Abstract
The nutrient-sensing hexosamine signaling pathway modulates the levels of O-linked N-acetylglucosamine (O-GlcNAc) on key targets impacting cellular signaling, protein turnover and gene expression. O-GlcNAc cycling may be deregulated in neurodegenerative disease, cancer, and diabetes. Studies in model organisms demonstrate that the O-GlcNAc transferase (OGT/Sxc) is essential for Polycomb group (PcG) repression of the homeotic genes, clusters of genes responsible for the adult body plan. Surprisingly, from flies to man, the O-GlcNAcase (OGA, MGEA5) gene is embedded within the NK cluster, the most evolutionarily ancient of three homeobox gene clusters regulated by PcG repression. PcG repression also plays a key role in maintaining stem cell identity, recruiting the DNA methyltransferase machinery for imprinting, and in X-chromosome inactivation. Intriguingly, the Ogt gene resides near the Xist locus in vertebrates and is subject to regulation by PcG-dependent X-inactivation. OGT is also an enzymatic component of the human dosage compensation complex. These ‘evo-devo’ relationships linking O-GlcNAc cycling to higher order chromatin structure provide insights into how nutrient availability may influence the epigenetic regulation of gene expression. O-GlcNAc cycling at promoters and PcG repression represent concrete mechanisms by which nutritional information may be transmitted across generations in the intra-uterine environment. Thus, the nutrient-sensing hexosamine signaling pathway may be a key contributor to the metabolic deregulation resulting from prenatal exposure to famine, or the ‘vicious cycle’ observed in children of mothers with type-2 diabetes and metabolic disease.
1. Introduction
The cycling of O-GlcNAc at serine and threonine residues is a nutrient-responsive, post-translational modification (PTM) that impacts target protein activity. The diverse set of proteins (over 600) that are regulated by this PTM are found both in the nucleus and cytoplasm and participate in many fundamental aspects of cellular homeostasis such as cell signaling, mRNA transcription, and protein stability [1, 2]. It has been twenty years since O-GlcNAc was first localized to transcription factors and chromatin [3-5]. This discovery suggested the possibility for nutrient-responsive control of the transcriptional machinery through O-GlcNAc modification.
Since those initial observations, our understanding of the role of O-GlcNAc cycling has matured. The enzymes responsible for the addition and removal of this modification have been identified and cloned in several model systems [6-9]. Additionally, upstream modulators that control the level of the donor sugar, UDP-N-acetylglucosamine, are also known [10-13]. Finally, a variety of inhibitors and sensors of O-GlcNAc cycling have been developed [14-16]. Armed with these tools, a number of laboratories have identified O-GlcNAc cycling as a key regulator of nutrient sensing (reviewed in [2]). Additionally, dysregulation of this pathway has a profound impact on diseases of nutrient sensing, such as type II diabetes, and neurodegeneration [17].
In this review, we will focus on the evolution of nutrient responsive O-GlcNAc cycling, detail the impact of O-GlcNAc cycling in the embryonic development of several model systems, and discuss its role in epigenetic programming of developmental fate.
2. Hexosamine signaling—a nutrient-responsive pathway evolves
O-GlcNAc cycling at serine and threonine residues is maintained by the action of OGT and the OGA, enzymes that add and remove O-GlcNAc, respectively. UDP-N-acetylglucosamine (UDP-GlcNAc) is the donor sugar for the transferase reaction. This reaction is the terminal step of the hexosamine biosynthetic pathway.
The hexosamine biosynthetic pathway is acutely sensitive to nutrient flux. Approximately 2-5% of total glucose is shunted into this pathway to produce UDP-GlcNAc. The hexosamine biosynthetic pathway integrates the nutrient status of the cell by utilizing glucose, acetyl-CoA, glutamine, and UTP to produce UDP-GlcNAc (Fig.1). Interestingly, glucosamine can rapidly increase the levels of UDP-GlcNAc by bypassing the rate-limiting enzyme in this pathway, glutamine:fructose-6-phosphate amidotransferase (GFAT) [10, 11]. OGT transmits this nutrient information throughout the cell by glycosylating target proteins. While UDP-GlcNAc is used throughout the secretory pathway as a building block for the synthesis of N-linked and O-linked glycans, as well as the assembly of GPI-anchors, nuclear and cytosolic O-GlcNAc-modified proteins appear to be particularly sensitive to physiological flux of the UDP-GlcNAc pools.
Figure 1.
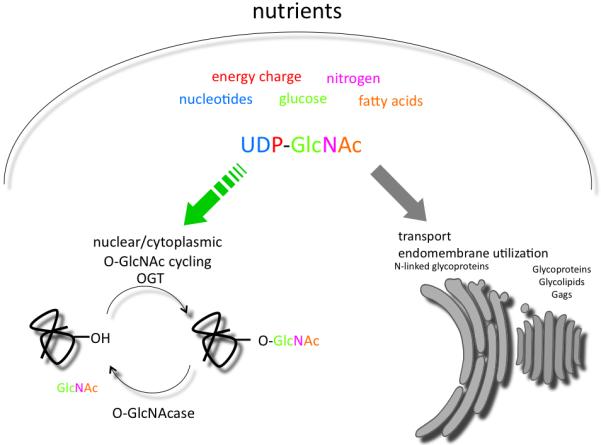
The nutrient-responsive hexosamine pathway. The concentration of UDP-GlcNAc is responsive to levels of the indicated precursors and serves as a sensor of nutrient status. Pools of UDP-GlcNAc are utilized in the nuclear/cytoplasmic compartment by O-linked GlcNAc transferase (OGT) or transported into the endoplasmic reticulum and golgi (shown in gray) for utilization in N-linked and O-linked glycoprotein biosynthesis and in the formation of glycosaminoglycans (GAGS). O-GlcNAc cycling is maintained by OGT and the O-GlcNAcase and is acutely sensitive to physiological flux of UDP-GlcNAc (green arrow).
