Abstract
Most cancers depend on a high rate of aerobic glycolysis for their continued growth and survival. Paradoxically, some cancer cell lines also display addiction to glutamine despite the fact that glutamine is a nonessential amino acid that can be synthesized from glucose. The high rate of glutamine uptake exhibited by glutamine-dependent cells does not appear to result solely from its role as a nitrogen donor in nucleotide and amino acid biosynthesis. Instead, glutamine plays a required role in the uptake of essential amino acid and in maintaining activation of TOR kinase. Moreover, in many cancer cells, glutamine is the primary mitochondrial substrate and is required to maintain mitochondrial membrane potential and integrity as well as support of the NADPH production needed for redox control and macromolecular synthesis.
Cancer cells can be addicted to glutamine
The advent of molecular cancer genetics diverted biologists away from studies of cancer cell metabolism. However, two recent sets of observations have begun to stimulate interest into how signal transduction is integrated with metabolism. First, several oncogenes and tumor suppressors have been linked to the regulation of metabolic processes [1–3]. Second, efficacy of several cancer therapies have been linked to their effects on metabolism [4–7]. One of the implications of these studies is the synergistic potential of combining pharmacologics that target signal transduction components with those that target metabolic pathways.
One of the oldest conundrums in cancer biology is Otto Warburg’s observation that cancers tended to take up more glucose and produce more lactic acid than normal tissue [8]. These observations led Warburg to hypothesize that cancer resulted from the regression of cells to the more primitive metabolism exhibited by proliferating single cell eukaryotes [9]. Recent studies have implicated oncogenic activation of glucose uptake as the cause of the “Warburg effect [2].” Constitutively activated components of phosphoinositide 3-kinase (PI3K) signaling can directly stimulate levels of glucose uptake and metabolism that exceed the cell’s capacity to use glucose in support of bioenergetic and macromolecular synthesis. When this occurs, the cancer cell secretes excess glycolytic metabolites in the form of lactic acid. In some tumors, this seemingly wasteful metabolism of glucose is mirrored by a similarly inefficient metabolism of glutamine [10, 11]. Such cancer cells, in fact, cannot survive in the absence of exogenous glutamine, and exhibit “glutamine addiction” [12]. Most cancer researchers have viewed the switch of glutamine from a nonessential to an essential amino acid (EAA) as an artifact of in vitro culture. However, recent studies have suggested that glutamine is a key substrate required for anabolic growth of mammalian cells. This review will address the role of glutamine in cell growth, the signaling pathways that regulate the cellular utilization of glutamine, and potential therapeutic strategies that might exploit the dependence of certain cancer cells on glutamine.
Glutamine provides nitrogen for protein and nucleotide synthesis
The growing cancer must synthesize nitrogenous compounds in the form of nucleotides and NEAAs. Glutamine is the obligate nitrogen donor in as many as three independent enzymatic steps for purine synthesis (phosphoribosylpyrophosphate (PRPP) amidotranferase, phosphoribosylformylglycinamidine (FGAM) synthetase, GMP synthetase) and in two independent enzymatic steps for pyrimidine synthesis (carbamoyl phosphate synthetase II, CTP synthetase) [13–15]. In these reactions, glutamine donates its amide (γ nitrogen) group and is converted to glutamic acid in the process.
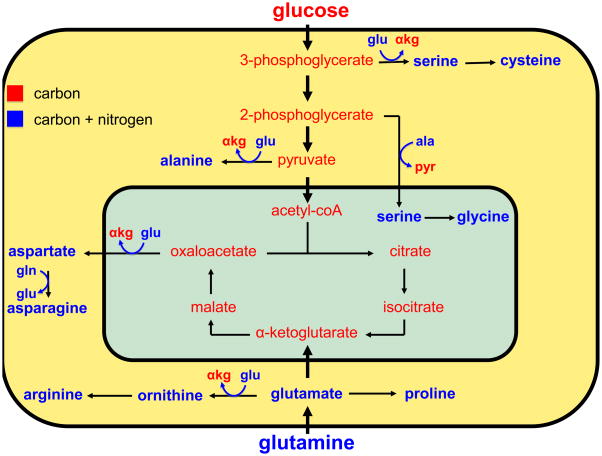
Glutamic acid is the primary nitrogen donor for the synthesis of the NEAAs [13, 15] (Figure 1). The transaminases transfer the amine group from glutamic acid (originally glutamine’s α nitrogen) to α-ketoacids. The α-ketoacids used to generate NEAAs are the carbon catabolites of glucose or glutamine: pyruvate, 3-phosphoglycerate, oxaloacetate, and glutamic acid gamma-semialdehyde, which are used to synthesize alanine, serine, aspartate, and ornithine, respectively. Serine is a precursor for glycine and cysteine biosynthesis, ornithine is a precursor for arginine biosynthesis, and aspartate is a precursor for asparagine biosynthesis (glutamine obligately donates its amide group to this reaction). Glutamic acid contributes its carbon skeleton and nitrogen to the synthesis of proline. Tyrosine is the only NEAA that is not derived from glucose or glutamine; it is directly produced from the EAA phenylalanine.
