Abstract
The indenoisoquinoline NSC724998 is a novel topoisomerase I (Top1) inhibitor entering Phase I clinical trials at the National Cancer Institute, USA. In this study, 2-D PAGE analysis was performed on nuclear lysates prepared from HCT-116 and A375 cells treated with 1µM NSC724998 for 24 hrs and the differentially regulated spots identified by LC-MS/MS. 114 protein spot differentials were identified, 66 from A375 cells and 48 from HCT-116 cells. Proteins related to apoptosis changed specifically in A375 cells, whereas proteins involved in the ubiquitin-proteasome system were highly enriched in treated HCT-116 cells. Importantly, 12 differentially expressed proteins (ETFA, HCC1, HNRCL, KAP1, NPM, NUCL, PRDX1, PRP19, PSB6, RAE1L, RU2A and SFRS9) were common to both cell lines. Western blotting and immunocytochemistry confirmed significant nuclear upregulation of both the proteasome subunit PSB6 and the transcriptional repressor KAP1. Interestingly, increased KAP1 polypeptide was accompanied by enhanced phosphorylation at Ser824. Similar to γH2AX, KAP1 phosphorylation was consistently enhanced in a panel of 12 cell lines and in A375 xenografts following NSC 724998 treatment. In summary, these data enhance our understanding of protein dynamics in the nucleus following DNA damage and provide an alternate marker (pKAP1) with potential for monitoring clinical responses to Top1 poisons.
Keywords: topoisomerase I, indenoisoquinoline, DNA damage, KAP1, proteasome, γH2AX, nuclear proteome
Introduction
Indenoisoquinolines are 2nd generation toposisomerase I (TopI) inhibitors with enhanced pharmacokinetic and activity profiles relative to the classical TopI poison, camptothecin 1, 2. Indenoisoquinolines were first identified as atypical TopI inhibitors after COMPARE analysis 3 showed a strong correlation between the NCI 60 cell screen activities of NSC 314622 and camptothecins 4. This initial finding prompted extensive structure-activity relationship (SAR) studies to define clinically relevant analogs 5, 6. Indenoisoquinolines have several properties that distinguish them from camptothecins; the absence of the labile hydroxylactone E-ring which imparts increased chemical stability 5, they target different DNA sequences than camptothecin, and the TopI-drug-DNA complexes have greater stability 4, 7. Importantly, indenoisoquinolines are also active at nanomolar concentrations, overcome multidrug resistance and have promising in vivo efficacy 8, 9. As a consequence, development has progressed to a point where indenoisoquinolines are undergoing evaluation in Phase I clinical trials. Currently, pharmacodynamic monitoring of indenoisoquinoline activity relies on quantitation of γH2AX, a phosphorylated histone associated with the presence of double-stranded DNA breaks (DSSBs) 10. However, there is still considerable scope to define other protein markers of activity and to further elucidate molecular mechanisms underlying drug-induced DNA damage 11.
Several detailed studies have been reported investigating the effects of camptothecin on the cellular proteome 12–14. These reports highlighted profound alteration in proteins related to the DNA damage response and apoptosis induction and emphasized the importance of organelle enrichment in improving detection of differentially expressed proteins. In this study, a proteomics platform based on 2D-PAGE and LC-MS/MS was used to generate a nuclear proteome for A375 and HCT-116 cells treated with the indenoisoquinoline, NSC 724998. Results showed that enrichment of this subcellular organelle uncovered previously unidentified changes implicated in DNA damage and repair. Furthermore, in A375 cells, proteins implicated in apoptosis were overrepresented whereas under identical conditions in less sensitive HCT-116 cells, elements of the ubiquitin-proteasome system showed altered expression in response to treatment, highlighting the potential role of the proteasome in DNA damage response and Top1-DNA complex degradation. 15–17 Lastly, we identify the Ser824 phosphorylated form of the transcriptional repressor KAP1 (transcription intermediary factor 1β) 18 as a conserved marker of drug activity. It is hoped that the data presented here will ultimately provide useful insights in support of indenoisoquinoline clinical development.
Materials and Methods
Materials
Indenoisoquinoline NSC 724998 was obtained from the Drug Synthesis and Chemistry Branch of the Developmental Therapeutics Program, NCI (Rockville, MD). All cell lines were from the DCTD Tumor Repository (Frederick, MD). Primary antibodies were purchased as follows: proteasome 20s β6, HCC1, Nucleolin, KAP1, Nucleophosmin (NPM), P84 and VDAC1 from Abcam (Cambridge, MA); P53 from Santa Cruz Biotechnology, Inc (Santa Cruz, CA); p-KAP1 from Bethyl (Montgomery, TX); ETFA from Proteintech Group (Chicago, IL); γH2Ax from Upstate-Millipore (Billerica, MA), tubulin, β-actin from Sigma (St. Louis, MO). Secondary HRP-conjugated antibodies were from Jackson Immunoresearch (West Grove, PA). Unless otherwise indicated all other chemicals and inhibitors were from Sigma (St. Louis, MO).
Cytotoxicity and cell viability
Cells were placed into each well of a 96-well plate 24 h before treatment. Sample or buffer control (10 µL) were added to the appropriate wells and the plates were incubated at 37°C in a humidified CO2 incubator for the times indicated in the figure legends. To determine protein synthesis, the serum-containing media was replaced with serum- and leucine-free RPMI containing 0.03 µCi of [14C]-leucine. Incubation continued for 2–3 h at 37°C. The cells were harvested onto glass fiber filters using a PHD cell harvester, washed with water, dried with methanol, and counted. The results are expressed as % [14C]-leucine incorporation into the control-treated cells. To assay for cell viability, 10 µL WST reagent were added to each well and the plate was incubated for 2–4 h followed by reading the absorbance of the formazan dye product at 450nm, using a microplate reader (Bio-Tek Instruments). Experiments were performed at least twice with triplicate determinations for each point and the data pooled. The IC50 was defined as the concentration of NSC 724998 required to inhibit protein synthesis or cell viability by 50% relative to control-treated cells.
Cell Cycle Analysis
Cells were treated for the times noted in the figure legends, harvested and washed once with PBS. The samples were resuspended in 5 mL PBS and 5 mL cold 70% ethanol was added dropwise. After 5 min incubation, the cells were centrifuged, resuspended in 10 mL cold 70% ethanol and stored at 4°C for 1 h. The cells were washed twice with 5 mL PBS and resuspended in 1 mL PBS containing 50 µg/mL propidium iodide (Molecular Probes, Eugene, OR) and 100 µg/mL RNase A (Sigma, St. Louis, MO). After 1 h at 37°C, cell cycle analysis was performed using the FL3-A channel on a FACScan flow cytometer.
Western Blotting
Cells were treated for the times noted in the figure legends, washed twice in PBS and lysed in RIPA-CHAPS buffer (50mM Tris-HCl, pH7.4, 1mM EDTA, 1% CHAPS, 1% deoxycholate, 1x complete protease inhibitor (Roche, Indianapolis, IN)). Lysates were sonicated, centrifuged to remove insoluble material and protein concentration determined using the Coomassie Plus Protein assay (Pierce, Rockford, IL) according to the manufacturer’s instructions. SDS-PAGE was performed on a 10% NUPAGE Bis-Tris gel with subsequent transfer to a PVDF membrane by electroblotting. Following overnight blocking in 4% milk/TBS, membranes were incubated with primary antibody for 2 h, washed several times in 4% milk/TBS and incubated with the secondary, peroxidase-conjugated antisera for 2 h. Bands were visualized using enhanced chemiluminescence reagents ECL (Amersham, GE Healthcare, Piscataway, NJ) according to the manufacturer’s protocol. Immunoblots were scanned using a Kodak Image Station 4000 Pro and captured using Kodak Molecular Imaging software (Carestream Health, New Haven CT).
Immunocytochemistry
Cells, grown on glass coverslips until 60–80% confluent, were treated with 1 µM NSC 724998 for 24 hours. Coverslips were then washed twice with PBS and fixed with 4% paraformaldehyde for 20 minutes, followed by blocking and permeabilization with the addition of PBS/4% BSA/0.2% saponin for 4 hours. Primary antibody was then added to cells in the same buffer followed by overnight incubation. Cells were rinsed 4 times with PBS/ 4% BSA/ 0.2% saponin and Alexa488-conjugated secondary antibody (Molecular Probes, Eugene OR) added (1 µg/ml final). After 2 hours incubation, slides were washed extensively in PBS, mounted in Prolong Gold (Invitrogen, Carlsbad CA) and visualized using a 63x objective on a DM-LB compound fluorescence microscope (Leica Microsystems, Bannockburn, IL) equipped with Bioquant software (Bioquant Corp., Nashville TN).
Immunohistochemistry
Paraffin embedding and staining of sections from A375 xenograft tumors was performed by the Pathology/Histotechnology Laboratory (PHL) at NCI-Frederick (Frederick, MD). In brief, γ-H2AX staining was performed using a Bond-Max autostainer (Leica Microsystems, Bannockburn, IL) with Heat-Induced Epitope Retrieval (HIER) in citrate buffer, and the following antibody incubation times; anti-γH2AX (Upstate-Millipore, Billerica MA) (1:100) 60 min, Streptavidin Alexa 488 conjugate (1:100) 30 min (Invitrogen, Carlsbad CA), DAPI counterstain (1:25,000 in PBS) 10 min. Stained sections were then mounted with Prolong Gold antifade reagent (Invitrogen, Carlsbad MA). For pKAP1 (Ser824) staining the following procedure was used; 2% Normal Goat Serum block 20 min (Vector Labs, Burlingame CA), pKAP-1 (Bethyl, Montgomery, TX), (1:250) 60 min, 2Ab Goat a/Rabbit, Alexa 488 conjugated (1:100) 30 min (Molecular Probes, Eugene OR), DAPI counterstain (1:25,000 in PBS) 10 min followed by mounting with Prolong Gold (Invitrogen, Carlsbad CA). Images were then captured using a 40x objective on a DM-LB compound fluorescence microscope (Leica Microsystems, Bannockburn, IL) equipped with Bioquant software (Bioquant Corp., Nashville TN).