2.1. Evolutionary conservation of the Hexosamine Signaling Pathway
The analogy between phosphorylation and O-GlcNAc modification is often made as these two signaling pathways share many common features and targets. However, these two signaling pathways differ in one important respect; where there are hundreds of kinases and phosphatases, most organisms contain only one OGT and one OGA. The exception appears to be vascular plants and mosses, each containing two OGTs; spindly (spy) and secret agent (sec) (reviewed in [18]). In animals, a single OGT appears to be the rule with zebrafish being the only exception. Zebrafish has two ogt genes termed ogta and ogtb. These genes do not appear to be the result of the teleost specific gene duplication event as other teleost fish genomes contain only one ogt gene [19, 20].
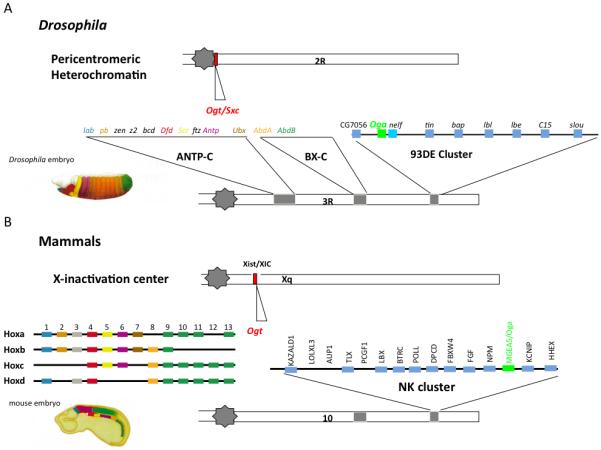
The genomic location of OGT encoding genes in some animals is also intriguing. In the fly, Ogt/sxc is present in a distinct heterochromatin-euchromatin boundary in the centromeric heterochromatin of chromosome 2R (41C) (Fig.2A). In humans (and all mammals), OGT is present on the X-chromosome at Xq13.1, quite close to the Xist locus and the X-inactivation center (XIC) (Fig.2B). The close proximity of Ogt genes to known heterochromatin boundaries may have significance for the regulation of its expression in mammals, as will be discussed in a later section.
Figure 2.
O-GlcNAc cycling is essential for PcG-mediated transcriptional repression. A) The three core polycomb repressive complexes found in Drosophila are shown. The mammalian (black text) and C. elegans (orange text) homologs are also listed. The yellow stars denote catalytic activity. Ph is modified by O-GlcNAc (blue hexagon). B) Epigenetic regulation is maintained by the concerted action of PcG and Trx. In Drosophila the PcG and Trx complexes bind to PRE/TREs (yellow). The two dashed lines represent conditions of repressed (left line) and active transcription (right line). The middle area represents ‘bivalent’ or ‘poised’ genes that are characterized by the competing activity of both complexes and exhibit both active and repressive chromatin marks. O-GlcNAc cycling at PcG/Trx or RNA PolII (or both) would impart a nutrient responsive element to the epigenetic machinery.
2.2. Ogt—single gene, many partners
An important question in the field is how a single enzyme can act on a vast array of tissues and on many different protein substrates in a regulated fashion. Part of the answer comes from the modular structure of the tetratricopeptide (TPR) domains in OGT. These domains produce a super helical structure containing an asparagine ladder that is critical for protein recognition [21]. Alternative splicing produces multiple isoforms, each with varying number of TPRs. This difference allows each isoform to modify a select subset of substrates [6, 7, 22]. Alternative splicing also targets OGT to different subcellular compartments [23]. Additionally, two-hybrid analysis using the TPR domain of OGT revealed a host of OGT-interacting proteins (OIP) that provide another layer of regulation by bridging the interaction of the enzyme with its with target substrates [24, 25].
2.3. Oga and the NK homeobox cluster
The OGA is also derived from a single gene in most metazoans known to encode this activity. Here again, splice variants exist which increase the diversity of transcripts encoded by this single gene. Interestingly, the presence of an identifiable O-GlcNAcase is not universally associated with the presence of an Ogt gene. As discussed elsewhere [2, 18] it is not clear whether this is due to gene loss in some organisms or acquisition of only one arm of the signaling pathway. Among sequenced genomes, the most distantly related O-GlcNAcase genes are from the trematode Schistosoma mansoni and nematodes such as Caenorhabditis elegans and Caenorhabditis briggsae.