Figure 1. Glucose and glutamine can provide the carbon and nitrogen for the synthesis of the nonessential amino acids.
High throughput glucose and glutamine (gln) uptake provide the growing cell with a large pool of carbon and nitrogen for the biosynthesis of the nonessential amino acids. Compounds containing carbon, but not nitrogen, are shown in red, whereas those containing carbon and nitrogen are shown in blue. Carbon precursors derived from glycolysis (3-phosphoglycerate, 2-phosphoglycerate, pyruvate) and glutaminolysis (oxaloacetate, glutamic acid γ-semialdehyde) serve as the carbon substrate for amino acid biosynthesis. Glutamine-derived glutamic acid (glu) donates its amine group to these carbon substrates to produce nonessential amino acids (serine, alanine, aspartate, ornithine) and α-ketoglutarate (αkg). Alanine (ala) serves as the amine donor to produce serine and pyruvate (pyr) in the mitochondrion for the synthesis of glycine. Glutamine provides the carbon and nitrogen for the synthesis of proline, ornithine, and arginine. Glutamine can also serve as a direct nitrogen donor in the synthesis of asparagine from aspartic acid.
Glutamine activates TORC1 signaling
Glutamine’s contribution to amino acid biosynthesis establishes it as a key ingredient for the protein translation needs of cancer cells. A further role for glutamine in cancer cell protein translation stems from observations that a master regulator of protein translation, the mammalian target of rapamycin complex 1 (mTORC1), is responsive to glutamine levels.
The evolutionarily conserved and rapamycin-sensitive mTORC1, a master regulator of cell growth, activates protein translation and inhibits macroautophagy in response to amino acid abundance and growth factor signaling among other stimuli [16]. In the presence of sufficient amino acids, growth factor signaling through the insulin-like growth factor (IGF)-PI3K-Akt or the extracellular signal-regulated kinase (ERK)- ribosomal protein S6 kinase (RSK) pathways activates mTORC1 through releasing the Ras homolog enriched in brain (RHEB) GTPase from repression by the tumor suppressors, tuberous sclerosis 1 (TSC1)–TSC2. The importance of sufficient amino acid levels for TORC1 activation was initially recognized in yeast, in which amino acid starvation recapitulated the effects of treatment with the TORC1 inhibitor, rapamycin, in suppressing TORC1-induced protein translation and inducing autophagy. In yeast, TORC1 is responsive to both glutamine and EAA levels [17].
Substantial evidence supports the role of amino acids in the activation of mTORC1-dependent signaling [18–22]. Of the EAAs, mTORC1-signaling appears to respond most acutely to leucine; however glutamine is also necessary for maximal mTOR activation for reasons that remained unknown until recently [18, 23]. To better understand the mechanism through which glutamine synergizes with EAAs to regulate mTORC1 activation, Nicklin et al. [24] studied a cell line in which mTORC1 activation was dependent on the presence of both glutamine and the EAAs. The authors found that a portion of the glutamine taken up through the SLC1A5 (also called ASCT2) glutamine importer was rapidly exported through the bidirectional amino acid transporter SLC7A5 (also called LAT1) in exchange for the uptake of extracellular EAAs. Knockdown of SLC1A5 in these cells impaired glutamine uptake and export, EAA uptake, and mTORC1 activation, suggesting that glutamine uptake and export is required for EAA activation of mTORC1.
Although glutamine is a critical contributor to the NEAA pool incorporated into newly translated proteins, this study demonstrates that a portion of the glutamine imported into cells is not utilized for anabolic metabolism. Rather, glutamine is shuttled out of the cell in exchange for EAAs that directly activate TORC1 for the initiation of protein translation and cell growth. Of course, these EAAs can also be directly incorporated into newly synthesized proteins. As such, glutamine’s uptake and subsequent export serves both as a signal to mTORC1 and a source of EAAs to promote protein translation.