Protein Preparation and Nuclei Extraction
A375 and HCT-116 cells at 60–70% confluence in T75 flasks containing 15ml RPMI-1640 were treated for 24 h with 1 µM indenoisoquinoline (NSC 724998) or control diluent. Cells were then washed twice with PBS, thoroughly drained, and lysed with buffer containing 7 M urea, 2 M thiourea, 4% (W/V) CHAPS, 1% v/v Pharmalyte 3–11 (GE Biosciences, Piscataway, NJ), 40 mM dithioerythritol (DTE), 50 mM Tris-HCl (pH 7.4) containing 1x protease inhibitor cocktail tablet. Cell lysates were sonicated twice for 10 sec and centrifuged to remove any insoluble material. For nuclear sample preparation, the cell pellets were resuspended with 1x hypotonic buffer (CelLytic NuCLEAR Extraction Kit, Sigma, St. Louis, MO), containing 1% protease inhibitor cocktail and incubated on ice for 20–30 min. The lysed cells were then homogenized using a glass Dounce homogenizer (Kontes, Fisher Scientific, Pittsburgh, PA) with 20 strokes on ice with frequent visualization to monitor progress. Following centrifugation at 2000g for 4 min at 4 °C to pellet the nucleus, the pellet was resuspended in 1 mL of 1x isotonic buffer, pelleted at 6000g for 2 mins, the supernatant removed, and washed 4 times, pelleting at 7000g for 2 mins each time. The dried pellet was lysed with the above lysis buffer and protein concentration determined. Following this, protein sample concentrations were validated by 1D-PAGE to ensure equal loading (see Supplementary Figure 1).
Two Dimensional Electrophoresis (2D-PAGE) and Image Analysis
For first dimension electrophoresis, 400 µg protein sample was adjusted to 450 µL with rehydration buffer containing 8 M urea, 2% (w/v) CHAPS, 25mM DTE, 1% v/v Pharmalyte 3–11 and 0.002% bromophenol blue, and applied to 24 cm Immobiline DryStrips (pH 3–11 NonLinear, Amersham GE Healthcare, Piscataway, NJ). Rehydration and isoelectric focusing (IEF) were performed in the Ettan IPGphor apparatus (Amersham, GE Healthcare, Piscataway, NJ) at 20°C, max. 80 µA per strip, according to the following program: 4 h at 0 V, 7 h at 30 V (rehydration); 1 h at 200 V, 1 h at 500 V, 1 h at 1000 V, then 8–12 h at 8000 V (IEF) until reaching 80–100 kVh. After isoelectrofocusing, the IEF strips were cut into three equal pieces, equilibrated for 20 min with shaking in 5 mL 50 mM Tris-HCl (pH 6.8), 6 M urea, 30% glycerol, 2% w/v SDS, and a trace of bromophenol blue containing 2% (w/v) DTE followed by 20 min in the same solution containing 2.5% w/v iodoacetamide in place of 2% DTE. For the second dimension, the strips were cut into 3 equal pieces and each piece placed on top of a NuPAGE 4–12% Bis-Tris ZOOM mini gel (Invitrogen, Carlsbad CA), sealed with 0.5% agarose in NuPAGE MES SDS running buffer and run for 40 min at 200V. The gels were washed with H2O, fixed in 7% acetic acid, 10% methanol for 30 min, and then stained overnight in SYPRO Ruby protein gel stain (Molecular Probes, Eugene OR). To decrease background fluorescence, the gels were destained in 7% acetic acid, 10% methanol for 30 min before imaging, using a Typhoon TRIO imager (GE Healthcare, Piscataway, NJ) set to a resolution of 100 µm. The images were analyzed with PDQuest software version 8.0.1 (Bio-Rad, Hercules CA), using the “automated spot detection and matching wizard” tool, and visually checking for undetected or incorrectly detected spots. After grouping the gels within a matched set, the density of each spot was normalized using the local regression mode and a paired t test with p ≤ 0.05 was applied for statistical analysis between control and treated groups. Three biological replicate experiments were performed and only those spots showing consistent modulation between all three replicates were selected for further analysis.
Gel Cutting and In-gel Tryptic Digestion of Proteins
Protein spots were excised from the SYPRO Ruby stained gels and transferred to a 96-well plate. Gel spots were washed twice with 100 mM ammonium bicarbonate, dehydrated with acetonitrile and dried in a SpeedVac concentrator SC110A (Savant, Fisher Scientific, Pittsburgh, PA). The dry gel spots were rehydrated with 50 mM ammonium bicarbonate buffer containing 12.5 ng/µL sequencing grade porcine trypsin (Promega, Madison, WI) for 45 min on ice. The buffer was then removed, replaced with 50 mM ammonium bicarbonate and digestion carried out at 37°C for 16 h. The supernatant was removed and the tryptic peptides were extracted first with 25 mM ammonium bicarbonate and thereafter with 5% formic acid for 20 min each. The supernatant was pooled with the combined extracts, dried in the SpeedVac concentrator and dissolved in 30 µL 1% formic acid, 5% acetonitrile prior to mass spectrometric analysis of 10µL aliquots.
Mass Spectrometry
The technique employed for protein identification involved microcapillary liquid chromatography - tandem mass spectrometry (LC-MS/MS). Trypsin digested protein samples were analyzed using a Finnigan LTQ ion trap mass spectrometer with an applied electrospray potential of 1.8 kV on a nanoelectrospray source, and capillary temperature of 200 °C. The IT (ion trap)-MS was operated in a data-dependant mode where each full MS scan was followed by three MS/MS scans, in which the three most abundant peptide molecular ions detected from the MS scan were dynamically selected for three subsequent MS/MS scans using a collision-induced dissociation (CID) energy of 35%. CID spectra were analyzed using SEQUEST in Bioworks version 3.3.1 SP1 (Thermo Electron Corp.), against the Swiss-Prot indexed human protein database of 14,306 protein entries in FASTA format (Feb, 2009 release). The variable modification of +57.021 Da and +15.995 Da were set for iodoacetamide alkylated cysteinyl, and for oxidized methionine residues, respectively. A mass tolerance of 1.0 AMU was used for mass measurements in the MS mode and 1.5 AMU for the MS/MS mode. Only peptides possessing tryptic termini (allowing for up to two internal missed cleavages) possessing Δ correlation scores (CN) ≥0.1 and charge-state-dependent cross-correlation (Xcorr) criteria, ≥2.0 for [M + H]1+ peptides, ≥2.2 for [M + 2H]2+ peptides, and ≥3.2 for [M + 3H]3+ peptides, were considered legitimate identifications. Only proteins identified by at least two tryptic peptides excluding the same sequence with different charge states were considered. Each sample was analyzed twice and only the proteins with Xcorr values >20 are listed in Supplementary Tables 1A and 1B.
Results
Establishing conditions for proteome analysis
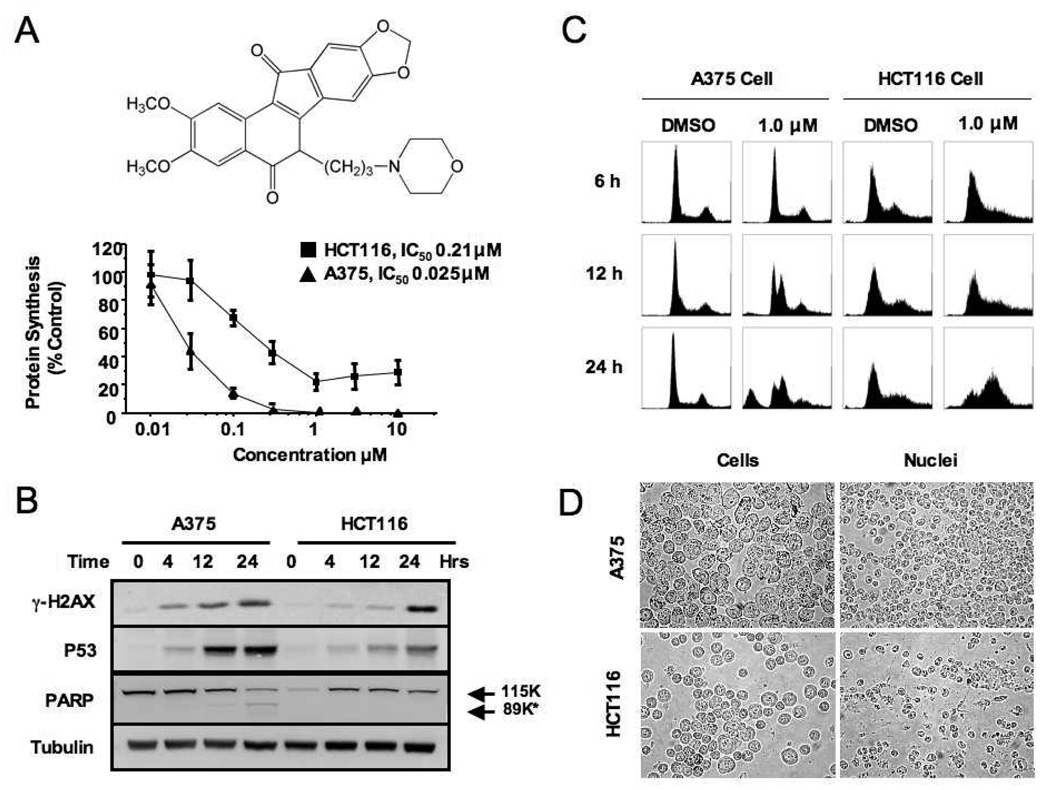
Two cell lines, A375 (melanoma) and HCT-116 (colon carcinoma) were selected for proteomic analysis based on their differential sensitivity to NSC 724998 in in vivo xenograft animal models; A375 tumors were very sensitive to treatment with NSC 724998 (structure illustrated in Figure 1A) while HCT-116 tumors were less sensitive. Several different assays were used to determine the relevant single-dose treatment point for in vitro proteomic analysis. The first assay, protein synthesis inhibition (Figure 1A), indicated that the two cell lines differed in sensitivity by one order of magnitude; A375 melanoma (IC50 25 nM) and HCT-116 colon carcinoma (IC50 210 nM). Cell viability assays using the cell proliferation reagent, WST-1, yielded similar results (IC50 0.2 and >10 µM, A375 and HCT-116 cells, respectively, see Supplementary Figure 2). Next, western blotting for DNA damage markers revealed that treatment with 1µM NSC 724998 enhanced γ-H2AX expression in both cell lines at 4 hr which then increased at 12 and 24 hrs (Figure 1B). In the third assay, cell cycle analysis (Figure 1C), A375 cells showed an S-phase arrest at 12 hrs of treatment followed by a strong induction of apoptosis at 24h whereas HCT-116 cells arrested in G2/M phase at 24h. The propensity of A375 cells to undergo apoptosis, as opposed to enter cell cycle arrest is inferred to be the basis for the marked difference in IC50, which is also supported by PARP cleavage (Figure 1B). Additionally, increasing the concentration of NSC 724998 still resulted in a G2-phase arrest for HCT-116 cells rather than apoptosis, indicating that the response is not concentration-dependent (Supplementary Figure 3). These results suggested that, although both cell lines undergo a DNA-damage response after treatment with NSC 724998, marked differences exist with respect to their cellular ‘fate’. On this basis, the decision was made to treat both cell lines with 1µM NSC 724998 for 24 hrs.