One interesting aspect of the evolution of O-GlcNAcase and hexosamine signaling is the common presence of the O-GlcNAcase gene MGEA5 in a highly conserved cluster of Nk homeobox genes in vertebrates (Fig.2B). This Nk gene cluster is thought to be quite ancient, possibly predating the evolutionary divergence of sponges, bilaterians and cnidarians [26]. The NK genes present in the cluster function primarily in mesoderm development. The NK gene cluster is one of three large clusters in vertebrates, the Hox cluster (specifying mostly ectoderm), and the Parahox cluster (specifying endoderm) being the other two [27]. Large regions of synteny can be observed from Drosophila and Anopheles to humans providing evidence for an 800 million year old conserved region in the human genome [28]. As shown in Figure 2A, Drosophila oga (CG5871) is adjacent to the gene encoding Negative Elongation Factor (NELF) and the two genes are flanked by the downstream homeobox gene CG7056 (Hhex, in mammals). The NK homeobox cluster in vertebrates also contains oga adjacent to the canonic members of this group [29] (Fig.2B). While duplications have resulted in four Hox clusters present on different chromosomes in vertebrates, Oga is present in only one of the clusters associated with the Ladybird1 (Lbx1) gene on chromosome 10 in humans (19 in mouse). In organisms as diverse as Drosophila and the mouse, Lbx genes play key roles in muscle and neural development [30, 31]. Similar findings have been reported in zebrafish and chicken. The genomic position of the Oga within the highly conserved Nk homeobox gene cluster has intriguing implications for its regulation and will be discussed section 7.1.
3. Hexosamine signaling is essential for vertebrate development
O-GlcNAc cycling is essential in all vertebrates examined to date; deletion of Ogt in the mouse, fish, or frog results in extreme developmental defects, most often leading to death of the developing embryo [20, 32, 33].
3.1 Mammalian development
It has been difficult to study to role of OGT in early development using the mouse model. Classical disruption using homologous recombination proved to be embryonic stem cell lethal, necessitating the use of targeted disruption by the Cre-lox system [34]. Targeted deletion of Ogt in the developing oocyte also was lethal with death occurring around day 5 post fertilization. Knocking out Ogt in targeted tissue led to profound changes in all cell types examined, such as T-cell apoptosis and fibroblast senescence [35]. Neuronal specific ablation resulted in smaller pups with aberrant locomotor activity. These animals did not nurse well and died approximately 10 days after birth [35]. Thus, understanding Ogt function in the mouse has been problematic, although results to date suggest essential and pleiotropic roles for this enzyme during development.
EMeg32 is essential for the efficient production of UDP-GlcNAc. This enzyme was identified in a screen for differentially expressed genes in the hematopoietic precursor cells [12]. EMeg32 is an essential gene; mice homozygous for the Emeg32 deletion died at embryonic day 7.5, with a pronounced delayed development [13]. Deletion of the gene encoding EMeg32 drastically reduced the intracellular pools of UDP-GlcNAc, but did not eliminate UDP-GlcNAc, as a salvage pathway [36] also maintains this nucleotide sugar. The reduction in UDP-GlcNAc disproportionately affected the levels of nuclear and cytoplasmic O-GlcNAc modified proteins versus N-linked glycosylated proteins found in the secretory pathway [13]. This disparity reinforces the idea that fluctuating levels of UDP-GlcNAc are preferentially sensed through the action of O-GlcNAc cycling in the nucleus and cytosol (Fig.1).
3.2. Zebrafish development
Ablation of Ogt in zebrafish also disrupts early embryonic development. Alternative splicing of Zebrafish ogta and ogtb produce four and two transcripts, respectively. These transcripts are differentially expressed throughout development and appear to glycosylate unique substrates in vitro [19, 20]. Variants from each gene were depleted using injected morpholinos. Injected embryos exhibited shortened body axis, smaller heads and small or absent eyes. Additionally, these embryos displayed enhanced apoptosis and delayed epiboly. These striking phenotypes were accompanied by modest changes in overall O-GlcNAc levels. Overexpression of human O-GlcNAcase, like depletion of Ogt, shifted the balance of O-GlcNAc cycling, resulting in a lower, but not depleted level of O-GlcNAc modified proteins. That such profound developmental changes are accompanied by modest changes in the O-GlcNAc pool suggests that zebrafish development is acutely sensitive to fluctuating levels of O-GlcNAc.
3.3. Xenopus development
A similar pattern of Ogt expression is found in the diploid frog Xenopus tropicalis. Like zebrafish, Ogt expression is largely restricted to the nervous system in Xenopus [37]. Morpholino-targeted depletion of Ogt produced embryos with a bifurcated axis (spina bifida) or abrupt truncation at the posterior end with little or no tail bud [33]. Unfortunately, a separate morpholino directed to ogt had no effect on development, suggesting either the second morpholino did not deplete OGT effectively, or the phenotype is nonspecific. However, in both zebrafish and the frog, Ogt is highly expressed in the nervous system and appears to become restricted to the brain later in development while also resulting in a shortened body axis in both organisms.
4. Hexosamine signaling in the nematode; an exception to the rules?
In the nematode C. elegans, O-GlcNAc cycling is not essential for development. Like most other metazoans, C. elegans has only one OGT and OGA. Since animals with presumptive null alleles for the transferase (ogt-1) or the removing enzyme (oga-1) are viable and fertile, this genetic system provides a unique opportunity to examine the consequences of a complete lack of O-GlcNAc or enhanced levels of O-GlcNAc. While these genes are non-essential for development, their loss produced significant changes in macronutrient stores, as well as altered pathways responsive to insulin signaling, such as dauer formation, longevity and the stress response [38-41]. These results suggest that O-GlcNAc cycling is impacting pathways regulated by insulin signaling and that elevated O-GlcNAc blunts the insulin signaling pathway (insulin resistance), while a lack of O-GlcNAc improves insulin signaling (insulin hypersensitivity).