Glutamine as a mitochondrial substrate
Although the unusually high glucose requirement of cancer cells was described in the 1920s, the essential glutamine requirement of proliferating cells was first highlighted by Harry Eagle in 1955. In his studies of the nutritional requirements of cell lines growing in vitro, Eagle observed that the glutamine consumption rate of many of the cell lines exceeded the consumption of any other amino acid by ten-fold [12]. The cell lines he studied could not proliferate in the absence of exogenous glutamine and many could not maintain their viability in the absence of glutamine. In 1971, Kovacevic and colleagues observed that glutamine’s carbons could be accounted for in carbon dioxide released by cells, providing evidence that glutamine could serve as a combustible fuel [10]. Subsequent experiments showed that certain cancer cell lines consume glutamine at a rate substantially higher than that of any other amino acid. In order to be oxidized, glutamine must first lose its amide group to form glutamic acid, which then loses its amine group to form the tricarboxylic acid (TCA) cycle metabolite, α-ketoglutarate. As mentioned, the enzymes involved in nucleotide biosynthesis contribute to the conversion of glutamine to glutamic acid. The enzyme glutaminase also contributes to this activity through the release of glutamine’s amide group as free ammonia. The generated glutamic acid can be converted into α-ketoglutarate through the cellular transaminases or through glutamic acid dehydrogenase, which catalyzes the NAD(P)+-dependent liberation of the amine as ammonia.
Recent studies have extended our understanding of glutamine’s contribution to the intermediary carbon metabolism of cancer cells. Real-time 13C NMR studies have shown that a significant fraction of glutamine carbon is converted into lactic acid and secreted from the cell [25]. This seemingly wasteful metabolism of glutamine is analogous to the cancer cell metabolism of glucose observed by Warburg and is consistent with the observation that the glutamine consumption rate exceeds its rate of incorporation into newly synthesized proteins by ten-fold [25, 26]. Conversion of glutamine into lactic acid requires the activity of malic enzyme, which oxidatively decarboxylates malic acid producing carbon dioxide, NADPH, and pyruvate. Nucleotide and lipid synthesis both depend on NAPDH for a source of reducing equivalents [1]. High throughput glutaminolysis through the provision of substrate for malic enzyme can provide a proliferating cell with a significant fraction of its NADPH needs [25].
Glutamine contributes to macromolecular synthesis in ways other than the production of NADPH. Real-time 13C NMR studies in a glioblastoma cell line have shown that glutamine contributes the majority of the cellular oxaloacetate pool [25]. Oxaloacetate is the obligate substrate that condenses with acetyl-coA to form citrate, which can donate acetyl-coA groups for the synthesis of cholesterol and fatty acids [27] as well as the modification of chromatin structure [28]. In providing the cancer cell with a source of oxaloacetate, glutamine provides anaplerosis, the refilling of the mitochondrial carbon pool. Replenishment of the mitochondrial carbon pool by glutamine provides the mitochondria with precursors for the maintenance of mitochondrial membrane potential and for the synthesis of nucleotides, proteins, and lipids. When first discovered, the high rate of aerobic glycolysis in cancer was felt to reflect mitochondrial dysfunction; however, a significant body of evidence supports that mitochondrial respiratory capacity is maintained in cancer [29]. The available data on mitochondrial glutamine metabolism in cancer cells supports the indispensable nature of mitochondrial metabolism to the physiology of many cancer cell types. In non-transformed, non-proliferative tissues, such as pancreas, liver, kidney, muscle, and brain, cells are reported to rely upon the activity of pyruvate carboxylase [30–33], for production of oxaloacetate through pyruvate carboxylation. This activity enables these cell types to use glucose for their anaplerotic needs. By contrast, 13C NMR studies do not support the presence of pyruvate carboxylase activity in several cancer cell types [25, 34]. The mechanism through which pyruvate carboxylase activity is suppressed in these cell types is an area of active investigation as is the connection between pyruvate carboxylase activity and glutamine dependence.
