interpeptide crosslink whose sequences are then identified by PepLynx.
For each MS/MS spectrum, a list of putative interlinked peptides was generated as described in the methods. Interlinked peptides were ranked by score from highest to lowest and the highest was accepted using a set of heuristics that included: (1) exceeding a base scoring threshold, (2) contribution of both peptides to the score, and (3) significantly larger score than the next best possibility. The PepLynx software algorithm automated the acceptance of interlinked peptides for each spectrum using the aforementioned heuristic thresholds, and if the heuristic thresholds were not met, rejected all putative interlinked peptides for that spectrum. To assure the confidence of the identified interlinked peptides, our protein sequence database contained two sets of proteins in addition to our target protein(s). Firstly, a SEQUEST18 analysis with Percolator19 validation was performed on our MS data and every protein detected in the crosslinking reaction was included in our protein database. Secondly, decoy protein sequences consisting of the shuffled sequences of all detected proteins were also added to our database. The first set of proteins insured that crosslinks involving contaminant proteins could be appropriately assigned. The second set of proteins (the decoy proteins) is known not to exist, thus any putative interlinked peptides containing sequence from the decoy proteins does not exist. Our software is capable of analyzing a database of up to several hundred proteins, making it suitable for the analysis of large complexes consisting of dozens of proteins, and also for the analysis of complex crosslinked samples produced by single-step affinity purification of protein complexes from crude protein extracts so long as the target proteins are a major constituent of the purification ant sufficient protein can be obtained.
High Throughput Crosslinking Analysis of Known Proteins
We validated our crosslinking and analysis methods by applying them to the analysis of five proteins of known atomic structure. These proteins were: The BRCA1/BARD1 RING-domain heterodimer, β-lactoglobulin, green fluorescent protein (GFP), lysozyme and ribonuclase A. We made our initial analysis very rapid in order to test our method’s potential in higher-throughput applications where time cannot be spent optimizing reaction and analysis conditions for each individual sample. All our reactions were therefore performed in a set volume (~ 0.25 mL) of PBS (pH 8) containing 0.88 mM BS3 and 100 μg of protein. All reactions were allowed to crosslink at room temperature for 5 hours before quenching with NH4HCO3. Each crosslinking reaction and tryptic digestion was done just once.
An initial MS run was carried out as described in the methods section using data dependent acquisition (DDA) for MS/MS. Crosslinks discovered for each protein during this initial run are shown in Table 1 and are labeled *. Because of the slow fragmentation scan speed and relatively low abundance of crosslinked peptides, targeted mass MS/MS was performed in subsequent runs using the list of Hardklör identified crosslinked features from the initial DDA analysis (Figure 1). Each crosslinked feature was sorted by m/z and retention time. The analyzer method was divided into 10 minute segments, and up to four m/z values were selected for fragmentation across each segment. If more than four crosslinked features existed for any 10 minute segment, then the analyzer method was repeated using the next set of four m/z values. Additional crosslinks discovered using data from the targeted mass MS/MS scans are shown in Table 1, and are labeled † to indicate the contribution of these additional MS runs to the data presented. The targeted mass MS/MS runs contributed to 48% of the total crosslinks discovered.
Table 1.