5. Hexosamine signaling is essential for Polycomb group repression
A growing body of evidence from the model systems Drosophila and C. elegans link hexosamine signaling to transcriptional repression typified by PcG proteins. In this section, we will summarize this body of evidence.
5.1. PcG and Hox gene regulation
Control of Hox gene regulation by PcG proteins and the trithorax (Trx) complex provides an extreme example of spatio-temporal control of gene expression. This precise gene regulation is responsible for the anterior-posterior segmentation in all bilaterian animals and is thought to be a major driving force in the evolution of body plan diversity [27]. PcG proteins were originally identified, and are best characterized and understood, in Drosophila as factors necessary to maintain Hox repression and prevent homeotic transformations [42, 43]. It is now appreciated that these repressive protein complexes are conserved throughout the animal and plant kingdoms and can act on a hundreds of genes controlling a plethora of developmental programs including stem cell fate, cancer development, X inactivation, and vernilization in plants [44-47]. While PcG proteins are comprised of a diverse group of proteins and regulate a variety of cellular programs, the unifying theme of PcG function is to act as an epigenetic regulator of cell fate and maintain cellular identity through many rounds of cell division.
5.2. PcG repressive complex
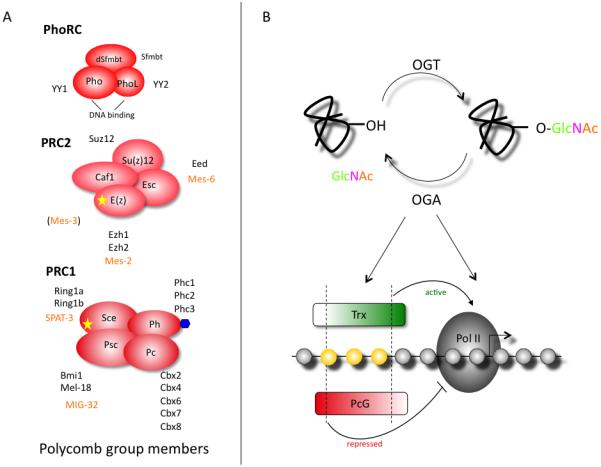
The PcG proteins act on the chromatin as a cooperative, mulitmeric complex known as the PcG repressive complex (PRC). These complexes come in several flavors and differ between species as to the number of proteins in each complex (Fig.3A). However, the basic organization and function is conserved. In Drosophila there are three main PcG repressive complexes: PRC1, PRC2, and PhoRC [48]. PRC2 is comprised of four core proteins: Enhancer of zeste (E(z)), Suppressor of zeste 12 (SU(z)12), Chromatin assembly factor 1 (Caf1), and extra sex combs (Esc). These proteins act in concert to trimethylate histone tails at lysine residue 27 on histone H3 (H3K27me3). This methyltransferase activity is found in the SET domain of Enhancer of zeste (E(z)). H3K27me3 is recognized by PRC1 through the chromodomain of Polycomb (Pc). The other components of PRC1 are Posterior sex combs (Psc), Polyhomeotic (Ph), and Sex combs extra (Sce). Sce is an E3 ubiquitin ligase that is responsible for the monoubiquitination of histone H2A at lysine 119. The methyltransferase activity of PRC2 and the ubiquitin ligase activity of PRC1 are both important for efficient PcG repression. In Drosophila, these complexes are recruited to the chromatin by regulatory elements called Polycomb Response Elements (PREs). While a similar response element is expected to be present in mammals, no such motif has yet been identified.
Figure 3.
The genes encoding enzymes of O-GlcNAc cycling are at discrete locations within the genomes of flies and mammals suggesting conserved regulation by higher order chromatin structure. OGT (red) is present near sites of heterochromatin-euchromatin boundaries on Chr2R in Drosophila (upper) and near the Xist locus in mammals (lower). The Oga gene (green) is present in the 93DE cluster of homeobox genes in the fly and in the orthologous NK cluster of homeobox genes in most vertebrates.
5.3. Ogt is super sex combs (Sxc): a homeotic gene in Drosophila
Two recent reports identify Ogt as the gene encoded by super sex combs (sxc), an essential component of PcG complexes in Drosophila [49, 50]. Ingham first described mutations in sxc in 1984 as causing a variety of homeotic transformations and death in pharate adults [51]. Interestingly, he showed that maternal stores of Sxc could suppress homeotic transformations due to sxc mutations. The lethal phenotype associated with sxc mutations was rescued by expression of Human Ogt [50]. This experiment clinches the role of Ogt as a PcG member and highlights the evolutionary conservation of PcG repression.
What role is Sxc/Ogt playing in PcG repression? Using ChIP-chip experiments, anti-O-GlcNAc antibodies bound over 1000 sites across the Drosophila genome, 490 of these sites were binding sites for the PcG factors Ph and/or PhoRc. The top 1% of O-GlcNAc sites were almost exclusively (111/114) bound by Ph and/or PhoRc [49]. The loss of Sxc/Ogt had little effect on the DNA binding capacity of other PcG proteins, as measured by ChIP-chip. Genes at these sites were derepressed, demonstrating that Ogt is essential for full repression by PcG. Interestingly, the H3K27me3 histone modification was unchanged at these sites. The requirement of Ogt activity in the rescue of the Sxc phenotype suggests that some of the proteins within PRC1, PRC2, and PhoRC complexes could be modified by O-GlcNAc. So far affinity chromatography to enrich O-GlcNAc modified proteins has only identified Ph as an O-GlcNAcylated substrate. However, detecting O-GlcNAc modification is notoriously difficult so this number may grow as more sensitive detection methods are applied to the PcG proteins.