Oncogenic levels of c-MYC regulate glutamine metabolism
As mentioned, glutamine is the obligate nitrogen donor for nucleotide synthesis. Five enzymatic steps in the synthesis of purines and pyrimidines use glutamine as a source of nitrogen. Recent studies using quantitative RT-PCR and chromatin-immunoprecipitation (ChIP) in multiple cell systems have suggested that c-MYC (Myc) binds and transactivates eleven genes involved in nucleotide biosynthesis [35]. Myc is a basic helix–loop–helix zipper (bHLHZ) protein that heterodimerizes with the small bHLHZ protein MAX and exerts both activating and repressing transcriptional effects [36]. Of the five enzymatic steps utilizing glutamine, three (PRPP amidotransferase, carbamoyl phosphate synthetase II, CTP synthetase) are directly regulated by Myc at the transcriptional level [37]. Carbamoyl phosphate synthetase II, a rate-limiting glutamine-dependent enzyme involved in pyrimidine synthesis, has also been found to be regulated via epidermal growth factor receptor (EGFR)-dependent mitogen-activated protein kinas (MAPK) phosphorylation [38] as well as caspase-dependent degradation [39].
Cancer cells take up and metabolize glucose and glutamine to a degree that far exceeds their needs for these molecules in anabolic macromolecular synthesis. Commonly occurring oncogenic signal transduction pathways initiated by receptor tyrosine kinases or Ras engage PI3K-Akt signaling to directly stimulate glycolytic metabolism [1, 2]. Oncogenic levels of Myc have recently been linked to increased glutaminolysis through a coordinated transcriptional program [26, 40, 41]. Myc-activation/amplification is one of the most common oncogenic events observed in cancer and is known to drive the progression of human lymphomas [42, 43], neuroblastoma [44] and small cell lung cancer [45]. Quantitative RT-PCR and ChIP experiments support Myc’s binding and transcriptional activation of two high affinity glutamine transporters: SLC38A5 (also called SN2) and SLC1A5 (ASCT2), the transporter required for glutamine-dependent mTORC1 activation [24, 26]. In addition to facilitating glutamine uptake, Myc promotes the metabolism of imported glutamine into glutamic acid and ultimately into lactic acid [26]. Whether the tendency of Myc to complement Ras and PI3K-Akt [46, 47] is related to the interdependence of glutamine and glucose metabolism in support of cell growth remains an open question.
The importance of glutamine metabolism for the MYC-amplified cell has been highlighted by a recent demonstration that Myc also influences post-transcriptional regulation of glutamine catabolism. In a screen for Myc-regulated mitochondrial proteins, Gao et al. [41] found that glutaminase protein levels were significantly upregulated in Myc-overexpressing cells. Yet the glutaminase mRNA expression level did not correlate with its increased protein levels, leading the authors to hypothesize that Myc regulates glutaminase through a post-transcriptional mechanism. Using an algorithm-based approach, the authors found that the microRNA miR-23a/b repressed the translation of glutaminase through binding its 3′ untranslated region (UTR). Notably, the authors had previously identified miR-23a/b as a strong target of Myc transcriptional repression. This study further links Myc overexpression to the cellular ability to catabolize glutamine into glutamic acid, thereby providing cells with a large pool of carbon for anaplerosis and NADPH production. Whether other oncogenic signaling pathways also contribute to the regulation of glutamine metabolism remains to be determined.
Myc-induced metabolic reprogramming triggers cellular dependency on exogenous glutamine as a source of carbon for the maintenance of the mitochondrial membrane potential and macromolecular synthesis. Indeed, glutamine depletion kills transformed cells in a Myc-dependent manner [26, 40]. The cell death induced by glutamine starvation can be rescued by the overexpression of Bcl-2, Bcl-xL, or a dominant negative form of caspase-9, implicating the intrinsic apoptotic pathway as the mechanism of cell death [26, 48]. Substitution of glutamine with a membrane-permeable form of α-ketoglutarate, pyruvate, or oxaloacetate also rescues this death [26, 48]. However, consistent with the importance of glutamine as an obligate source of nitrogen, neither overexpression of anti-apoptotic proteins nor addition of TCA cycle intermediates can rescue the proliferation defect observed in glutamine-starved cells [26]. Nevertheless, glutamine depletion of Myc-transformed cells leads to a profound reduction in the levels of TCA cycle metabolites despite abundant extracellular availability of glucose, supporting the importance of glutamine in the maintenance of mitochondrial anaplerosis [48]. These findings suggest Myc transformation might also suppress the ability of tumor cells to use glucose as an anapleurotic substrate perhaps through upregulation of LDH-A.