Figure 1.
Establishing conditions for proteome analysis. A) Upper, the structure of the non-camptothecin topoisomerase (Top1) inhibitor indenoisoquinoline NSC 724998. Lower, Protein synthesis assay to determine the activity of NSC 724998 following 24 hrs incubation for HCT-116 and A375 cells. B) Western blotting of whole cell lysates (WCL) from A375 and HCT-116 cells treated with 1uM NSC 724998 for markers of DNA damage and apoptosis. C) Cell cycle analysis of A375 and HCT-116 cells treated with 1µM NSC 724998 for 24 hrs. D) Phase contrast images of nuclei isolated from A375 and HCT-116 cells after Dounce homogenization and differential centrifugation.
Proteome Analysis
Our initial strategy involved proteomic analysis of both whole cell lysate (WCL) and the nuclear fraction of A375 and HCT-116 cells. However, due to the negligible difference between control and treated cells in the proteome of the WCL (data not shown), the decision was made to focus on generating a nuclear proteome. Following treatment, control and treated A375 and HCT-116 cells were disrupted by Dounce homogenization and nuclei purified by centrifugation, as described in the Methods section. The purity of isolated nuclei was confirmed using phase contrast microscopy (Figure 1D). Control and treated nuclear lysates from 3 separate biological replicates were then subjected to 2D-PAGE analysis. In this study, a ‘mini’ two-dimensional gel format, in which the 24 cm isoelectric focusing nonlinear strip (pH 3–11) was divided into three equal pieces and applied to three 4–12% Bis-Tris Zoom gels, was chosen for its high-throughput, simplicity in handling and reproducibility. Following the 2nd dimension, the gels were stained with SYPRO Ruby and analyzed for differential expression using PDQuest software. Over 300 spots were identified within each mini gel, where the intensity of spots was normalized by densitometry within each gel. The protein spots with significant changes in intensity between corresponding fractions (paired t-test, p < 0.05) in a consistent direction (increase or decrease) among three experiments were chosen for protein identification. A representative two-dimensional gel composite from an A375 experiment is shown in Supplementary Figure 4.
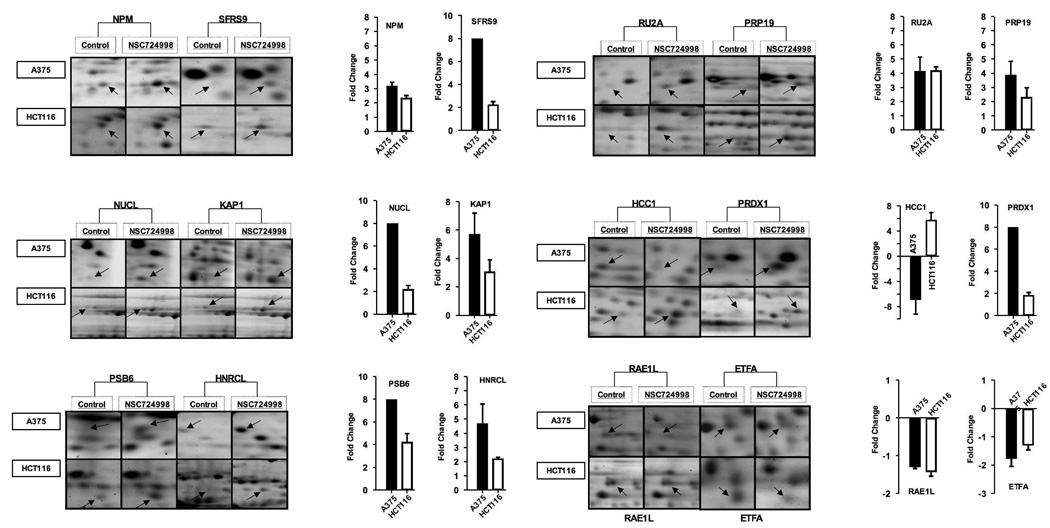
Analysis of gel sets showed that NSC 724998 altered 76 spots in A375 cells and 48 spots in HCT-116. These spots were excised, digested with trypsin, and subjected to nanospray microcapillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) for protein identification. This work culminated in the identification of 114 differentially regulated protein spots (66 in A375; 48 in HCT-116), where 96 were found to be unique after removal of redundant entries. A list of identified proteins associated with the identified peptide sequences, Xc score, Mr, PI, coverage and other MS/MS related data are shown in Supplementary Table 1. Among these, 12 proteins were conserved in both cell lines with 9 proteins (NPM, SFRS9, NUCL, KAP1B, PSB6, HNRCL, RU2A, PRP19, and PRDX1) up-regulated, 2 proteins (RAE1L, ETFA) down-regulated and one protein (HCC1) down-regulated in A375 and up-regulated in HCT-116 cells. Magnified views of spot changes for these conserved proteins with quantitation histograms are shown in Figure 2.
Figure 2.
Magnified view of gel images for 12 modulated proteins identified in both cell lines. Fold change values for spot intensities were determined using PDQuest software using data from 3 replicate gels. Standard error was not calculated for the protein spots with differentials over 8 fold.
Gene Ontology Analysis
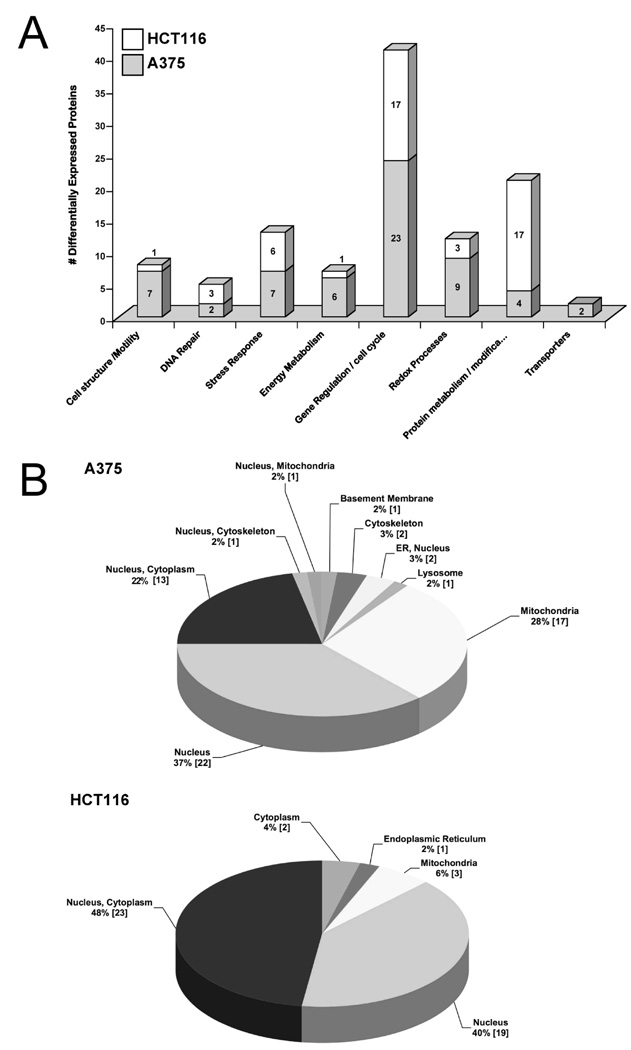
Differentially expressed proteins were broadly subdivided according to molecular function and subcellular localization by cross-referencing the PANTHER (http://www.pantherdb.org/) and SWISSPROT (http://www.expasy.ch) databases. Results were tabulated and are shown in Table 1, with graphical representation shown in Figure 3. In terms of molecular function, the subdivisions showing the greatest degree of change was gene regulation / cell cycle (23/60 proteins for A375 and 17/48 proteins for HCT-116) and protein metabolism/modification for HCT-116 cells (17/48 proteins (Figure 3A). As regards subcellular localization, 35 (58%) or 42 (88%) of the differentially modulated proteins identified in A375 or HCT-116 cells, respectively had either nuclear or nuclear/cytoplasmic localization (Figure 3B). For both cell lines, mitochondrial sub-cellular localization was the next highest value (28% A375 and 6% for HCT-116). It is noteworthy that apoptosis is associated with translocation of mitochondrial proteins to the nucleus. Therefore, the higher level of apoptosis in A375 cells provides a likely explanation for the increased levels of mitochondrial proteins found in the nuclei of these cells. Overall, these results provide independent verification that the proteomes are enriched for nuclear proteins.