From these experiments it appears that Ogt plays an integral role in the ability of the PcG proteins to repress genes appropriately, presumably acting downstream of PRC2 binding and H3K27 trimethylation. This leaves the door open for several points of regulation by O-GlcNAc cycling (Fig.3B). The O-GlcNAc modification of Ph could alter the activity of PRC1 and disturb histone H2A ubiquitination. Or perhaps loss of Sxc/Ogt disrupts the balance of the Trx and PcG complexes; note that the Trx complex is thought of as an anti-repressor, instead of an activator. O-GlcNAc modification in the SET domain of the Trithorax-related (SET) domain protein MLL5 was shown to alter its methyltransferase activity [52], providing another potential mechanism for influence of on gene expression. Finally, O-GlcNAc cycling could alter the activity of RNA Pol II at the promoter of these marked genes. The C-terminal Domain (CTD) of RNA Pol II is O-GlcNAc-modified in other organisms and is conserved in Drosophila [5, 53].
5.4. O-GlcNAc marks the promoters of numerous Caenorhabditis elegans genes
In parallel with studies in Drosophila on Sxc/Ogt, we have carried out ChIP-chip experiments using anti-O-GlcNAc antibodies and whole genome C. elegans tiling arrays [41]. These studies were controlled genetically by utilizing null alleles of ogt-1 and oga-1. We found that over 800 promoters were marked by O-GlcNAc, and that a number of transcriptional regulators associated with PcG repression were among those most heavily marked genes (Fig.4). Many of the genes identified are associated with innate immunity, longevity, and stress. In particular, genes found in the TGFβ, insulin, and MAPK pathways were heavily represented (Fig.4C). These classes of genes were also deregulated when the transcriptome of the O-GlcNAc cycling mutant strains were analyzed by expression arrays. Some 1,400 of the 13,000 expressed genes in the worm were deregulated in the ogt-1 null allele. Over 200 genes were altered in the O-GlcNAcase (oga-1) knockout strain. The genes deregulated in the O-GlcNAc cycling mutants include genes in the insulin and MAPK pathways and microRNAs regulating dauer, stress, longevity and innate immunity. Taken together, these findings suggest that O-GlcNAc, while not essential in the nematode, may play a critical role in transcriptional regulation.
Figure 4.
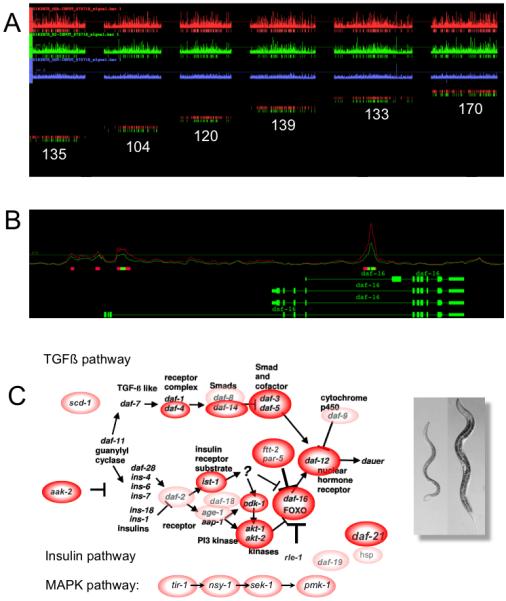
Whole genome analysis of O-GlcNAc cycling on promoters and altered gene expression in viable C. elegans mutants. A) Identification of 800 genes whose promoters are marked by O-GlcNAc using ChIP-on-chip tiling arrays. Strains analyzed were an oga-1 knockout (top row, red), wildtype (middle row, green) and ogt-1 knockout (bottom row, blue). The number of marks on each linkage group from the oga-1 knockout (red) and wildtype (green) is shown below. B) A representative gene from chromosome 1, daf-16, shows that O-GlcNAc is restricted to promoters. The O-GlcNAc signal is higher in fed (red line) than in starved (green line) worms, suggesting that the O-GlcNAc mark is nutrient-responsive. C.) Genes marked by O-GlcNAc (red and pink circles) are enriched in the TGFβ, insulin signaling, and MAPK pathways. A dauer worm (right panel) is shown next to a normal L4 larvae (left panel).
6. O-GlcNAc cycling and the implications for early development in vertebrates and nematodes
The discovery of Ogt as the Sxc PcG protein, and the presence of O-GlcNAc at the promoter of C. elegans genes, has cast the role of O-GlcNAc cycling in a new light. It is well established that O-GlcNAc cycling impacts a multitude of signaling pathways that coordinate nutrient sensing, growth and the stress response. It is now clear that O-GlcNAc cycling can act at the level of the chromatin through PcG to epigenetically regulate cell fate. Given this link, it is worth re-examining the vertebrate models of O-GlcNAc cycling. Are the lethal phenotypes seen in the Ogt mouse model, as well as the profound changes in the frog and zebrafish Ogt morpholino knockdown, consistent with altered PcG repression in the developing embryo?