Targeting Glutamine Addiction: Past, Present, and Future
A wide variety of human cancer cell lines have shown sensitivity to glutamine starvation, including those derived from pancreatic cancer, glioblastoma multiforme, acute myelogenous leukemia, and small cell lung cancer [49]. Experiments in the 1950s showed that the compounds 6-diazo-5-oxo-L-norleucine (L-DON) and azaserine, isolated from a species of Streptomyces, have significant activity as glutamine analogues [50]. Later, another glutamine analogue, acivicin, was also isolated. Research on the glutamine dependency of cell lines in vitro spurred the testing of these compounds as therapeutics. Pre-clinical testing of all three agents showed a significant cytotoxic effect against certain tumor types both in culture and in mouse xenograft models [14]. Although all three of these agents showed clinical activity, their use was discontinued due to dose-limiting neurotoxicity, gastrointestinal toxicity, and myelosuppression [14]. All three compounds show their greatest activity in inhibiting the glutamine-dependent enzymatic steps in nucleotide biosynthesis [51]. In fact, acivicin has only a minor effect on glutaminolysis, while significantly reducing glutamine-dependent nucleotide biosynthesis. These studies demonstrate that glutamine mimetics are unduly toxic. Current investigation seeks to preserve the glutamine metabolism required for normal tissue physiology while impairing the glutamine addiction of cancer (Figure 2).
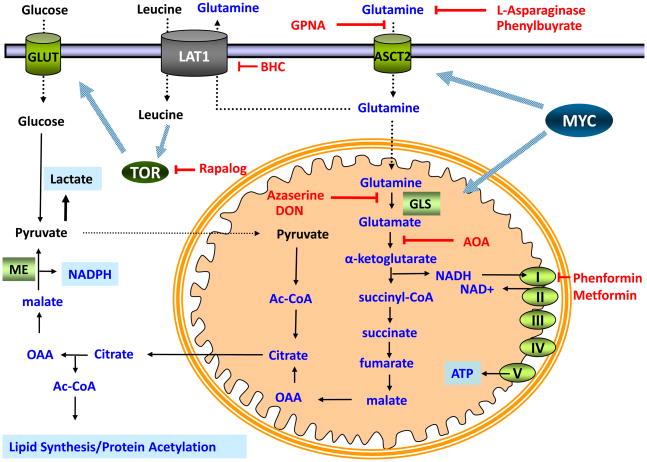
Figure 2. Pharmacologic Targets in Glutamine Metabolism.
Myc enables cancer cells to maximize glutamine uptake from the extracellular space through upregulation of the glutamine importer, ASCT2. Uptake of glutamine can be suppressed both by depletion of glutamine from the extracellular space with L-asparaginase and phenylbutyrate or through inhibition of ASCT2-dependent uptake with L-γ-glutamyl-p-nitroanilide (GPNA). Once glutamine enters the cell, it can be metabolized through glutaminolysis to provide NADPH or exported to facilitate TOR kinase activation. Myc enables conversion of glutamine into glutamic acid via upregulation of glutaminase (GLS), an enzyme whose activity can be inhibited by treatment with 6-diazo-5-oxo-L-norleucine (L-DON) and Azaserine. Transamination of glutamic acid to α-ketoglutarate can be inhibited with amino-oxyacetic acid (AOA). Mitochondrial metabolism of α-ketoglutarate leads to the production of citrate, its cleavage into oxaloacetate (OAA), reduction of oxaloacetate into malate, and oxidation of malate to pyruvate via malic enzyme (ME) to produce pyruvate and NADPH. The high rate of glutamine metabolism through successive steps into oxaloacetate establishes glutamine as the primary anaplerotic substrate. This metabolism requires the regeneration of NAD+ through the electron transport chain, a process that can be inhibited with the biguanides, phenformin and metformin. Glutamine’s use as a substrate for the amino acid exchanger LAT1 can be suppressed with 2-aminobicyclo-(2,2,1)heptanecarboxylic acid (BCH) treatment and TOR activity can be suppressed by rapamycin treatment.
Suppressing cancer cell glutamine uptake
Numerous studies have detected an upregulation of high affinity glutamine transporters in cancer [52]. One primary example, SLC1A5 (ASCT2), is a direct target of the Myc oncoprotein [26], and is upregulated in a host of cancers [52]. L-γ-glutamyl-p-nitroanilide (GPNA), one of a panel of SLC1A5 inhibitors [53], can inhibit glutamine uptake and inhibit glutamine-dependent mTOR activation in vitro [24].
