Abstract
Autosomal recessive nonsyndromic deafness is one of the most frequent forms of inherited hearing impairment. Over 30 autosomal recessive nonsyndromic hearing loss loci have been mapped, and 15 genes have been isolated. Of the over 30 reported autosomal recessive nonsyndromic hearing loss (NSHL) loci, the typical phenotype is prelingual non-progressive severe to profound hearing loss with the exception of DFNB8, which displays postlingual onset and DFNB13, which is progressive. In this report we describe a large inbred kindred from a remote area of Pakistan, comprising six generations and segregating autosomal recessive nonsyndromic prelingual deafness. DNA samples from 24 individuals were used for genome wide screen and fine mapping. Linkage analysis indicates that in this family the NSHL locus, (DFNB35) maps to a 17.54 cM region on chromosome 14 flanked by markers D14S57 and D14S59. Examination of haplotypes reveals a region that is homozygous for 11.75 cM spanning between markers D14S588 and D14S59. A maximum two-point LOD score of 5.3 and multipoint LOD score of 7.6 was obtained at marker D14S53. The interval for DFNB35 does not overlap with the regions for DFNA9, DFNA23 or DFNB5.
Keywords: autosomal recessive nonsyndromic hearing loss, DFNA9, DFNA23, DFNB5, DFNB35, 14q24.1—14q24.3 prelingual, Pakistan
Introduction
Deafness is a relatively common sensory disorder, with approximately 1 in 800 children born with serious permanent hearing impairment. In addition, a large proportion of older adults suffer from progressive hearing loss.1 Hearing impairment can be associated with other clinical features as part of distinct syndrome, but in most cases (70%) it is the sole clinical manifestation.2 Nonsyndromic hearing loss (NSHL) is the most genetically heterogeneous trait known.3 Over 70 loci have been reported and total of 27 genes have been identified.4 It is estimated that ≤85% of nonsyndromic hearing loss is caused by autoso-mal recessive defects.5 For nonsyndromic autosomal recessive deafness, the hearing impairment usually has prelingual onset, involves all the frequencies and is severe to profound.3
To date over 30 loci for autosomal recessive hearing loss have been mapped and 15 genes (CDH23 [MIM 605516], CLDN14 [MIM 605608], GJA1 [MIM 121014], GJB2 [MIM 121011], GJB6 [MIM 604418], MYO7A [MIM 276903], MYO15 [MIM 602666], OTOA [MIM 607038], OTOF [MIM 603681], SLC26A4 [MIM 605646], STRC [MIM 606440], TCM1 [MIM 606706], TECTA [MIM 602574], TMPRSS3 [MIM 605511], USH1C [MIM 605242]) have been isolated. This extreme genetic heterogeneity suggests that there are many different processes that can malfunction within the inner ear to cause hearing loss.6
Materials and methods
Family history
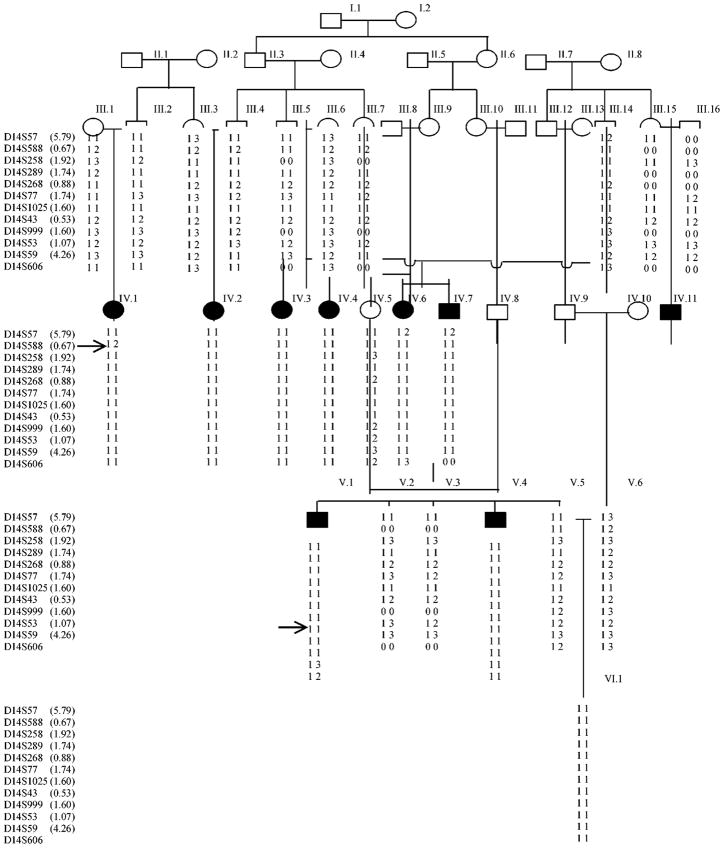