Much work has been done to understand the role of PcG proteins in vertebrate development. Deletion of the PRC2 proteins in the mouse is lethal with most embryos arresting just after gastrulation (Reviewed in [44] see also[54-56]). Deletion of PRC1 members is less severe, resulting in anteroposterior transformations and changes in hematopoietic precursors [57, 58]. The exception is Ring1A/B. Like the PRC2 members, deletion of Ring1A/B in the mouse causes a gastrulation defect [59]. EZH2 (Enhancer of zeste homolog) regulates actin cytoskeletal dynamics and contributes to the embryonic arrest in knockout models [55]. A similar dysfunction of the actin cytoskeleton was seen in zebrafish embryos with morpholino depletion of Ogt [20]. Such similar phenotypes are interesting and may suggest, but do not prove, an essential role for Ogt in PcG-regulated development. By modulating O-GlcNAc in ES cells, either genetically or with inhibitors, it should be possible to separate the potential influence of O-GlcNAc cycling on essential upstream transcription factors (Oct4/sox2/Nanog) from its role in the maintenance of repression by PcG that is necessary for early development.
The C. elegans model provided the first genetic link between insulin signaling and O-GlcNAc cycling [38-41]. In these studies, no overt change in development was noted. This raises the question: does O-GlcNAc cycling influence PcG silencing in C. elegans? Functional analogs to both PRC2 and PRC1 have been identified in C. elegans (Fig.3, orange type). The PRC2-like complex is composed of the gene products from mes-2, mes-3, and mes-6[60]. These maternal effect sterile (mes) genes are termed “grandchildless” as the offspring are sterile [61]. Additionally, mes mutations also result in defects in anteroposterior axis patterning and neuroblast migration [62]. MES-2 contains a SET domain and, in complex with MES-3 and MES-6, provides the repressive H3K27 methylation mark. The activity of this complex is essential for silencing of genes on autosomes and repression of the X chromosome in the developing germ cells [63, 64].
Only recently has the functional PRC1 analog in C. elegans been identified [65]. It is composed of MIG-32 and SPAT-3A (Fig.3A). Mutants of mig-32 and spat-3A are defective in H2A ubiquitination and have some mild anteroposterior axis patterning defects that overlap with the PRC2 mes mutants. However in contrast to the grandchildless mes mutants, mig-32 and spat-3A mutants are viable and fertile [65]. These mild phenotypes associated with mig-32 and spat-3A mutations suggest that PRC1 activity (H2A ubiquitination) is not essential for full silencing by the PRC2 complex in C. elegans. Therefore, unlike Drosophila and vertebrates, PRC1-mediated ubiquitination is not essential for Hox gene regulation in C. elegans. In Drosophila, O-GlcNAc cycling may very well act on PRC1 activity; it will be of interest to see if this relationship is conserved in C. elegans.
7. Hexosamine signaling and epigenetics
Epigenetic effects may be modulated by environmental factors leading to changes in gene expression distinct from those encoded in the genome. Perhaps the most notable example of this kind of environmental influence is the modulation of Agouti coat colors in the offspring of mothers given a dietary supplementation [66]. Other epigenetic mechanisms must surely exist and one intriguing possibility is that hexosamine signaling may be able to exert epigenetic effects under certain circumstances. In this final section, we will propose mechanisms by which such epigenetic transmission might take place and how it may be manifested in human disease.
7.1. An O-GlcNAc link to X-inactivation
As detailed in previous sections, O-GlcNAc cycling appears to be essential for PcG repression in Drosophila and is likely to play a similar, conserved role in vertebrates. OGT has been found to be a component of the mammalian dosage compensation complex suggesting that O-GlcNAc cycling may play a role in this process [67]. The process of X-inactivation in mammals involves the random inactivation of one of the two X-chromosomes and the adjustment of transcription from the single active X to equal that of the autosomes [68, 69]. Recently it has been demonstrated that PcG action is essential for dosage compensation and X-inactivation in vertebrates [70, 71]. Thus, the levels of O-GlcNAc could influence the process of dosage compensation through modulation of PcG function.
Interestingly, the gene encoding Ogt in mammals is placed very close to the Xist locus (Fig.2B), the gene encoding the RNA involved in X-inactivation. Even more intriguingly, Ogt is one of a small number of genes that are highly regulated in embryonic stem cells during the process of X-inactivation, owing to its close proximity to Xist [68]. The timing of X-inactivation of these closely linked genes appears to be related to their distance from the Xist locus. Thus, a key modulator of PcG repression, OGT, is itself highly regulated during the process of X-inactivation in mammals. This finding suggests that a feedback mechanism may exist regulating the early expression of Ogt to mediate the subsequent process of X-inactivation.
Another feedback loop involves the regulation of OGT and its sister enzyme, OGA. ChIP-chip experiments in Drosophila identify the NK locus as a target of PcG [72]. If PcG regulates the expression of the NK locus, then OGT could control the expression of Oga, which is located within this cluster. Thus, OGT acting through PcG would repress Oga expression and bias the cell towards sustained O-GlcNAc levels. Improper or incomplete X-inactivation is associated with a number of diseases including autoinflammatory disease [73, 74]. Subtle changes in the levels of OGT/OGA, through mechanisms just mentioned, would alter cellular O-GlcNAc levels. These changes might have profound effects on subsequent development and stem-cell identity in lineages, such as those of the hematopoietic system involved in immune-cell function (Fig.5).
Figure 5.