Table 1.
Nuclear proteins differentially regulated after NSC 724998 treatment
| A375 | |||||||
|---|---|---|---|---|---|---|---|
| Bilogical Process | ID | Protein Name | Swissprot | Spot# | Molecular Function | Celluar Component | Mean FC T/C(n=3) |
| Cell structure /Motility | ANXA2 | Annexin A2 | P07355 | 5 | Calcium binding | Cytoskeleton | Appear |
| CHCH3 | Coiled-coil-helix-coiled-coil-helix domain protein 3 | Q9NX63 | 52 | Protein binding | Mitochondrion | 0.30±0.15 | |
| COF1 | Cofilin-1 | P23528 | 54 | Actin assembly | Nucleus, Cytoplasm | 0.44 ±0.33 | |
| LAMC1 | Laminin gamma-1 chain precursor | P11047 | 1 | Cell adhesion, migration | Basement membrane | Disappear | |
| LMNA | Lamin-A/C (70 kDa lamin) | P02545 | 32 | Structural molecule activity | Nucleus | Appear | |
| LMNA | Lamin-A/C (70 kDa lamin) | P02545 | 60 | Structural molecule activity | Nucleus | 1.97 ±0.68 | |
| SAP3 | Ganglioside GM2 activator precursor | P17900 | 13 | Sphingolipid activator | Lysosome | Appear | |
| TPM1 | Tropomyosin 1 alpha chain | P09493 | 8 | Stabilizing cytoskeleton | Cytoskeleton | Appear | |
| DNA Repair | PRP19* | Pre-mRNA-splicing factor 19 | Q9UMS4 | 36 | DNA binding, Protein ubiquination | Nucleus | 3.91 ±1.64 |
| RBBP4 | Retinoblastoma-binding protein 4 | Q09028 | 23 | Chromatin regulator | Nucleus | 4.82±2.77 | |
| Energy Metabolism | ALDOA | Fructose-bisphosphate aldolase A | P04075 | 49 | Fructose, actin binding | Nucleus, Cytoskeleton | 0.76 ±0.1 |
| ATPO | ATP synthase O subunit | P48047 | 69 | ATP synthesis | Mitochondrion | 4.34 ±1.68 | |
| ETFA* | Electron transfer flavoprotein subunit alpha | P13804 | 45 | Electron carrier activity | Mitochondrion | 0.59±0.14 | |
| FUMH | Fumarate hydratase | P07954 | 62 | Fumarate metabolism | Mitochondrion | 4.45±2.23 | |
| MDHM | Malate dehydrogenase | P40926 | 70 | Oxidoreduction | Mitochondrion | 2.54 ±0.58 | |
| THIL | Acetyl-CoA acetyltransferase) | P24752 | 46 | Transferase | Mitochondrion | 0.58±0.23 | |
| Gene Regulation / Cell Cycle | H2B1A | Histone H2B type 1-A | Q96A08 | 74 | Transcription regulation | Nucleus | 0.28±0.13 |
| H4 | Histone H4 | P62805 | 28 | Transcription regulation | Nucleus | Appear | |
| HCC1* | Nuclear protein Hcc-1 | P82979 | 24 | Transcription, Translation regulation | Nucleus | 0.19 ±0.12 | |
| HNRCL* | Heterogeneous nuclear ribonucleoprotein C-like 1 | O60812 | 20 | RNA binding | Nucleus | 4.70 ±2.38 | |
| HNRH1 | Heterogeneous nuclear ribonucleoprotein H | P31943 | 22 | mRNA processing, splicing | Nucleus | 0.36 ±0.18 | |
| HNRPD | Heterogeneous nuclear ribonucleoprotein D0 | Q14103 | 39 | mRNA processing, Transcription regulation | Nucleus | Appear | |
| HNRPL | Heterogeneous nuclear ribonucleoprotein L | P14866 | 60 | mRNA processing, splicing | Nucleus | 1.97 ±0.68 | |
| HNRPM | Heterogeneous nuclear ribonucleoprotein M | P52272 | 73 | mRNA processing, splicing | Nucleus | 5.12 ±1.77 | |
| MATR3 | Matrin-3 | P43243 | 61 | Transcription regulation, RNA binding | Nucleus | 2.90 ±0.53 | |
| NDKA | Nucleoside diphosphate kinase A | P15531 | 59 | Nucleotide binding | Nucleus, Cytoplasm | 0.14 ±0.10 | |
| NHPX | NHP2-like protein 1 | P55769 | 58 | mRNA processing, splicing | Nucleus | 0.25±0.28 | |
| NUCL* | Nucleolin (Protein C23) | P19338 | 11 | Transcriptional elongation | Nucleus | Appear | |
| PTRF | Polymerase I and transcript release factor | Q6NZI2 | 14 | Transcription regulation | Nucleus, Cytoplasm | Appear | |
| RAE1L* | mRNA-associated protein mrnp 41 | P78406 | 48 | mRNA export factor | Nucleus, Cytoplasm | 0.77 ±0.04 | |
| ROA1 | Heterogeneous nuclear ribonucleoprotein A1 | P09651 | 33 | mRNA processing, splicing | Nucleus, Cytoplasm | 4.40 ±0.96 | |
| ROA1 | Heterogeneous nuclear ribonucleoprotein A1 | P09651 | 72 | mRNA processing, splicing | Nucleus, Cytoplasm | 2.03 ±0.40 | |
| ROAA | Heterogeneous nuclear ribonucleoprotein A/B | Q99729 | 37 | mRNA binding | Nucleus | 4.60 ±1.75 | |
| ROAA | Heterogeneous nuclear ribonucleoprotein A/B | Q99729 | 38 | mRNA binding | Nucleus | 4.77 ±3.11 | |
| RU17 | U1 small nuclear ribonucleoprotein 70 kDa | P08621 | 64 | mRNA processing, splicing | Nucleus | 1.96 ±0.61 | |
| RU2A* | U2 small nuclear ribonucleoprotein A' | P09661 | 26 | mRNA processing, splicing | Nucleus | 4.19 ±1.67 | |
| RU2A* | U2 small nuclear ribonucleoprotein A' | P09661 | 29 | mRNA processing, splicing | Nucleus | Appear | |
| SFRS9* | Splicing factor, arginine/serine-rich 9 | Q13242 | 4 | mRNA processing, splicing | Nucleus | Appear | |
| SSBP | Single-stranded DNA-binding protein | Q04837 | 54 | Single-stranded DNA-binding | Mitochondrion | 0.44 ±0.33 | |
| THOC6 | THO complex subunit 6 homolog | Q86W42 | 44 | DNA binding | Nucleus | 0.54 ±0.33 | |
| KAP1/TIF1B* | Transcription intermediary factor 1-beta | Q13263 | 40 | Transcription regulation | Nucleus | Appear | |
| WDR74 | WD repeat protein 74 | Q6RFH5 | 50 | DNA binding | Nucleus | 0.41±0.21 | |
| Protein metabolism / modification | PSB6* | Proteasome subunit beta type 6 precursor | P28072 | 15 | Protein degradation | Nucleus, Cytoplasm | Appear |
| RM46 | 39S ribosomal protein L46 | Q9H2W6 | 21 | Ribosomal protein | Mitochondrion | 0.51 ±0.31 | |
| RS21 | 40S ribosomal protein S21 | P63220 | 57 | Ribosomal protein | Nucleus, Cytoplasm | 0.26 ±0.36 | |
| UBIQ | Ubiquitin | P62988 | 10 | Protein ubiquitination | Nucleus, Cytoplasm | Appear | |
| Redox Processes | ACADM | Medium-chain specific acyl-CoA dehydrogenase | P11310 | 42 | Oxidoreduction | Mitochondrion | 0.59 ±0.35 |
| ADRO | NADPH:adrenodoxin oxidoreductase | P22570 | 63 | Oxidoreduction | Mitochondrion | 2.51±0.36 | |
| G3P | Glyceraldehyde-3-phosphate dehydrogenase | P04406 | 72 | Oxidoreduction | Nucleus, Cytoplasm | 2.03 ±0.40 | |
| DHE3 | Glutamate dehydrogenase 1 | P00367 | 41 | Oxidoreduction | Mitochondrion | 0.88 ±0.23 | |
| GLRX5 | Glutaredoxin-related protein 5 | Q86SX6 | 7 | Electron carrier activity | Mitochondrion | Appear | |
| NDUA8 | NADH dehydrogenase [ubiquinone] 1 alpha subunit 8 | P51970 | 65 | Electron carrier activity | Mitochondrion | 2.09±0.13 | |
| NDUAD | NADH dehydrogenase [ubiquinone] 1 alpha subunit 13 | Q9P0J0 | 25 | Electron transport | Nucleus, Mitochondrion | 2.89±1.72 | |
| MTDC | Methylenetetrahydrofolate dehydrogenase | P13995 | 47 | Oxidoreduction | Mitochondrion | 0.36±0.06 | |
| PRDX1* | Peroxiredoxin-1 | Q06830 | 34 | Oxidoreduction | Nucleus, Cytoplasm | Appear | |
| Stress Response | CALR | Calreticulin precursor | P27797 | 35 | Chaperone, DNA binding | ER, Nucleus | Appear |
| CH10 | 10 kDa heat shock protein | P61604 | 67 | Chaperone | Mitochondrion | Appear | |
| CH60 | 60 kDa heat shock protein | P10809 | 19 | Chaperone | Mitochondrion | 3.64 ±2.11 | |
| GRP78 | 78 kDa glucose-regulated protein precursor | P11021 | 18 | Chaperone | ER, Nucleus | Appear | |
| HSP7C | Heat shock cognate 71 kDa protein | P11142 | 16 | Chaperone | Nucleus, Cytoplasm | Appear | |