Crosslinks revealed by a rapid analysis using BS3. Distances between crosslinked lysine ε-amino groups and α-carbon groups measured directly from PDB reference structures are shown next to lysine ε-amino distances after side chain repacking calculations using Rosetta. Reference PDBs were: 1jm7.pdb (model 1) for BRCA1/BARD1; 1bsq.pdb for β-lactoglobulin; 2hgd.pdb for GFP; 2lym.pdb for lysozyme and 3rsp.pdb for ribonuclase A. BS3 and DSS can span a maximum distance of 11.4 Å between the amine groups on the lysine side chains (according to the manufacturer). Theoretically, this distance constraint can still be met if the α-carbons of the lysines are within 24 Å. Side chain repacking calculations using Rosetta allowed us to determine lysine NZ-NZ distances after minimization. The final column of this table indicates the analysis needed to discover each crosslink. Crosslinks marked (*) were found using a single DDA MS run and a fully automated analysis. Crosslinks marked (†) were discovered through additional targeted mass MS/MS runs performed using the list of Hardklör identified crosslinked features from the initial DDA analysis but still using a fully automated analysis. Crosslinks marked (‡) were discovered through the use of a less stringent scoring cutoff during automated software analysis followed by manual validation of the data. Results from two additional crosslinking reactions are also shown in this table. BARD1/BRCA1 low (1/50th of previous) BS3 concentration results are labeled (LC) while BARD1/BRCA1 standard concentration is labeled (SC). GFP crosslinked with DSS is labeled (DSS) while GFP crosslinked with the standard BS3 is labeled (BS3).
| Protein Name | Crosslinked Lysine 1 | Crosslinked Lysine 2 | Lysine αC- αC distance from PDB | Lysine NZ-NZ distance from PDB | Lysine NZ- NZ distance after minimization | Analysis used for discovery |
|---|---|---|---|---|---|---|
| BRCA1/BARD1 | A20 | B110 | 15.3 | 11.2 | 9.21 | †SC |
| BRCA1/BARD1 | A38 | A70 | 9.10 | 16.1 | 9.99 | †LC |
| BRCA1/BARD1 | A45 | A70 | 11.3 | 9.60 | 9.98 | †LC |
| BRCA1/BARD1 | A50 | A56 | 10.8 | 15.9 | 9.89 | *LC |
| BRCA1/BARD1 | A50 | A70 | 11.1 | 11.1 | 9.89 | †SC/*LC |
| BRCA1/BARD1 | A56 | A70 | 12.2 | 17.8 | 10.0 | †SC/† LC |
| β-lactoglobulin | A47 | A69 | 13.4 | 16.9 | 10.1 | ‡ |
| β-lactoglobulin | A77 | A91 | 12.6 | 11.2 | 8.89 | * |
| β-lactoglobulin | A135 | A141 | 10.1 | 12.0 | 9.91 | * |
| β-lactoglobulin | A138 | A141 | 5.43 | 7.82 | 7.73 | * |
| GFP | A101 | A131 | 11.4 | 11.4 | 9.69 | * BS3/*DSS |
| GFP | A101 | A166 | 10.4 | 8.35 | 8.16 | †BS3 |
| GFP | A107 | A126 | 5.20 | 9.17 | 7.20 | * BS3/*DSS |
| GFP | A107 | A162 | 15.2 | 15.6 | 9.59 | †BS3/*DSS |
| GFP | A156 | A162 | 9.80 | 12.1 | 9.84 | †DSS |
| lysozyme | A13 | A33 | 15.7 | 21.5 | 15.7 | † |
| lysozyme | A33 | A116 | 13.3 | 22.0 | 14.2 | † |
| lysozyme | A13 | A116 | 19.5 | 24.8 | 19.8 | * |
| ribonuclase A | A31 | A37 | 7.87 | 14.6 | 9.21 | * |
| ribonuclase A | A37 | A41 | 8.75 | 13.9 | 7.15 | * |
Our analyses also detected crosslinks involving random decoy sequence as follows: BRCA1/BARD1 low concentration analysis resulted in 2 random decoy hits; β-lactoglobulin analysis resulted in 1 random decoy hit. GFP BS3 analysis resulted in 1 random decoy hit. GFP DSS resulted in 1 random decoy hit. Lysozyme resulted in 1 random decoy hit.