Suppressing glutamine-dependent anaplerosis
Studies using the transaminase inhibitor amino-oxyacetic acid (AOA) have suggested that the major route through which glutamine-derived carbon enters the TCA cycle in Myc-transformed cells is through transamination [54]. These studies suggested that glutamic acid dehydrogenase, the principal enzyme converting glutamic acid into α-ketoglutarate in pancreas, is not the rate-determining step in proliferating cells. Recent data suggests that AOA may in fact be a promising cancer therapeutic. AOA treatment produced a cytostatic effect on the growth of a breast cancer cell line in a mouse xenograft model without any obvious dose-limiting toxicities [55]. AOA treatment has also shown a cytotoxic effect on a glutamine-dependent MYC-amplified glioblastoma cell line in vitro while having no significant effect on a paired Myc-deficient line [26]. These studies suggest that selective inhibition of one component of glutamine metabolism (glutamate transamination) might recapitulate the anti-cancer effects but not the nonspecific toxicity of wholesale inhibition of glutamine metabolism.
Inhibiting Complex I
Glutamine-dependent cancer cells undergo a dramatic metabolic reprogramming such that mitochondria are reprogrammed to produce anabolic precursors from glutamine. The entry and flux of glutamine through the TCA cycle requires the continual regeneration of mitochondrial NAD+ through the activities of the mitochondrial electron transport chain. It has been a longstanding hypothesis that the biguanides activate AMPK through inhibition of the mitochondrial respiratory chain [56–59]. Metformin has shown promise in slowing the growth of cancer cells in vitro and xenografted tumors in vivo [60–63]. Epidemiological studies also show a reduced incidence of cancer in patients treated with metformin compared to diabetics treated with other modalities [64]. However, therapeutic levels of metformin exhibit the greatest mitochondrial effects in the liver, further studies are needed to determine the extent to which in vivo efficacy of these agents is due to liver-dependent effects, such as the lowering of blood glucose concentration and/or tumor-specific effects on glutamine metabolism.
Targeting glutamine-dependent mTOR activation
As previously noted, a portion of the glutamine imported into cancer cells via SLC1A5 is directly exported through the SLC7A5–SLC3A2 (LAT1–4f2hc) complex. The subsequent import of EAAs, such as leucine, activates the mTORC1 kinase. In vitro treatment of cancer cells with GPNA, an inhibitor of SLC1A5, or with 2-aminobicyclo-(2,2,1) heptanecarboxylic acid (BCH), an inhibitor of SLC7A5–SLC3A2 blocks the glutamine-dependent activation of mTORC1 and induces autophagy [24]. The in vivo utility of this approach for the treatment of glutamine-addicted/mTORC1-dependent cancers is under investigation.
Enzymatic lowering of blood glutamine levels
L-asparaginase (Elspar, [Merck & Co. Inc.], Oncaspar [Enzan Inc.]) hydrolyzes asparagine into aspartic acid and ammonia, and is a cornerstone of the treatment of pediatric acute lymphoblastic leukemia (ALL) [65]. This treatment is believed to be successful because ALL cells are incapable of synthesizing asparagine de novo. L-asparaginase also possesses significant glutaminase activity, capable of hydrolyzing glutamine to glutamic acid and ammonia [66]. L-asparaginase significantly depletes glutamine levels and studies have confirmed that the success of treatment correlates with glutamine-depletion [49, 66]. However, L-asparaginase treatment in adults has shown significant toxicity [67]. More work is needed to clarify the potential role for L-asparaginase therapy for the treatment of childhood and adult glutamine-addicted cancers.
An alternative agent that could be used to deplete plasma glutamine is phenylbutyrate (Buphenyl, [Ucyclyd Pharma], Ammonaps [Swedish Orphan]), an FDA approved pharmacologic for the treatment of hyperammonemia in patients with acute liver failure or with congenital urea cycle disorders [68]. Pharmacologic doses of phenylbutyrate lead to a significant depletion of plasma glutamine levels [69]. In humans, phenylbutyrate spontaneously breaks down to form phenylacetate, which is conjugated with glutamine by the hepatic enzyme phenylacetyl Coenzyme A: glutamine acyltransferase to yield phenylacetylglutamine. This latter product is then excreted in the urine [70].
Concluding Remarks
Increasing evidence suggests that oncoproteins can directly reprogram tumor cell metabolism, rendering the cells addicted to certain nutrients in a way non-transformed cells are not. However, whether alterations in cancer metabolism can be safely targeted therapeutically remains to be determined. In this review, the importance of glutamine metabolism for cancer growth and viability was highlighted and the possibility of developing therapies that can exploit glutamine metabolism for therapeutic gain was considered. Undoubtedly, like other targeted therapies being developed, therapies directed against glutamine metabolism will be most effective in tumors that display glutamine-dependence. Whether glutamine-derivatives can be utilized to investigate tumor metabolism in vivo in a manner analogous to the way the glucose analog, fluorodeoxyglucose for positron emission tomography (FDG-PET) has developed, is currently under investigation. If successful, such studies will help establish once and for all whether glutaminolysis is a tissue culture artifact or a potential Achilles’ heel of cancer cells.