| HSP7C | Heat shock cognate 71 kDa protein | P11142 | 17 | Chaperone | Nucleus, Cytoplasm | 3.65 ±1.03 | |
| NPM* | Nucleolar phosphoprotein B23 | P06748 | 3 | Chaperone | Nucleus | 3.22±0.40 | |
| NPM* | Nucleolar phosphoprotein B23 | P06748 | 15 | Chaperone | Nucleus | Appear | |
| PPIA | Peptidyl-prolyl cis-trans isomerase A | P62937 | 74 | Chaperone | Nucleus, Cytoplasm | 0.28±0.14 | |
| Transporters | NXF1 | Nuclear RNA export factor 1 | Q9UBU9 | 73 | mRNA transport | Nucleus, Cytoplasm | 5.12 ±1.77 |
| TIM13 | Translocase subunit Tim13 | Q9Y5L4 | 56 | Protein Transport | Mitochondrion | 0.42 ±0.28 | |
| HCT116 | |||||||
| Biological Process | ID | Protein Name | Swissprot | Spot# | Molecular Function | Celluar Component | Mean-fold T/C(n=3) |
| Cell structure /Motility | LMNB2 | Lamin-B2 | Q03252 | 2 | Structural molecule activity | Nucleus | 0.74 ±0.10 |
| DNA Repair | PRP19* | Pre-mRNA-splicing factor 19 | Q9UMS4 | 29 | DNA binding, Protein ubiquination | Nucleus | 2.34 ±1.11 |
| RFA2 | Replication protein A 32 kDa subunit | P15927 | 27 | Single-stranded DNA, protein binding | Nucleus | Appear | |
| RFA3 | Replication factor-A protein 3 | P35244 | 3 | Single-stranded DNA, protein binding | Nucleus | Appear | |
| Energy Metabolism | ETFA* | Electron transfer flavoprotein subunit alpha | P13804 | 33 | Electron carrier activity | Mitochondrion | 0.79 ±0.17 |
| Gene Regulation / Cell Cycle | CT011 | Protein C20orf11 | Q9NWU2 | 8 | Protein binding | Nucleus, Cytoplasm | Appear |
| ELOC | Transcription elongation factor B polypeptide 1 | Q15369 | 4 | Transcription, Ubiquitin-conjugation | Nucleus | 2.21 ±0.34 | |
| EXOS2 | Exosome complex exonuclease RRP4 | Q13868 | 33 | Exonuclease, rRNA processing | Nucleus | 0.79±0.17 | |
| EXOS5 | Exosome complex exonuclease RRP46 | Q9NQT4 | 44 | Exonuclease, rRNA processing | Nucleus | 0.20±0.19 | |
| FUBP2 | Far upstream element-binding protein 2 | Q92945 | 30 | mRNA processing, Transcription regulation | Nucleus, Cytoplasm | 3.31 ±2.44 | |
| HCC1* | Nuclear protein Hcc-1 | P82979 | 22 | Transcription, Translation regulation | Nucleus | 5.78±1.99 | |
| HNRPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | P07910 | 11 | RNA binding | Nucleus | 2.24 ±0.13 | |
| HNRCL* | Heterogeneous nuclear ribonucleoprotein C-like 1 | O60812 | 12 | RNA binding | Nucleus | 1.84 ±0.12 | |
| IMDH2 | Inosine-5'-monophosphate dehydrogenase 2 | P12268 | 28 | Oxidoreduction | Nucleus, Cytoplasm | 1.62 ±0.45 | |
| MAX | Protein max (Myc-associated factor X) | P61244 | 5 | Transcription regulation | Nucleus | 4.26 ±1.25 | |
| NUCL* | Nucleolin (Protein C23) | P19338 | 16 | Transcriptional elongation | Nucleus | 2.23 ±0.51 | |
| PCNA | Proliferating cell nuclear antigen | P12004 | 10 | Regulation of DNA replication | Nucleus | 2.37±0.27 | |
| PININ | Pinin | Q9H307 | 17 | mRNA processing, Transcription regulation | Nucleus | 3.09±1.43 | |
| RAE1L* | mRNA-associated protein mrnp 41 | P78406 | 45 | mRNA processing, transport | Nucleus, Cytoplasm | 0.71 ±0.08 | |
| RU2A* | U2 small nuclear ribonucleoprotein A' | P09661 | 40 | mRNA processing, splicing | Nucleus | 4.21 ±0.41 | |
| SFRS9* | Splicing factor, arginine/serine-rich 9 | Q13242 | 24 | mRNA processing, splicing | Nucleus | 2.28 ±0.41 | |
| KAP1/TIF1B* | Transcription intermediary factor 1-beta | Q13263 | 17 | Transcription regulation | Nucleus | 3.09 ±1.43 | |
| Protein metabolism / modification | CSN8 | COP9 signalosome complex subunit 8 | Q99627 | 6 | Regulator of the ubiquitin conjugation | Nucleus, Cytoplasm | Appear |
| PRS10 | 26S protease regulatory subunit S10B | P62333 | 43 | Transcription, protein degradation | Nucleus, Cytoplasm | 2.54 ±1.07 | |
| PRS6A | 26S protease regulatory subunit 6A | P17980 | 14 | Transcription, protein degradation | Nucleus, Cytoplasm | 3.91 ±2.91 | |
| PRS6B | 26S protease regulatory subunit 6B | P43686 | 13 | Transcription, protein degradation | Nucleus, Cytoplasm | Appear | |
| PRS8 | 26S protease regulatory subunit 8 | P62195 | 42 | Transcription, protein degradation | Nucleus, Cytoplasm | 1.26 ±0.27 | |
| PSA3 | Proteasome subunit alpha type 3 | P25788 | 7 | Protein binding, degradation | Nucleus, Cytoplasm | 2.94±1.50 | |
| PSA4 | Proteasome subunit alpha type 4 | P25789 | 41 | Protein binding, degradation | Nucleus, Cytoplasm | 5.43 ±1.78 | |
| PSA5 | Proteasome subunit alpha type 5 | P28066 | 9 | Protein binding, degradation | Nucleus, Cytoplasm | 5.59±1.81 | |
| PSA6 | Proteasome subunit alpha type 6 | P60900 | 21 | Protein binding, degradation | Nucleus, Cytoplasm | Appear | |
| PSA7 | Proteasome subunit alpha type 7 | O14818 | 40 | Protein binding, degradation | Nucleus, Cytoplasm | 4.21 ±0.41 | |
| PSB2 | Proteasome subunit beta type 2 | P49721 | 19 | Protein degradation | Nucleus, Cytoplasm | Appear | |
| PSB4 | Proteasome subunit beta type 4 precursor | P28070 | 18 | Protein degradation | Nucleus, Cytoplasm | Appear | |
| PSB5 | Proteasome subunit beta type 5 precursor | P28074 | 39 | Protein degradation | Nucleus, Cytoplasm | Appear | |
| PSB6* | Proteasome subunit beta type 6 precursor | P28072 | 5 | Protein degradation | Nucleus, Cytoplasm | 4.26 ±1.25 | |
| PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | P55036 | 15 | Protein binding, degradation | Nucleus, Cytoplasm | Appear | |
| PSDE | 26S proteasome non-ATPase regulatory subunit 14 | O00487 | 23 | Protein binding, degradation | Nucleus, Cytoplasm | Appear | |
| PSME3 | Proteasome activator complex subunit 3 | P61289 | 26 | Protein binding, proteasome activation | Nucleus, Cytoplasm | Appear | |
| Redox Processes | HEM6 | Coproporphyrinogen III oxidase, | P36551 | 34 | Oxidoreduction | Mitochondrion | 0.43 ±0.36 |
| P5CR1 | Pyrroline-5-carboxylate reductase 1 | P32322 | 31 | Oxidoreduction | Mitochondrion | 0.68 ±0.05 | |
| PRDX1* | Peroxiredoxin-1 | Q06830 | 38 | Oxidoreduction | Cytoplasm | 1.88 ±0.37 | |
| Stress Response | ENPL | Heat shock protein 90 kDa beta member 1 | P14625 | 1 | Chaperone | Endoplasmic reticulum | Disappear |
| HSP74 | Heat shock 70 kDa protein 4 | P34932 | 16 | Chaperone | Cytoplasm | 3.09 ±1.43 | |
| NPM* | Nucleolar phosphoprotein B23 | P06748 | 10 | Chaperone | Nucleus | 2.37±0.27 | |
| PARK7 | Protein DJ-1 (Oncogene DJ1) | Q99497 | 20 | Chaperone | Nucleus, Cytoplasm | 2.62 ±0.73 | |
| PPIH | Peptidyl-prolyl cis-trans isomerase H | O43447 | 37 | mRNA splicing, Chaperone | Nucleus | 1.26±0.30 | |
| TCPB | T-complex protein 1 subunit beta | P78371 | 29 | Chaperone | Nucleus, Cytoplasm | 2.34 ±1.11 | |
Proteins conserved between cell lines
Figure 3.
Gene ontology analysis. Protein identifications for all 114 differentially regulated protein spots (66 from A375 cells and 48 from HCT-116 cells) were manually interrogated using PANTHER (http://www.pantherdb.org/) and SWISSPROT (http://www.expasy.ch) databases and results subdivided according to A) molecular function and B) subcellular localization.