When developing our analysis software our aim was to reduce or remove the need for manual validation of crosslinks while confirming our data agreed with published structures. Analyses run fully automatically with a stringent score cutoff are marked * or † in Table 1. To determine whether we could increase our discovery rate by allowing manual validation, we also ran our software with a less stringent cutoff and performed manual validation on the output. For all our analyses, this approach added just one new crosslink (see row marked ‡ in Table 1). The algorithm presented here was thus very effective, and essentially removed the need for manual analysis of our data. It is clear from the data that targeted mass MS/MS significantly contributed to the amount of data obtained from most samples. Because each step of the analysis can be automated by software algorithms, it is possible to envision a real-time application that can detect interlinked peptides by their isotopic signature during data acquisition. Interlinked peptides can then be selectively targeted for MS/MS analysis without the need to construct follow-up methods using predetermined mass targets.
Additional Crosslinking Analysis to Increase Observed Crosslinks
Having done an initial rapid analysis using single protein samples and a minimal number of MS runs, we performed two additional crosslinking reactions under different conditions to determine whether this would increase the number of crosslinks we could detect. These reactions were BRCA1/BARD1 using 1/50th of the previous concentration of BS3 crosslinker and GFP using the hydrophobic homologue of BS3, called DSS.
The two BRCA1/BARD1 reactions yielded 6 unique crosslinks. Three of these were detected under standard conditions and 5 under low crosslinker concentrations. Two crosslinks were common to both analyses (see standard concentration [SC] and low concentration [LC] data in Table 1). The two GFP reactions yielded 5 unique crosslinks of which 3 were common to both reactions (see GFP [BS3] and [DSS] data in Table 1). From these data it was apparent that performing additional crosslinking reactions under varying conditions does increase the number of observed crosslinks.
Crosslink Validation Using Published Protein Structures
Many authors have validated the accuracy of their crosslink assignments by analyzing proteins of known structure. 9–11, 20–22 Based on the distance between residues observed to have been crosslinked, assignments either agree or disagree with published atomic structures. The crosslinkers used in this study, DSS and BS3, are amine reactive and have a spacer arm of 11.4 Å. The cutoff for the distance allowed between the crosslinked lysine ε-amino groups is thus 11.4 Å. Lysine side chains have significant intrinsic flexibility, however, so a strict distance cutoff between lysine ε-amino groups could be misleading in certain cases. To get around this problem many authors have instead used a cutoff of 24 Å between lysine α-carbon groups (the carbon backbone of a protein is generally less flexible). To span this distance, the full extension of the crosslinker’s spacer arm and a specific orientation of the side chains of both the lysine residues is required.11, 23 In some cases, although the distance between two α-carbon groups may be within 24 Å, their ε-amino groups might not be able to come within 11.4 Å without a conformational change in the carbon backbone of the protein. In order to address this difficulty, we explored the structural feasibility of lysine-lysine crosslinking in a given PDB structure using Rosetta’s fixed backbone side-chain refinement protocol. Within this protocol, the potential crosslinking of a pair of lysines is tested by sampling sidechain rotamers from the Dunbrack rotamer library with moves kept or rejected based on the standard Rosetta full atom energy function supplemented with a distance constraint of 10Å on the pair of lysine NZ atoms.24 This establishes a framework for approximating side-chain flexibility while preventing steric clashes and maintaining physically realistic side-chain interactions. We consider crosslinking possible if the two NZ atoms are within 11.4Å at the end of the calculation. Rotamer sampling can fail to satisfy this restraint in one of two ways, either when no rotamers exist that could span the geometry set by the backbone or when the rotamers required to do so would increase the energy significantly by introducing, for example, steric clashes that cannot be resolved by the movement of other sidechains. This method aims to provide a more informed prediction as to whether a specific crosslink is likely to be possible given the published structural information available. We used this method both to analyze our own crosslinking results and to compare our data with previously published crosslinking data on the same proteins.