Before the onset of the study, approval was obtained from the Quaid-I-Azam University Institutional Review Board. Informed consent was obtained from all family members who participated in the study. The pedigree structure is based upon interviews with multiple family members. Pedigree 2 is from a rural region of Pakistan (Figure 1). It provides convincing evidence for an autosomal recessive mode of inheritance. Family members rarely marry outside the kindred, and consequently consanguineous unions are common. It is likely for such a kindred that the hearing impaired individuals are homozygous for the same mutation. All affected individuals regardless of age displayed profound hearing loss affecting all frequencies, implying that hearing impairment in this family is not progressive. All of the affected individuals underwent examination for defects in outer ear morphology, mental retardation and other clinical features that could indicate that hearing impairment was syndromal. There was no evidence in this kindred that hearing impairment belonged to a syndrome or that there is gross vestibular involvement.
Figure 1.
Drawing of pedigree 2 which segregates DFNB35. The sexes of some of the family members have been changed to protect the anonymity of the family. The genetic map distance in centimorgans (cM) are given in parenthesis next to the marker name. Black symbols represent individuals with hearing impairment due to DFNB35. Clear symbols represent unaffected individuals. Haplotypes for the most closely linked STRPs are shown below each symbol.
Extraction of genomic DNA and genotyping
Venous blood samples were obtained from 24 family members including nine individuals who are hearing impaired. Genomic DNA was extracted from whole blood following a standard protocol,7 quantified by spectrophoto-meteric readings at optical density 260 and diluted to 40 ng/ul for PCR amplification. PCR reactions for microsatellite markers were performed according to standard procedure in a total volume of 25 ul with 40 ng genomic DNA and of 50 ug of each primer, 200 mM dNTP, 1X PCR buffer (Life Technology). PCR was carried out for 35 cycles: 95°C for 1 min, 57°C for 1 min, 72°C for 1 min in a thermal cycler (Perkin Elmer). PCR products were resolved on 8% nonde-naturing polyacrylamide gel and genotypes were assigned by visual inspection.
Linkage analysis
Pair wise linkage study was carried out using MLINK of the FASTLINK computer package.8 Due to the size of the pedigree and the number of non-founders the pedigree was split in half in order to carry out the multipoint linkage analysis using the GENEHUNTER computer program.9 For the analysis an autosomal recessive mode of inheritance with complete penetrance and a disease allele frequency of 0.001 was assumed.
To elucidate the gene defect in this family, we initially searched for linkage by using polymorphic markers mapped within 25 autosomal recessive non-syndromic deafness loci listed on the Hereditary Hearing loss homepage (http://dnalab-www.uia.ac.be/dnalab/hhh/). When linkage to known candidate loci was excluded, a genome-wide scan was initiated. An initial genome-wide screen with microsatellite markers spaced approximately at 20 cM intervals was conducted by use of the DNA from five of the affected individuals (IV-1, IV-3, IV-8, V-1 and VI-1).