Validation of conserved changes
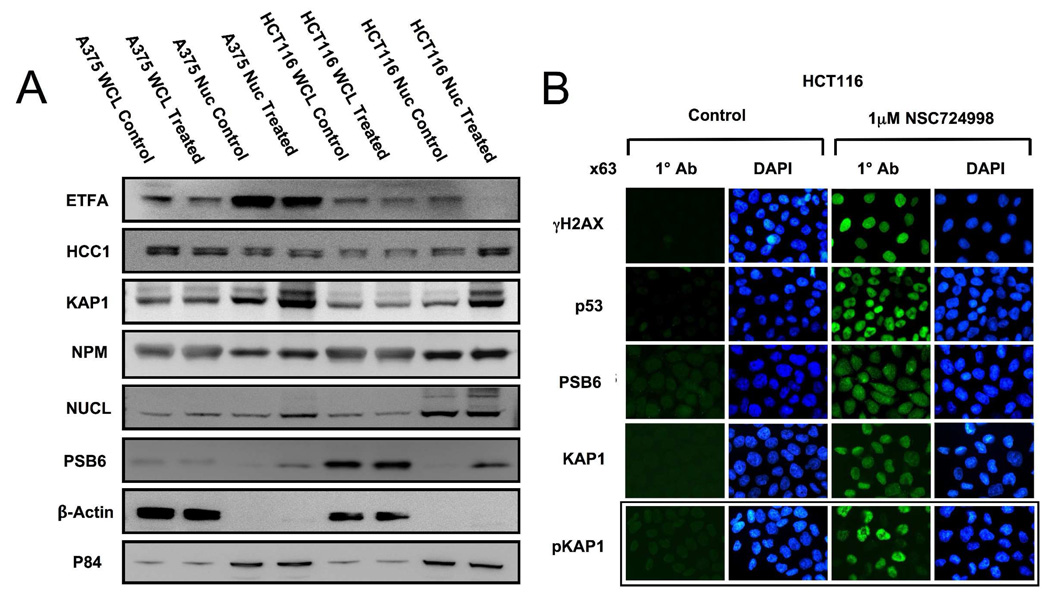
From the 12 conserved proteins identified by 2D-PAGE, 6 with available antibodies (NPM, PSB6, ETFA, KAP1, HCC1, and NUCL) were selected for further validation. As shown in Figure 4A, PSB6, KAP1, NPM and NUCL were clearly up-regulated in the nucleus of treated A375 nuclear samples, while ETFA was down-regulated and HCC1 showed no change in protein levels. For HCT-116 cells, PSB6, KAP1 and HCC1 were upregulated in the nucleus following NSC 724998 treatment, while EFTA decreased and no change was observed for NPM and NUCL. Interestingly, those proteins modulated in the nucleus were either less modulated or not modulated at all in the whole cell lysate. Overall, these results confirmed data from the 2D-PAGE analysis to a large degree. Following this, immunocytochemistry was performed on HCT-116 cells using antibodies against PSB6 and KAP1, two proteins that exhibited the greatest degree of change in both cell lines (Figure 4B). Antibodies against γH2AX and p53 were included as controls. Results confirm that increased levels of nuclear localized γH2AX, p53, PSB6 and KAP1 were observed after NSC 724998 treatment. Lastly, the appearance of a higher molecular weight KAP1 band in treated nuclei (Figure 4A) led us to speculate that a phosphorylation state change may be responsible for this shift. Therefore, cells were also stained with an anti-phospho-KAP1 (Ser824) antibody. Results demonstrated significant increases in nuclear pKAP1 staining with treatment.
Figure 4.
Validation of conserved changes. A) Western blotting of total cellular and nuclear lysates from A375 and HCT-116 cells treated with 1µM NSC 724998 for 24 hr with antibodies against six conserved proteins (ETFA, HCC1, KAP1, NPM1, NUCL and PSB6). Control antibodies for cytoplasmic (β-actin) and nuclear compartments (p84) were included as controls. B) Immunocytochemistry of HCT-116 cells treated as described in Methods and stained with antibodies against γH2AX, p53, PSB6, KAP1 and pKAP1. The nuclear counterstain DAPI was added in the final wash step and cells visualized using a LEICA DM-LB compound microscope with a 63x objective.
Phospho-Ser824 KAP1 is a conserved marker of in vitro and in vivo NSC 724998 activity
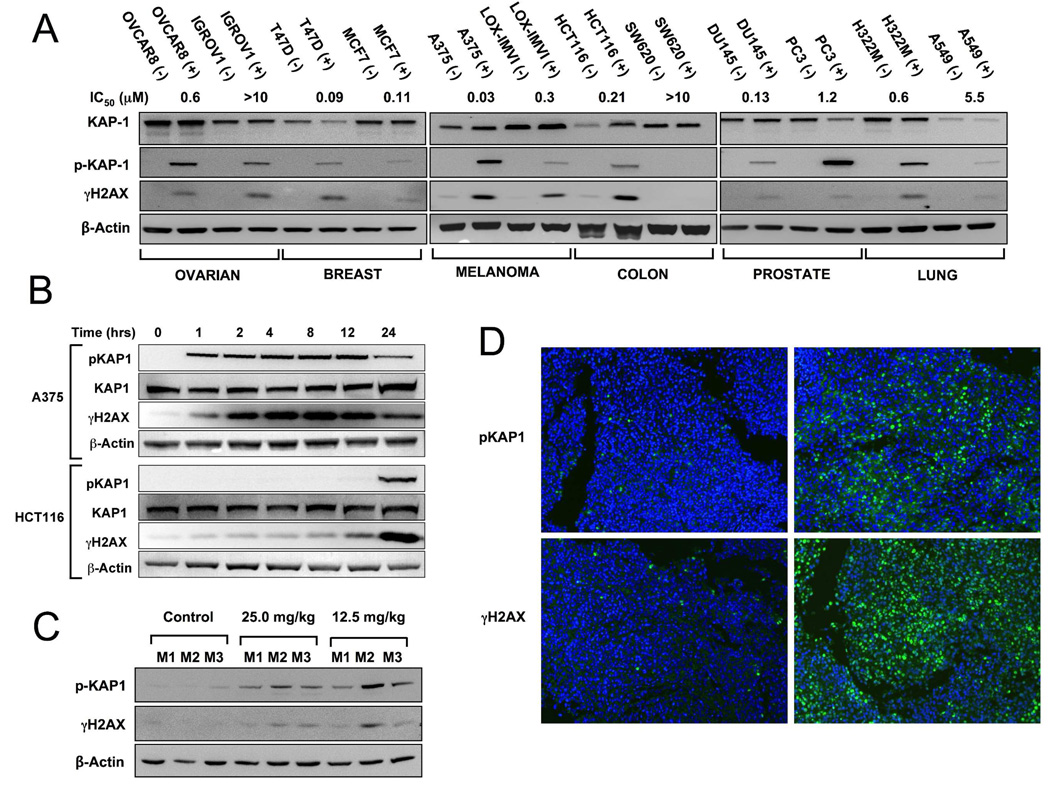
Data demonstrating that the phosphorylated form of KAP1 was qualitatively induced in A375 and HCT-116 cells after NSC 724998 treatment was then extended to a panel of 12 cell lines. Results shown in Figure 5A show that pKAP1 is absent in untreated cells and consistently induced in treated cells, in a manner similar to that observed for γH2AX. Additionally for each tumor type with the exception of the prostate cancer pair, the level of pKAP1 correlates with sensitivity to NSC 724998. Interestingly, levels of KAP1 polypeptide itself were not consistently elevated across the panel of lines. Similarities in the induction of γH2AX and pKAP1 were also evident from a time course in A375 and HCT-116 cells (Figure 5B) as well as in a dose response assay in A375 cells (Supplementary Figure 5). Next, western blotting was performed using lysates from A375 tumor xenografts collected 24 hr after treatment with either 12.5 or 25 mg/kg NSC 724998 (Figure 5C). Results show that for both γH2AX and pKAP1 significant increases in both markers are evident after 24 hr exposure. Lastly, staining of formalin fixed paraffin embedded (FFPE) sections taken from NSC 724998 treated animals demonstrated increases in pKAP1-specific staining after treatment (Figure 5D), indicating that this antigen may be suitable for evaluating clinical responses to NSC 724998.
Figure 5.
Phospho-Ser824 KAP1 is induced in vivo and in vitro following NSC 724998 exposure. A) Whole cell lysate was prepared from a diverse panel of 12 cell lines treated with 1µM NSC 724998 for 24 hours and blotted for expression of KAP1, pKAP1, γH2AX and β-actin. IC50 values generated using the C14-leucine viability assay are noted. B) A375 and HCT116 cells were treated with 1µM NSC 724998 for the indicated length of time, lysate prepared and probed for expression of KAP1, pKAP1, γH2AX and β-actin. C) Tumor lysate from A375 xenografts exposed to 12.5 or 25 mg/kg NSC 724998 for 24 hours (3 mice (M) per group) was probed for expression of pKAP1, γH2AX and β-actin. D) Tumors from NSC 724998 treated or control A375 xenografts were subjected to FFPE, sections cut and stained for expression of pKAP1 and γH2AX (Alexa488 staining protocol).
Discussion
As topoisomerase I inhibitors, indenoisoquinolines trap Top1 into cleavage complexes (TOP1ccs), generating replication-mediated DNA double-strand breaks (DSSB) and triggering a variety of cellular responses 1. These responses involve cell cycle control, DNA repair, and apoptosis, where effects can be both cell-type and dose specific 1. This study, which involved proteome analysis of nuclei from NSC 724998 treated HCT-116 and A375 cells, was an attempt to define specific nuclear targets of drug activity. Results from proteome analysis identified 96 non-redundant differentially regulated proteins, where 12 were conserved between cell lines. Subsequent gene ontology analysis confirmed that for both cell lines, proteins with a predicted nuclear localization were significantly enriched, providing overall confidence in the purification strategy. Broadly speaking, two major trends were evident between A375 and HCT-116 proteomes. Significant numbers of proteins implicated in apoptosis induction were identified in nuclei from treated A375 cells, whereas in HCT-116 cells proteins involved in the ubiquitin-proteasome system were over-represented.