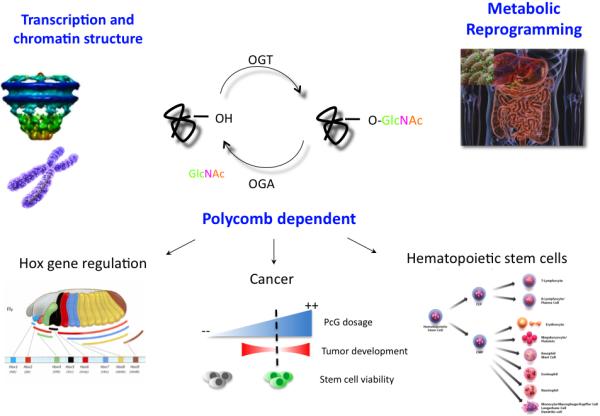
A summary of the multiple mechanisms by which O-GlcNAc cycling may impact development, stem cell fate, and metabolism. O-GlcNAc, which modifies the nuclear pore complex may function to organize the higher order structure of chromatin. X-inactivation contributes to the transcriptional regulation of OGT as described in the text. O-GlcNAc also directly modifies components of the transcription machinery and acts through polycomb repression. The resultant transcriptional effects may be manifested in changes in stem cell viability where PcG dosage is critically important (see text). In certain cell lineages such as the hematopoietic stem cell, these cell fate decisions may lead to changes in the immune system. The nutrient-responsive, fine-tuning of signaling pathways and changes in stem cell fate could contribute to the metabolic reprogramming seen in certain diet-induced maladies.
8. Conclusions
Given the large number of targets of PcG repression in vertebrates, including the many Hox genes discussed above, it is not surprising that PcG-mediated gene regulation during the prenatal period could have a profound impact on the developing fetus. Since O-GlcNAc is a nutritionally responsive modification and impacts PcG function, its effects may be amplified if exerted early in the developmental program. Two examples of the intrauterine environment programming the metabolic fate of the newborn are the epigenetic changes detected in people conceived during the Dutch famine or Dutch Hunger Winter of 1944-1945 and the ‘vicious cycle’ of type-2 diabetes perpetuated in the offspring of affected individuals [75-77]. The hexosamine pathway provides one means of explaining how prenatal exposure to excess nutrients in the intrauterine environment could play a role in the metabolic programming of the offspring.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Love D, Hanover J. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- [2].Hanover JA, Krause M, Love D. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kelly W, Hart G. Glycosylation of chromosomal proteins: localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell. 1989;57:243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- [4].Jackson S, Tjian R. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc Natl Acad Sci U S A. 1989;86:1781–1785. doi: 10.1073/pnas.86.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kelly W, Dahmus M, Hart G. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- [6].Lubas W, Frank D, Krause M, Hanover J. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- [7].Kreppel L, Blomberg M, Hart G. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- [8].Heckel D, Comtesse N, Brass N, Blin N, Zang K, Meese E. Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum Mol Genet. 1998;7:1859–1872. doi: 10.1093/hmg/7.12.1859. [DOI] [PubMed] [Google Scholar]
- [9].Wells L, Gao Y, Mahoney J, Vosseller K, Chen C, Rosen A, Hart G. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J Biol Chem. 2002;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- [10].Marshall S, Bacote V, Traxinger R. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- [11].Marshall S, Bacote V, Traxinger R. Complete inhibition of glucose-induced desensitization of the glucose transport system by inhibitors of mRNA synthesis. Evidence for rapid turnover of glutamine:fructose-6-phosphate amidotransferase. J Biol Chem. 1991;266:10155–10161. [PubMed] [Google Scholar]
- [12].Boehmelt G, et al. Cloning and characterization of the murine glucosamine-6-phosphate acetyltransferase EMeg32. Differential expression and intracellular membrane association. J Biol Chem. 2000;275:12821–12832. doi: 10.1074/jbc.275.17.12821. [DOI] [PubMed] [Google Scholar]
- [13].Boehmelt G, et al. Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 2000;19:5092–5104. doi: 10.1093/emboj/19.19.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Macauley M, Whitworth G, Debowski A, Chin D, Vocadlo D. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- [15].Dorfmueller H, Borodkin V, Schimpl M, Shepherd S, Shpiro N, Van Aalten DM. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J Am Chem Soc. 2006;128:16484–16485. doi: 10.1021/ja066743n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim E, Kang D, Love D, Hanover J. Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr Res. 2006;341:971–982. doi: 10.1016/j.carres.2006.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lazarus B, Love D, Hanover J. O-GlcNAc cycling: implications for neurodegenerative disorders. Int J Biochem Cell Biol. 2009;41:2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Olszewski N, West C, Sassi S, Hartweck L. O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sohn K, Do S. Transcriptional regulation and O-GlcNAcylation activity of zebrafish OGT during embryogenesis. Biochem Biophys Res Commun. 2005;337:256–263. doi: 10.1016/j.bbrc.2005.09.049. [DOI] [PubMed] [Google Scholar]
- [20].Webster D, Teo C, Sun Y, Wloga D, Gay S, Klonowski K, Wells L, Dougan S. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jinek M, Rehwinkel J, Lazarus B, Izaurralde E, Hanover J, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- [22].Lazarus B, Love D, Hanover J. Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology. 2006;16:415–421. doi: 10.1093/glycob/cwj078. [DOI] [PubMed] [Google Scholar]
- [23].Love D, Kochan J, Cathey R, Shin S, Hanover J. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116:647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- [24].Iyer S, Hart G. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- [25].Cheung WD, Sakabe K, Housley M, Dias W, Hart G. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Larroux C, Fahey B, Degnan S, Adamski M, Rokhsar D, Degnan B. The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol. 2007;17:706–710. doi: 10.1016/j.cub.2007.03.008. [DOI] [PubMed] [Google Scholar]
- [27].Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- [28].Danchin E, Pontarotti P. Statistical evidence for a more than 800-million-year-old evolutionarily conserved genomic region in our genome. J Mol Evol. 2004;59:587–597. doi: 10.1007/s00239-004-2648-1. [DOI] [PubMed] [Google Scholar]