Acknowledgments
We apologize to those colleagues whose work we were unable to cite due to limited space. We thank Patrick S. Ward, Dr. Brian Keith, Dr. Justin R. Cross, and Tamar S. Wise for critical reading of the manuscript as well as Batya Wise for help with figure production. This work was supported by grants from the Cancer Research Institute and National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Pouyssegur J. Tumor Cell Metabolism: Cancer’s Achilles’ Heel. Cancer Cell. 2008;13:472. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Dang CV, et al. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 4.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR Signaling Network for Cancer Therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zong WX, et al. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P, et al. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburg O, et al. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 10.Kovacevic Z. The pathway of glutamine and glutamate oxidation in isolated mitochondria from mammalian cells. Biochem J. 1971;125:757–763. doi: 10.1042/bj1250757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2009;29:313. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 13.Donald Voet JGV. Biochemistry. John Wiley & Sons, Inc; 1995. [DOI] [PubMed] [Google Scholar]
- 14.Ahluwalia GS, et al. Metabolism and action of amino acid analog anti-cancer agents. Pharmacology & Therapeutics. 1990;46:243. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 15.Young VR, Ajami AM. Glutamine: The Emperor or His Clothes? J Nutr. 2001;131:2449S–2459. doi: 10.1093/jn/131.9.2449S. [DOI] [PubMed] [Google Scholar]
- 16.Wullschleger S, et al. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Crespo JL, et al. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci U S A. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 19.Blommaart EF, et al. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 20.Fox HL, et al. Amino acids stimulate phosphorylation of p70S6k and organization of rat adipocytes into multicellular clusters. Am J Physiol Cell Physiol. 1998;274:C206–213. doi: 10.1152/ajpcell.1998.274.1.C206. [DOI] [PubMed] [Google Scholar]
- 21.Xu G, et al. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J Biol Chem. 1998;273:28178–28184. doi: 10.1074/jbc.273.43.28178. [DOI] [PubMed] [Google Scholar]
- 22.Dann SG, Thomas G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Letters. 2006;580:2821. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- 23.Krause U, et al. Signalling pathways and combinatory effects of insulin and amino acids in isolated rat hepatocytes. Eur J Biochem. 2002;269:3742–3750. doi: 10.1046/j.1432-1033.2002.03069.x. [DOI] [PubMed] [Google Scholar]
- 24.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rafael MS, et al. Energy metabolism in tumor cells. FEBS Journal. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 30.Gibala MJ, et al. Anaplerosis of the citric acid cycle: role in energy metabolism of heart and skeletal muscle. Acta Physiologica Scandinavica. 2000;168:657–665. doi: 10.1046/j.1365-201x.2000.00717.x. [DOI] [PubMed] [Google Scholar]
- 31.Hasan NM, et al. Impaired Anaplerosis and Insulin Secretion in Insulinoma Cells Caused by Small Interfering RNA-mediated Suppression of Pyruvate Carboxylase. J Biol Chem. 2008;283:28048–28059. doi: 10.1074/jbc.M804170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassel B. Carboxylation and anaplerosis in neurons and glia. Mol Neurobiol. 2000;22:21–40. doi: 10.1385/MN:22:1-3:021. [DOI] [PubMed] [Google Scholar]
- 33.Owen OE, et al. The Key Role of Anaplerosis and Cataplerosis for Citric Acid Cycle Function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 34.Forbes NS, et al. Estradiol stimulates the biosynthetic pathways of breast cancer cells: detection by metabolic flux analysis. Metab Eng. 2006;8:639–652. doi: 10.1016/j.ymben.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Liu YC, et al. Global Regulation of Nucleotide Biosynthetic Genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bush A, et al. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes & Development. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graves LM, et al. Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature. 2000;403:328. doi: 10.1038/35002111. [DOI] [PubMed] [Google Scholar]
- 39.Huang M, et al. Caspase-Dependent Cleavage of Carbamoyl Phosphate Synthetase II during Apoptosis. Molecular Pharmacology. 2002;61:569–577. doi: 10.1124/mol.61.3.569. [DOI] [PubMed] [Google Scholar]