While 95% of the crosslinks we discovered are within the generic 24 Å cutoff between lysine α-carbons, only 40% have a distance of 11.4 Å or less between the crosslinked lysine ε-amino groups in the crystal structure (Table 1). After allowing amino acid side chain flexibility, 17 of the 20 crosslinks (85%) are within the 11.4 Å cutoff (see Table 1). Manual examination of the MS spectra for the three crosslinks outside the cutoff indicated that they were indeed observed (data not shown) and thus do not represent a problem on the part of our algorithm or MS analysis, but are more likely to have been the result of non-specific protein-protein interactions or aggregation in the test tube. The propensity of lysozyme to aggregate during crosslinking was confirmed to be higher than for the other proteins tested as crosslinking reactions performed at 10X protein concentration resulted in a soluble product for all proteins except lysozyme, which formed an insoluble precipitate after just 10 minutes of crosslinking at that concentration (data not shown). Based on comparison to published structures our method appears to have performed very well, as all detected either matched published structures or could be explained by biochemical artifacts which would be common to any crosslinking analysis method. Such artifacts could potentially be removed by gel filtration or SDS-PAGE purification of crosslinking products prior to MS analysis, however this is beyond the scope of the current work.
We used Rosetta to explore the published PDB structures for each protein and identify all possible crosslinks. Based on this modeling, we observed approximately 20% of the number of possible crosslinks (see Table S1 for detailed data). Lack of lysine reactivity could prevent a structurally possible crosslink from actually forming. We tested lysine reactivity by looking for dead-end peptides in our MS data. Dead-end peptides appear to be more common than crosslinked peptides and are thus a convenient means of determining whether a given lysine is reactive or not. Peptides containing lysines with a dead-end crosslinker were identified by database searching with SEQUEST using differential modifications. The search was performed twice, first using a 155.0946 dalton mass differential (NH4HCO3 quenched BS3), then repeated using a 156.0786 dalton mass differential (hydrolyzed BS3). The search parameters allowed for multiple dead-ends and multiple lysine miscleavages. This analysis (which we performed on all our data except for our low concentration BARD1/BRCA1 and our GFP DSS data) indicated that of the 98 lysine-lysine distances within 11.4 Å after minimization, both lysines were observed to be reactive in just 43 cases (Table S1). Given these data, we observed closer to 40% of the number of possible crosslinks.
Analysis of Previously Published Crosslink Data
Previous crosslinking studies on the proteins we analyzed in the current work vary in which crosslinks were detected and also in how many crosslinks were detected. In order to understand how other published crosslinking methods compare to our own in terms of both the number of crosslinks detected and also in the proportion of those detected that agree with the published structures, we used our modified version of Rosetta to reanalyze previously published crosslinking data. Table 2 summarizes all the crosslinks found during the current work along with crosslinks published by others on the same proteins where such data is available (see Table S2 for full details). Based on this analysis our method is comparable to other current methods in both sensitivity and accuracy. In addition our method is fast and requires no manual validation of the output, making it ideal for general use.
Table 2.
Summary of previously published crosslinking data on proteins studied in this paper (see Table S2 for full details).
| Total crosslinks observed | Total that fit published structure | % in agreement with published structure | References for these crosslinks |
|---|---|---|---|
| BRCA1/BARD1 | |||
| 6 | 6 | 100 | This paper |
| β-lactoglobuiln | |||
| 4 | 4 | 100 | 10 |
| 4 | 4 | 100 | 9 |
| 4 | 4 | 100 | This paper (inc A70) |
| GFP | |||
| 5 | 5 | 100 | This paper |
| Lysozyme | |||
| 2 | 0 | 0 | 10 |
| 3 | 0 | 0 | This paper |
| 2 | 1 | 50 | 9 |
| Ribonuclease A | |||
| 2 | 1 | 50 | 10 |
| 2 | 2 | 100 | 22 |
| 2 | 2 | 100 | 9 |
| 2 | 2 | 100 | 21 |
| 2 | 2 | 100 | This paper |
Lysozyme was the only protein where significant incongruity was reported amongst all authors. This is consistent with our suggestion that lysozyme has a propensity to aggregate during crosslinking.