Specific examples of apoptosis-associated proteins altered in A375 nuclei include ferredoxin reductase (ADRO/FDXR), which was upregulated, and transfers electrons from NADPH to cytochrome P450 via ferredoxin in mitochondria. Expression of this protein is regulated by p53 and has been shown, in certain cell types, to sensitize cells to apoptosis after challenge with DNA damaging agents by enhancing ROS production 19. This observation is noteworthy given that we also identify increased levels of HSP10 and HSP60 (CH10/CH60). These heat shock proteins stabilize a protein called fragile histidine triad gene product (Fhit), which itself increases the stability of ADRO/FDXR and promotes apoptosis 20. Increased levels were also observed for cell death-regulatory protein GRIM19 (NDUAD). This subunit of mitochondrial complex I potentiates apoptosis induced by agents such as IFN-β and retinoic acid 21. Decreased levels of peptidylprolyl isomerase A (cyclophilin A - PPIA) also deserve mention given that this molecule has been shown to be a pro-survival factor able to prevent hypoxia and cisplatin induced apoptosis through suppression of ROS 23.
Levels of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (G3P) were seen to increase in A375. Recent evidence supports the notion that G3P is involved in a variety of critical nuclear pathways in addition to apoptosis, where G3P is over-expressed and accumulates in the nucleus 24. Another metabolic enzyme, malate dehydrogenase 1 (MDHM) was similarly upregulated. This enzyme also serves as a transcriptional regulatory protein that directly induces p53-dependent apoptosis and cell cycle arrest by binding to p53-responsive elements in the promoter of downstream genes during conditions such as glucose deprivation 25. Conversely, Glutamate dehydrogenase 1 (DHE3) levels declined in A375 cells. This metabolic enzyme appears to be a cell survival factor given that inhibiting DHE levels using siRNA resulted in induction of cellular apoptosis 26.
Focusing on HCT-116, the proteasome is a ubiquitous multi-protein complex whose primary function is to degrade polyubiquitylated proteins 31. However, growing evidence implicates direct engagement of a nuclear ubiquitin-proteasome system (UPS) in DNA repair, replication, transcription and nuclear quality control 32, 33. The importance of the ubiquitin-proteasome system in the response to other Top1 poisons (e.g. camptothecin) has already been established 34. Although we observed changes in proteasome PSB6 subunits in both cell lines, many other subunits show changes exclusive to HCT-116 cells. Increased expression was seen for several 20S alpha subunits (PSA3-7) and beta forms (PSB2, 4–6). In addition, we observed increased expression of 26s regulatory ATPase subunits PRS6A, PRS6B, PRS8 and PRS10B along with the non-ATPase 26 regulatory subunits PSMD4 and PSDE. Increased levels were seen for PSME3, the gamma subunit of the 11S proteasome regulator along with CSN4 (COP9), which interacts with the 26s proteasome lid and regulates proteolysis 35. Therefore, it appears that in HCT-116 cells the composition of the nuclear proteasome is significantly altered after treatment with NSC 724998. It is interesting to speculate that proteasome subunit adaptations after drug treatment in HCT-116 cells serve to increase the efficiency of Top1cc clearance, given that the observed changes could confer an altered spectrum of substrate binding and degradation. Conversely, changes in proteasome composition may also reflect mechanism of transcriptional regulation 32.
HCT116 restricted changes also point to a DNA damage response and checkpoint arrest. Replication factors RFA2 and RFA3, which were upregulated, are phosphorylated by ATR upon DNA damage and play an important role in DNA excision repair. Interestingly, these proteins are found to co-localize with the γH2AX at sites of DNA damage 36. Proliferating cell nuclear antigen (PCNA), which was similarly upregulated, is a critical component of DNA replication machinery, with additional roles in DNA repair, cell cycle arrest and apoptosis 37. Lastly, increased expression of recognized cell survival factors PARK7 and TCPB suggest that HCT-116 is activating pathways involved in the prevention of programmed cell death 38 39.
The most important observations with respect to NSC 724998 pharmacodynamic marker discovery was that changes in twelve proteins were conserved between lines. Not surprisingly, most of the identified proteins were directly implicated in DNA damage responses. NPM (nucleophosmin) accumulates within the nucleolus after DNA damage and has been shown to interact with PARP-1 and PARP-2 40. PRP19 (pre-mRNA splicing factor 19) has an important role in DNA double-strand break (DSB) repair and premRNA splicing 41. NUCL (nucleolin), is nucleolar phosphoprotein involved primarily in the synthesis and maturation of ribosomes 42. NUCL is also implicated in the response to DNA damage given that in camptothecin or UV treated cells NUCL translocates from the nucleolus to the nucleoplasm, where it binds replication protein A and inhibits DNA replication initiation in a process that is p53-dependent 43. KAP1 (KRAB-associated protein 1), also known as TIF1-beta (transcriptional intermediary factor 1-β), mediates transcriptional repression through complexing with KRAB-domain transcription factors and increasing KRAB-mediated repression 44. KAP1 is also involved in maintaining chromatin condensation and is a substrate of the DNA damage-sensing kinase ATM 45.
Another protein showing a conserved change was HNRCL (heterogeneous nuclear ribonucleoprotein core protein C-like 1), which belongs to a family of abundant proteins (hnRNPS) implicated in mRNA processing, splicing control and the DNA damage response 46, 47. Similarly, proteins implicated in RNA metabolism including RU2A/SNRPA1 (small nuclear ribonucleoprotein polypeptide A), SFRS9 (splicing factor, arginine/serine-rich 9), and RAE1L (RNA export 1, S.pombe homolog) showed elevated expression after treatment. These changes likely reflect changes in overall RNA splicing, export and quality control in the nucleus that are commonly observed following DNA damage 48, 49.
Results obtained for HCC1 (cytokine induced protein 29 kD/RNA binding motif protein 39) are particularly interesting given the reciprocal changes observed between A375 and HCT-116 nuclei. HCC1 is a component of the nuclear matrix believed to be a transcriptional co-repressor and has recently been shown to have growth inhibitory properties, being able to arrest cells in G2/M and inhibit S-phase 50. It is interesting to speculate that increased levels of HCC1 in HCT-116 are responsible to some extent for G2/M arrest in this line. PRDX1 (peroxiredoxin 1) belongs to a family of antioxidant enzymes, and has itself been linked to regulation of c-myc and the DNA damage response 51. Finally, ETFA (electron transfer flavoprotein α-subunit), catalyzes the initial step of the mitochondrial fatty acid beta-oxidation. Although this protein is well characterized, and may have a role in apoptosis, the basis for altered expression in this proteome remains unresolved.
In the next phase of the project, changes in several conserved proteins were validated by western blotting. For the six proteins analyzed, results showed that the changes were restricted to nuclei and were largely similar to the 2D PAGE results. The KAP-1 data was especially interesting given that this protein is a substrate of ATM. Double-stranded DNA break formation as a consequence of replication fork collapse activates ATM (and other PI3-like kinases, ATR and DNA-PK), which leads to the phosphorylation of H2AX and other substrates including KAP1, culminating in transcriptional repression. The appearance, after treatment with NSC 724998, of a higher molecular weight form of KAP1 led us to investigate whether a phosphorylation state change was responsible for this shift. Immunocytochemistry showed that indeed, pKAP(Ser824) was significantly elevated after treatment. This trend was reproduced in total cell lysate from a panel of 12 cell lines and in tumors from A375 xenografts. The significance of this finding was that increases in pKAP1 closely mirrored those observed for γH2AX in all experiments. A time course of pKAP1 and γH2AX induction after NSC 724998 provided further support for this hypothesis, as did monitoring the induction of both antigens in A375 xenografts following treatment. Importantly, immunohistochemistry of paraffin-embedded section taken from treated A375 xenografts confirmed that the pKAP1 antibody used in this study could be used to study prevailing levels of DNA damage in archival specimens.
Conclusion
The clinical success of Top1/2 inhibitors has driven continued efforts to refine this class of drugs in terms of chemical stability and biological efficacy. Although indenoisoquinolines such as NSC 724998 have similarities with camptothecins, they are sufficiently divergent (e.g. DNA sequence specificity) to warrant independent proteomic study. In focusing our attention on the nucleus, we have defined several events that would otherwise have been hidden. This has allowed us to implicate the ubiquitin-proteasome as a possible basis for resistance to this agent. Also, preliminary data showing that pKAP1 has potential as a biomarker for NSC 724998 is encouraging but should be viewed in context, because other ATM substrates may have equal virtue. Given that phosphorylation state changes represent optimal treatment biomarkers, quantitative nuclear phosphoproteomics would be a logical next step for the unbiased identification of pharmacodynamic alternatives to γH2AX. Once the optimal set of phospho-state changes have been identified, proteomic technologies such as multiple reaction monitoring (MRM) may finally make it feasible to rapidly and sensitively quantify phosphopeptide biomarkers in patient samples as a means of evaluating clinical responses 52.
Supplementary Material
Figure 6.
Purified nuclei represent an ideal starting material for proteomic studies focusing on the effects of DNA damaging agents. In this report, cells were treated with Indenoisoquinoline NSC 724998, a 2nd generation topoisomerase I (TOP1) inhibitor followed by isolation of nuclei and identification of differentially regulated proteins by 2D-PAGE and LC-MS/MS. The identified changes have the potential to function as clinical biomarkers for this, and other, DNA damaging agents.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported [in part] by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute. NCI-Frederick is accredited by AAALACi and follows the Public Health Service Policy on the Care and Use of Laboratory Animals. All animals used in this research project were cared for and used humanely according to the following policies: The U.S. Public Health Service Policy on Humane Care and Use of Animals (1996); the Guide for the Care and Use of Laboratory Animals (1996); and the U.S. Government Principles for Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (1985).