- 40.Yuneva M, et al. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taub R, et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwab M, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 45.Nau MM, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 46.Land H, et al. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 47.Kauffmann-Zeh A, et al. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 48.Yuneva M, et al. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu MC, et al. Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase. Int J Cancer. 1978;22:728–733. doi: 10.1002/ijc.2910220615. [DOI] [PubMed] [Google Scholar]
- 50.Ovejera AA, et al. Efficacy of 6-diazo-5-oxo-L-norleucine and N-[N-gamma-glutamyl-6-diazo-5-oxo-norleucinyl]-6-diazo-5-oxo-norleucine against experimental tumors in conventional and nude mice. Cancer Res. 1979;39:3220–3224. [PubMed] [Google Scholar]
- 51.Griffiths M, et al. The role of glutamine and glucose analogues in metabolic inhibition of human myeloid leukaemia In vitro. International Journal of Biochemistry. 1988;25:1749. doi: 10.1016/0020-711x(88)90303-5. [DOI] [PubMed] [Google Scholar]
- 52.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: Partners in crime? Seminars in Cancer Biology. 2005;15:254. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Esslinger CS, et al. N[gamma]-Aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorganic & Medicinal Chemistry. 2005;13:1111. doi: 10.1016/j.bmc.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 54.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. J Biol Chem. 1984;259:6215–6221. [PubMed] [Google Scholar]
- 55.Thornburg J, et al. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Research. 2008;10:R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidoff F. Effects of guanidine derivatives on mitochondrial function. I. Phenethylbiguanide inhibition of respiration in mitochondria from guinea pig and rat tissues. J Clin Invest. 1968;47:2331–2343. doi: 10.1172/JCI105918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dykens JA, et al. Biguanide-induced mitochondrial dysfunction yields increased lactate production and cytotoxicity of aerobically-poised HepG2 cells and human hepatocytes in vitro. Toxicol Appl Pharmacol. 2008;233:203–210. doi: 10.1016/j.taap.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 58.Wang DS, et al. Involvement of organic cation transporter 1 in the lactic acidosis caused by metformin. Mol Pharmacol. 2003;63:844–848. doi: 10.1124/mol.63.4.844. [DOI] [PubMed] [Google Scholar]
- 59.El-Mir MY, et al. Dimethylbiguanide Inhibits Cell Respiration via an Indirect Effect Targeted on the Respiratory Chain Complex I. Journal of Biological Chemistry. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 60.Zakikhani M, et al. Metformin Is an AMP Kinase-Dependent Growth Inhibitor for Breast Cancer Cells. Cancer Res. 2006;66:10269–10273. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 61.Buzzai M, et al. Systemic Treatment with the Antidiabetic Drug Metformin Selectively Impairs p53-Deficient Tumor Cell Growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 62.Sahra IB, et al. The antidiabetic drug metformin exerts an antitumoral effect in vitro and in vivo through a decrease of cyclin D1 level. Oncogene. 2008;27:3576. doi: 10.1038/sj.onc.1211024. [DOI] [PubMed] [Google Scholar]
- 63.Hirsch HA, et al. Metformin Selectively Targets Cancer Stem Cells, and Acts Together with Chemotherapy to Block Tumor Growth and Prolong Remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans JMM, et al. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narta UK, et al. Pharmacological and clinical evaluation of l-asparaginase in the treatment of leukemia. Critical Reviews in Oncology/Hematology. 2007;61:208. doi: 10.1016/j.critrevonc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 66.Avramis VI, Panosyan EH. Pharmacokinetic/pharmacodynamic relationships of asparaginase formulations: the past, the present and recommendations for the future. Clin Pharmacokinet. 2005;44:367–393. doi: 10.2165/00003088-200544040-00003. [DOI] [PubMed] [Google Scholar]
- 67.Lessner HE, et al. Phase II study of L-asparaginase in the treatment of pancreatic carcinoma. Cancer Treat Rep. 1980;64:1359–1361. [PubMed] [Google Scholar]
- 68.Enns GM, et al. Survival after Treatment with Phenylacetate and Benzoate for Urea-Cycle Disorders. N Engl J Med. 2007;356:2282–2292. doi: 10.1056/NEJMoa066596. [DOI] [PubMed] [Google Scholar]
- 69.Thibault A, et al. Phase I study of phenylacetate administered twice daily to patients with cancer. Cancer. 1995;75:2932–2938. doi: 10.1002/1097-0142(19950615)75:12<2932::aid-cncr2820751221>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 70.Thibault A, et al. A Phase I and Pharmacokinetic Study of Intravenous Phenylacetate in Patients with Cancer. Cancer Res. 1994;54:1690–1694. [PubMed] [Google Scholar]