Results
In the course of screening 180 markers, eight genomic regions were found to be homozygous in all the five affected subjects. Upon testing the rest of the family members linkage to seven of these regions was excluded. All hearing impaired individuals were found to be homozygous at marker D14S53 that is located at 86.29 cM on chromosome 14 according to the Marshfield map.10 In addition, a two- point LOD score of 4.3 at θ=0.00 was obtained for marker D14S53.
For fine mapping 15 additional markers were selected from the Marshfield map;10 13 of these markers, D14S57, D14S1069, D14S119, D14S588, D14S258, D14S251, D14S289, D14S277, D14S268, D14S77, D14S43 and D14S999 are proximal to D14S53 and two of the markers D14S59 and D14S1008 are distal to D14S53. All 24 family members were genotyped for these markers. Probably due to the high degree of consanguinity six markers, D14S289, D14S1025, D14S119, D14S1069, D14S251 and D14S277 were non-informative. The data was reanalysed using two-point and multipoint linkage analysis. The results of the two-point linkage analysis are presented in Table 1. A maximum two-point LOD score of 5.3 and multipoint LOD score of 7.6 were obtained for marker D14S53 at 86.29 cM, with a 3.0 unit support interval from marker D14S57 (68.82 cM) to D14S59 (87.36 cM), an interval of 17.54 cM. The 14q haplo-type for pedigree 2 is presented in Figure 1. Affected individual IV.1, is not homozygous at marker D14S588. Since no phase information is available for IV.1 mother (III.1.) it is not possible to discern if a recombination event occurred between marker D14S588 and D14S258 in the gamete she passed to her son IV.1 or a more historic recombination event took place. A recombination event occurred at individual V.1 between markers D14S59 and D14S53, mapping DFNB35 centromeric to D14S59. Combining this information indicates that DFNB35 maps to an 11.75 cM between markers D14S588 and D14S59.
Table 1.
Two point LOD score results between the DFNB35 locus and chromosome 14 markers
| Marker | LOD score at θ= | ||||||
|---|---|---|---|---|---|---|---|
| 0.00 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | |
| D14S57 | 0.39 | 0.36 | 0.78 | 0.81 | 0.60 | 0.35 | 0.14 |
| D14S588 | 2.43 | 2.35 | 2.07 | 1.72 | 1.09 | 0.57 | 0.19 |
| D14S258 | 3.35 | 3.25 | 2.86 | 2.40 | 1.53 | 0.80 | 0.28 |
| D14S289 | 1.25 | 1.20 | 1.05 | 0.86 | 0.53 | 0.27 | 0.10 |
| D14S268 | 4.56 | 4.44 | 3.95 | 3.35 | 2.20 | 1.18 | 0.39 |
| D14S77 | 4.06 | 3.96 | 3.57 | 3.08 | 2.12 | 1.22 | 0.46 |
| D14S1025 | 0.85 | 0.82 | 0.71 | 0.57 | 0.34 | 0.16 | 0.05 |
| D14S43 | 4.64 | 4.53 | 4.05 | 3.46 | 2.32 | 1.27 | 0.44 |
| D14S999 | 4.17 | 4.06 | 3.59 | 3.00 | 1.89 | 0.93 | 0.27 |
| D14S53 | 5.53 | 5.39 | 4.85 | 4.16 | 2.80 | 1.53 | 0.51 |
| D14S59 | −Infinity | 2.16 | 2.40 | 2.14 | 1.38 | 0.67 | 0.20 |
| D14S606 | −Infinity | 0.93 | 1.27 | 1.15 | 0.71 | 0.32 | 0.09 |
Discussion
Three other loci for hearing loss have been localized to 14q: (1) DFNA9 responsible for postlingual progressive sensori-neural hearing loss was mapped to chromosome 14q12 –13 in large kindred with autosomal dominant hearing loss;11 (2) DFNA23, an autosomal dominant locus was mapped to 14q21 – q22, in a Swiss German kindred were hearing impaired members presented with prelingual neurosensory and conductive hearing loss;12 and (3) DFNB5, an autosomal recessive locus mapped to 14q in a consanguineous Indian kindred with severe to profound hearing impairment.13 Only the COCH gene for DFNA9 has been identified.14 The DFNB35 locus is telomeric to the DFNA23 locus and both DFNB5 and DFNA9 are centromeric to the DFNA23 locus. The region for DFNB35 does not overlap with DFNB5, DFNA9 and DFNA23, thus indicating the presence of a novel gene.