- [29].Wotton K, Weierud F, Dietrich S, Lewis K. Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates. BMC Evol Biol. 2008;8:171. doi: 10.1186/1471-2148-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schafer K, Braun T. Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat Genet. 1999;23:213–216. doi: 10.1038/13843. [DOI] [PubMed] [Google Scholar]
- [31].Gross M, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- [32].Shafi R, Iyer S, Ellies L, O’Donnell N, Marek K, Chui D, Hart G, Marth J. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kenwrick S, Amaya E, Papalopulu N. Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev Dyn. 2004;229:289–299. doi: 10.1002/dvdy.10440. [DOI] [PubMed] [Google Scholar]
- [34].Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- [35].O’Donnell N, Zachara N, Hart G, Marth J. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schacter H. In: Glycoconjugates. Pigman D, Horowitz MI, editors. Academic Press; New York: 1978. pp. 87–181. [Google Scholar]
- [37].Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev. 1998;77:95–141. doi: 10.1016/s0925-4773(98)00115-4. [DOI] [PubMed] [Google Scholar]
- [38].Hanover J, Forsythe M, Hennessey P, Brodigan T, Love D, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci U S A. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Forsythe M, Love D, Lazarus BD, Kim E, Prinz W, Ashwell G, Krause M, Hanover J. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci U S A. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lee J, Kim K, Lee J, Paik Y. Regulation of Dauer formation by O-GlcNAcylation in Caenorhabditis elegans. J Biol Chem. 2010;285:2930–2939. doi: 10.1074/jbc.M109.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Love D, Ghosh S, Mondoux M, Fukushige T, Wang P, Wilson M, Iser W, Wolkow CA, Krause M, Hanover JA. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci U S A. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lewis E. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- [43].Jürgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155. [Google Scholar]
- [44].Kerppola T. Polycomb group complexes--many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- [46].Sparmann A, Van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- [47].Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- [48].Schwartz Y, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- [49].Gambetta M, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- [50].Sinclair D, Syrzycka M, Macauley M, Rastgardani T, Komljenovic I, Vocadlo D, Brock H, Honda B. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci U S A. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ingham P. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–823. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- [52].Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder R, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- [53].Comer F, Hart G. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–7852. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- [54].Shumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- [55].O’Carroll D, Erhardt S, Pagani M, Barton S, Surani M, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pasini D, Bracken A, Jensen M, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Van Der Lugt NM, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- [58].Takihara Y, et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124:3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- [59].Voncken J, Roelen B, Roefs M, De Vries S, Verhoeven E, Marino S, Deschamps J, Van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Strome S. In: Wombook. Community T C e R, editor. WormBook; 2005. doi/10.1895/wormbook.1.9.1. [Google Scholar]
- [61].Capowski E, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ross J, Zarkower D. Polycomb group regulation of Hox gene expression in C. elegans. Dev Cell. 2003;4:891–901. doi: 10.1016/s1534-5807(03)00135-7. [DOI] [PubMed] [Google Scholar]
- [63].Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- [64].Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- [65].Karakuzu O, Wang D, Cameron S. MIG-32 and SPAT-3A are PRC1 homologs that control neuronal migration in Caenorhabditis elegans. Development. 2009;136:943–953. doi: 10.1242/dev.029363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dolinoy D, Huang D, Jirtle R. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mendjan S, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- [68].Lin H, Gupta V, Vermilyea M, Falciani F, Lee J, O’Neill LP, Turner B. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Oliver B. Sex, dose, and equality. PLoS Biol. 2007;5:e340. doi: 10.1371/journal.pbio.0050340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang J, Mager J, Chen Y, Schneider E, Cross J, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- [71].Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer K, Wang H, De La Cruz CC, Otte A, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- [72].Schwartz Y, Kahn T, Nix D, Li X, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- [73].Hewagama A, Patel D, Yarlagadda S, Strickland F, Richardson B. Stronger inflammatory/cytotoxic T-cell response in women identified by microarray analysis. Genes Immun. 2009;10:509–516. doi: 10.1038/gene.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Brooks W. X Chromosome Inactivation and Autoimmunity. Clin Rev Allergy Immunol. 2009 doi: 10.1007/s12016-009-8167-5. [DOI] [PubMed] [Google Scholar]
- [75].Heijmans B, Tobi E, Stein A, Putter H, Blauw G, Susser E, Slagboom P, Lumey L. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pettitt D, Jovanovic L. The vicious cycle of diabetes and pregnancy. Curr Diab Rep. 2007;7:295–297. doi: 10.1007/s11892-007-0047-x. [DOI] [PubMed] [Google Scholar]
- [77].Stern M, Haffner S. Type II diabetes and its complications in Mexican Americans. Diabetes Metab Rev. 1990;6:29–45. doi: 10.1002/dmr.5610060102. [DOI] [PubMed] [Google Scholar]