Conclusions
We have successfully developed a novel method for mass spectrometry based analysis of crosslinked proteins. We validated our method using five proteins/complexes of known structure and have shown that, along with the accompanying software, our method is capable of rapidly and accurately identifying inter- and intra-protein crosslinks with no manual data analysis. Our software allows the automated analysis of a database of up to several hundred proteins, making it suitable for the analysis of large protein complexes and complex crosslinked samples along with a large number of decoy proteins. The rapid workflow and absence of manual data-processing requirements make this method ideal for high-throughput studies and will do much to advance the use of protein crosslinking in structural biology. The software can be obtained from: http://proteome.gs.washington.edu/software/peplynx/
Supplementary Material
Acknowledgments
The authors would like to thank Jan Seebacher and Oliver Rinner for help and discussion regarding protein crosslinking and MS analysis. This work was supported by P41 RR011823 and R01 GM40506 to T.N. Davis.
References
- 1.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98 (8):4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437 (7062):1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 3.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403 (6770):623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 4.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415 (6868):141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 5.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415 (6868):180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 6.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J Mass Spectrom. 2003;38 (12):1225–37. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 7.Ihling C, Schmidt A, Kalkhof S, Schulz DM, Stingl C, Mechtler K, Haack M, Beck-Sickinger AG, Cooper DM, Sinz A. Isotope-labeled cross-linkers and Fourier transform ion cyclotron resonance mass spectrometry for structural analysis of a protein/peptide complex. J Am Soc Mass Spectrom. 2006;17 (8):1100–13. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Muller DR, Schindler P, Towbin H, Wirth U, Voshol H, Hoving S, Steinmetz MO. Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal Chem. 2001;73 (9):1927–34. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 9.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5 (4):315–8. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. Protein cross-linking analysis using mass spectrometry isotope-coded cross-linkers and integrated computational data processing. J Proteome Res. 2006;5 (9):2270–82. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 11.Huang BX, Kim HY, Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J Am Soc Mass Spectrom. 2004;15 (8):1237–47. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q, Doneanu CE, Shaffer SA, Adman ET, Goodlett DR, Nelson SD. Identification of the interactions between cytochrome P450 2E1 and cytochrome b5 by mass spectrometry and site-directed mutagenesis. J Biol Chem. 2006;281 (29):20404–17. doi: 10.1074/jbc.M601785200. [DOI] [PubMed] [Google Scholar]
- 13.El-Shafey A, Tolic N, Young MM, Sale K, Smith RD, Kery V. “Zero-length” cross-linking in solid state as an approach for analysis of protein-protein interactions. Protein Sci. 2006;15 (3):429–40. doi: 10.1110/ps.051685706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao X, Afonso C, Fenselau C. Dissection of proteolytic 18O labeling: endoprotease-catalyzed 16O-to-18O exchange of truncated peptide substrates. J Proteome Res. 2003;2 (2):147–52. doi: 10.1021/pr025572s. [DOI] [PubMed] [Google Scholar]
- 15.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal Chem. 2008;80 (22):8799–806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8 (10):833–7. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 17.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection and MS/MS, spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal Chem. 2007;79 (15):5620–32. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 19.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4 (11):923–5. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 20.Lee YJ, Lackner LL, Nunnari JM, Phinney BS. Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J Proteome Res. 2007;6 (10):3908–17. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 21.Pearson KM, Pannell LK, Fales HM. Intramolecular cross-linking experiments on cytochrome c and ribonuclease. A using an isotope multiplet method. Rapid Commun Mass Spectrom. 2002;16 (3):149–59. doi: 10.1002/rcm.554. [DOI] [PubMed] [Google Scholar]
- 22.Schilling B, Row RH, Gibson BW, Guo X, Young MM. MS2Assign automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J Am Soc Mass Spectrom. 2003;14 (8):834–50. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 23.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10 (7):1293–304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misura KM, Chivian D, Rohl CA, Kim DE, Baker D. Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc Natl Acad Sci U S A. 2006;103 (14):5361–6. doi: 10.1073/pnas.0509355103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.