Footnotes
Supporting Information Available: This material is available free at http://pubs.acs.org.
References
- 1.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6(10):789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 3.Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, Boyd MR. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J Natl Cancer Inst. 1989;81(14):1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 4.Kohlhagen G, Paull KD, Cushman M, Nagafuji P, Pommier Y. Protein-linked DNA strand breaks induced by NSC 314622, a novel noncamptothecin topoisomerase I poison. Mol Pharmacol. 1998;54(1):50–58. doi: 10.1124/mol.54.1.50. [DOI] [PubMed] [Google Scholar]
- 5.Strumberg D, Pommier Y, Paull K, Jayaraman M, Nagafuji P, Cushman M. Synthesis of cytotoxic indenoisoquinoline topoisomerase I poisons. J Med Chem. 1999;42(3):446–457. doi: 10.1021/jm9803323. [DOI] [PubMed] [Google Scholar]
- 6.Cushman M, Jayaraman M, Vroman JA, Fukunaga AK, Fox BM, Kohlhagen G, Strumberg D, Pommier Y. Synthesis of new indeno[1,2- c]isoquinolines: cytotoxic non-camptothecin topoisomerase I inhibitors. J Med Chem. 2000;43(20):3688–3698. doi: 10.1021/jm000029d. [DOI] [PubMed] [Google Scholar]
- 7.Antony S, Jayaraman M, Laco G, Kohlhagen G, Kohn KW, Cushman M, Pommier Y. Differential induction of topoisomerase I-DNA cleavage complexes by the indenoisoquinoline MJ-III-65 (NSC 706744) and camptothecin: base sequence analysis and activity against camptothecin-resistant topoisomerases I. Cancer Res. 2003;63(21):7428–7435. [PubMed] [Google Scholar]
- 8.Antony S, Agama KK, Miao ZH, Takagi K, Wright MH, Robles AI, Varticovski L, Nagarajan M, Morrell A, Cushman M, Pommier Y. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67(21):10397–10405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 9.Antony S, Kohlhagen G, Agama K, Jayaraman M, Cao S, Durrani FA, Rustum YM, Cushman M, Pommier Y. Cellular topoisomerase I inhibition and antiproliferative activity by MJ-III-65 (NSC 706744), an indenoisoquinoline topoisomerase I poison. Mol Pharmacol. 2005;67(2):523–530. doi: 10.1124/mol.104.003889. [DOI] [PubMed] [Google Scholar]
- 10.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teicher BA. Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem Pharmacol. 2008;75(6):1262–1271. doi: 10.1016/j.bcp.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Yu LR, Johnson MD, Conrads TP, Smith RD, Morrison RS, Veenstra TD. Proteome analysis of camptothecin-treated cortical neurons using isotope-coded affinity tags. Electrophoresis. 2002;23(11):1591–1598. doi: 10.1002/1522-2683(200206)23:11<1591::AID-ELPS1591>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Dai Z, Sadee W, Hancock WS. A pharmacoproteomics study of the cancer cell line EKVX using capillary-LC/MS/MS. Mol Pharm. 2006;3(5):566–578. doi: 10.1021/mp060002b. [DOI] [PubMed] [Google Scholar]
- 14.Yu Y, Wang LS, Shen SM, Xia L, Zhang L, Zhu YS, Chen GQ. Subcellular proteome analysis of camptothecin analogue NSC606985-treated acute myeloid leukemic cells. J Proteome Res. 2007;6(9):3808–3818. doi: 10.1021/pr0700100. [DOI] [PubMed] [Google Scholar]
- 15.Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61(15):5926–5932. [PubMed] [Google Scholar]
- 16.Desai SD, Liu LF, Vazquez-Abad D, D'Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272(39):24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 17.von Mikecz A. The nuclear ubiquitin-proteasome system. J Cell Sci. 2006;119(Pt 10):1977–1984. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- 18.Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J Mol Biol. 2009;385(3):800–810. doi: 10.1016/j.jmb.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu G, Chen X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21(47):7195–7204. doi: 10.1038/sj.onc.1205862. [DOI] [PubMed] [Google Scholar]
- 20.Okumura H, Ishii H, Pichiorri F, Croce CM, Mori M, Huebner K. Fragile gene product, Fhit, in oxidative and replicative stress responses. Cancer Sci. 2009;100(7):1145–1150. doi: 10.1111/j.1349-7006.2009.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angell JE, Lindner DJ, Shapiro PS, Hofmann ER, Kalvakolanu DV. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J Biol Chem. 2000;275(43):33416–33426. doi: 10.1074/jbc.M003929200. [DOI] [PubMed] [Google Scholar]
- 22.Jung H, Seong HA, Ha H. Direct interaction between NM23-H1 and macrophage migration inhibitory factor (MIF) is critical for alleviation of MIF-mediated suppression of p53 activity. J Biol Chem. 2008;283(47):32669–32679. doi: 10.1074/jbc.M806225200. [DOI] [PubMed] [Google Scholar]
- 23.Choi KJ, Piao YJ, Lim MJ, Kim JH, Ha J, Choe W, Kim SS. Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatininduced cell death. Cancer Res. 2007;67(8):3654–3662. doi: 10.1158/0008-5472.CAN-06-1759. [DOI] [PubMed] [Google Scholar]
- 24.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95(1):45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 25.Lee SM, Kim JH, Cho EJ, Youn HD. A nucleocytoplasmic malate dehydrogenase regulates p53 transcriptional activity in response to metabolic stress. Cell Death Differ. 2009;16(5):738–748. doi: 10.1038/cdd.2009.5. [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Sudderth J, Dang T, Bachoo RG, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockwin LH, Bumke MA, Yu SX, Webb SP, Collins JR, Hollingshead MG, Newton DL. Proteomic analysis identifies oxidative stress induction by adaphostin. Clin Cancer Res. 2007;13(12):3667–3681. doi: 10.1158/1078-0432.CCR-07-0025. [DOI] [PubMed] [Google Scholar]
- 28.Valencia CA, Ju W, Liu R. Matrin 3 is a Ca2+/binding protein cleaved by caspases. Biochem Biophys Res Commun. 2007;361(2):281–286. doi: 10.1016/j.bbrc.2007.06.156. [DOI] [PubMed] [Google Scholar]
- 29.Buzza MS, Zamurs L, Sun J, Bird CH, Smith AI, Trapani JA, Froelich CJ, Nice EC, Bird PI. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280(25):23549–23558. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- 30.Chua BT, Volbracht C, Tan KO, Li R, Yu VC, Li P. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol. 2003;5(12):1083–1089. doi: 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- 31.Adams J. The proteasome: structure, function, and role in the cell. Cancer Treat Rev. 2003;(29 Suppl 1):3–9. doi: 10.1016/s0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 32.Vlachostergios PJ, Patrikidou A, Daliani DD, Papandreou CN. The Ubiquitin-Proteasome System in cancer, a major player in DNA Repair. Part 1: Posttranslational regulation. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlachostergios PJ, Patrikidou A, Daliani DD, Papandreou CN. The Ubiquitin-Proteasome System in cancer, a major player in DNA Repair. Part 2: Transcriptional regulation. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motegi A, Murakawa Y, Takeda S. The vital link between the ubiquitin-proteasome pathway and DNA repair: impact on cancer therapy. Cancer Lett. 2009;283(1):1–9. doi: 10.1016/j.canlet.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33(12):592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24(5):1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochem Soc Trans. 2009;37(Pt 3):605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 38.Vasseur S, Afzal S, Tardivel-Lacombe J, Park DS, Iovanna JL, Mak TW. DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc Natl Acad Sci U S A. 2009;106(4):1111–1116. doi: 10.1073/pnas.0812745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abe Y, Yoon SO, Kubota K, Mendoza MC, Gygi SP, Blenis J. p90 ribosomal S6 kinase and p70 ribosomal S6 kinase link phosphorylation of the eukaryotic chaperonin containing TCP-1 to growth factor, insulin, and nutrient signaling. J Biol Chem. 2009;284(22):14939–14948. doi: 10.1074/jbc.M900097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118(Pt 1):211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 41.Lu X, Legerski RJ. The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem Biophys Res Commun. 2007;354(4):968–974. doi: 10.1016/j.bbrc.2007.01.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13(14):1911–1922. [PubMed] [Google Scholar]
- 43.Daniely Y, Dimitrova DD, Borowiec JA. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol Cell Biol. 2002;22(16):6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282(50):36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 46.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66(7):1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haley B, Paunesku T, Protic M, Woloschak GE. Response of heterogeneous ribonuclear proteins (hnRNP) to ionising radiation and their involvement in DNA damage repair. Int J Radiat Biol. 2009;85(8):643–655. doi: 10.1080/09553000903009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131(5):915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 49.Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, Bird G, Bentley D, Bertrand E, Kornblihtt AR. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137(4):708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Fukuda S, Pelus LM. Growth inhibitory effect of Hcc-1/CIP29 is associated with induction of apoptosis, not just with G2/M arrest. Cell Mol Life Sci. 2005;62(13):1526–1527. doi: 10.1007/s00018-005-5093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egler RA, Fernandes E, Rothermund K, Sereika S, de Souza-Pinto N, Jaruga P, Dizdaroglu M, Prochownik EV. Regulation of reactive oxygen species, DNA damage, and c-Myc function by peroxiredoxin 1. Oncogene. 2005;24(54):8038–8050. doi: 10.1038/sj.onc.1208821. [DOI] [PubMed] [Google Scholar]
- 52.Unwin RD, Griffiths JR, Whetton AD. A sensitive mass spectrometric method for hypothesis-driven detection of peptide post-translational modifications: multiple reaction monitoring-initiated detection and sequencing (MIDAS) Nat Protoc. 2009;4(6):870–877. doi: 10.1038/nprot.2009.57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.