At present, 26 genes and 161 Expressed sequence tags (ESTs) mapped to 17.54 cM region on chromosome 14q. This region contain genes, which include splicing factor SRP40-1, Presenilin 1, numb homologue, v-fos FBJ and transforming growth factor, beta 3, however, these genes are not strong candidates for DFNB35. Identifying the DFNB35 locus is the first-step in isolating the gene responsible for hearing loss in pedigree 2.
Acknowledgments
We wish to thank the family members for their invaluable participation and cooperation. The work was funded by Quaid-I-Azam University Research Fund, the American Hearing Research Foundation and the NIH – National Institute of Deafness and other Communication Disorders grant DC03594. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403.
References
- 1.Davis AC. Hearing in Adults. London: Whurr; 1995. [Google Scholar]
- 2.Bergstrom L, Hemenway WG, Downs MP. A high risk registry to find congenital deafness. Otolaryngol Clin North Am. 1971;4:369–399. [PubMed] [Google Scholar]
- 3.Van Camp G, Willems PJ, Smith RJH. Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet. 1997;60 :758–764. [PMC free article] [PubMed] [Google Scholar]
- 4.Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. 2002 August; URL: http://dnalab-www.uia.ac.be/dnalab/hhh/
- 5.Cohen M, Gorlin R. Epidemiology, etiology, and genetic patterns. In: Gorlin R, Toriello H, Cohen M, editors. Hereditary Hearing Loss and its Syndromes. Oxford: Oxford University Press; 1995. pp. 9–21. [Google Scholar]
- 6.Heller S, Hudspeth AJ. Two deaf mice, two deaf mice: murine candidate genes pinpoint the genetic bases of nonsyndromic hearing loss in humans. Nat Med. 1998;4:560–561. doi: 10.1038/nm0598-560. [DOI] [PubMed] [Google Scholar]
- 7.Grimberg J, Nawoschik S, Bellusico L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from Blood. Nucleic Acid Res. 1989;17 :8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottingham RW, Jr, Indury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 9.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 10.Broman KW, Murray JC, Scheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolis EN, Yandavi N, Nadol JB, et al. A gene for non-syndromic autosomaldominant progressive postlingual sensorineural hearing loss maps to chromosome 14q12 – 13. Hum Mol Genet. 1996;5 :1047–1050. doi: 10.1093/hmg/5.7.1047. [DOI] [PubMed] [Google Scholar]
- 12.Salam AA, Hafner FM, Linder TE, Spillmann T, Schinzel AA, Leal SM. A novel locus (DFNA23) for prelingual autosomal dominant nonsyndromic hearing loss maps to 14q21-q22 in Swiss German kindred. Am J Hum Genet. 2000;66:1984–1988. doi: 10.1086/302931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukushima K, Ramesh A, Srisailapathy CR, et al. Consanguineous nuclear families used to identify a new locus for recessive non-syndromic hearing loss on 14q. Hum Mol Genet. 1995;4:1643–1648. doi: 10.1093/hmg/4.9.1643. [DOI] [PubMed] [Google Scholar]
- 14.Robertson NG, Lu L, Heller S, et al. Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat Genet. 1998;20:299–303. doi: 10.1038/3118. [DOI] [PubMed] [Google Scholar]