Abstract
Molecular interactions between protein complexes and DNA carry out essential gene regulatory functions. Uncovering such interactions by means of chromatin-immunoprecipitation coupled with massively parallel sequencing (ChIP-Seq) has recently become the focus of intense interest. We here introduce QuEST (Quantitative Enrichment of Sequence Tags), a powerful statistical framework based on the Kernel Density Estimation approach, which utilizes ChIP-Seq data to determine positions where protein complexes come into contact with DNA. Using QuEST, we discovered several thousand binding sites for the human transcription factors SRF, GABP and NRSF at an average resolution of about 20 base-pairs. MEME-based motif analyses on the QuEST-identified sequences revealed DNA binding by cofactors of SRF, providing evidence that cofactor binding specificity can be obtained from ChIP-Seq data. By combining QuEST analyses with gene ontology (GO) annotations and expression data, we illustrate how general functions of transcription factors can be inferred.
INTRODUCTION
Chromatin immunoprecipitation (ChIP) has become an important assay for the genome-wide study of protein-DNA interactions and gene regulation1–3. In a typical ChIP experiment, protein complexes that contact DNA are crosslinked to their binding sites, the chromatin is sheared into short fragments, and then the specific DNA fraction that interacts with the protein of interest is isolated by means of immunoprecipitation (IP). A genome-wide readout of the protein binding sites is produced either by hybridization of the DNA pool to a tiling array (ChIP-chip4) or by end-sequencing of millions of different DNA fragments (ChIP-Seq5–9). In higher organisms, particularly mammals, ChIP-chip data tend to have low resolution and are often quite noisy10, two shortcomings that ChIP-Seq promises to surmount. As a consequence, ChIP-chip is being rapidly displaced by ChIP-Seq in genome-wide discovery of mammalian transcription factor binding sites.
The goal of ChIP-Seq data analyses is to find those genomic regions that are enriched in a pool of specifically precipitated DNA fragments. Regions of high sequencing read density are referred to as “peaks” to evoke the visual impression of many reads mapping to a specific region compared to few reads mapping to the genomic background. The output of software implementing peak-finding methodology is a list of “peak calls” that comprises the genomic locations of sites inferred to be occupied by the protein. To date, studies that have presented ChIP-Seq data5,6 used peak finding methodology that heuristically quantifies read density but does not take full advantage of certain important properties of the data such as the directionality of sequencing reads. The growing importance of ChIP-Seq demands development of rigorous and transparent statistical approaches that fully leverage the inherent advantages of ChIP-Seq.
We here describe QuEST (Quantitative Enrichment of Short Tags), a new ChIP-Seq data analysis method that is based on realistic statistical modeling of the ChIP-Seq experimental approach. QuEST generates peak calls with substantial power and resolution by leveraging key attributes of the sequencing data, such as directionality of reads and the size of fragments that were sequenced (which, importantly, is estimated from the data themselves rather than provided by the user). QuEST achieves the desired balance between sensitivity and specificity by calculating false-discovery rates (FDR) from controls that are routinely conducted as part of ChIP experiments. Underlying QuEST’s statistical framework is the Kernel Density Estimation approach11 (KDE), which facilitates aggregation of signal originating from densely packed sequencing reads at the transcription factor binding sites, leading to statistically robust peak calls.
To demonstrate the power and resolution of analyses facilitated by QuEST, we generated ChIP-Seq data for three functionally different human transcriptional regulatory proteins that have well-defined binding specificities and regulatory roles. GABP (GA-binding protein) and SRF (serum response factor) are thought to function primarily as transcriptional activators12–18, whereas NRSF (neuron-restrictive silencer factor) is a transcriptional repressor19,20. We apply QuEST to these data as part of a larger work flow that also includes MEME-based motif discovery and, in the case of SRF, identification of co-motifs that are indicative of cofactor interactions. Finally, the ChIP-Seq data are analyzed in conjunction with microarray results and GO terms to provide further insight into the function of the factors.
RESULTS
Analytical Framework
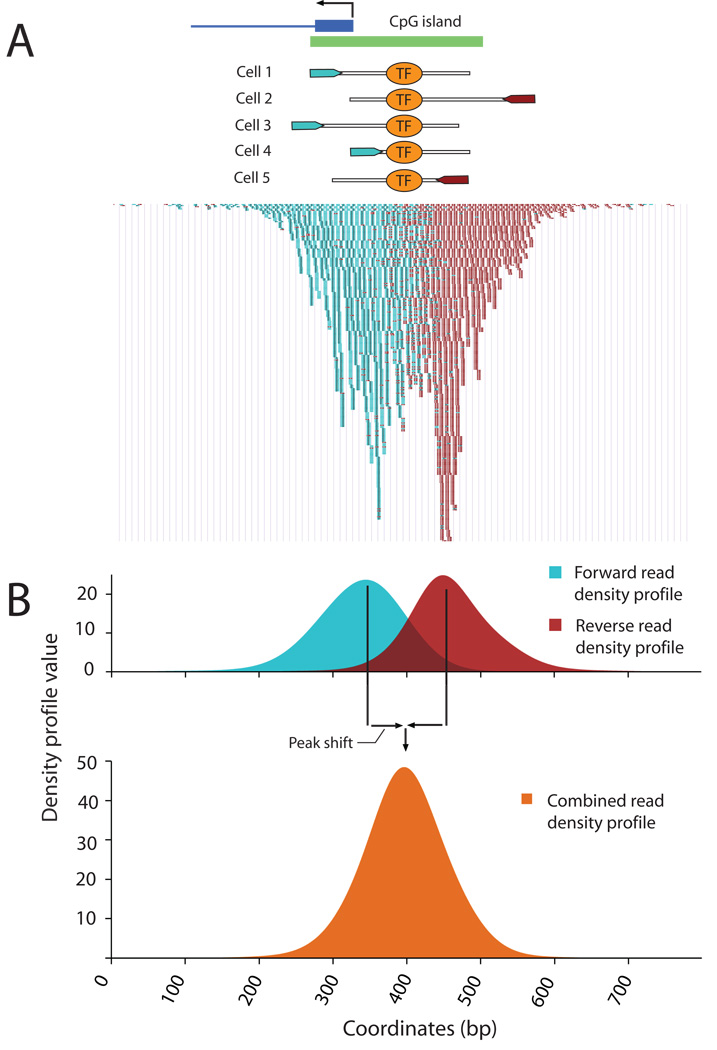
QuEST requires data in the form of genome coordinates (‘tags’) obtained from mapping several million sequencing reads to a reference genome. Tags from forward and reverse reads cluster on opposite sides of the transcription factor binding site (TFBS; Fig. 1A) This is because sequencing proceeds from one end of the fragment towards its middle in a strand-specific manner, which leads to an underrepresentation of tags in the immediate proximity of the TFBS.
Figure 1.
QuEST’s representation of ChIP-Seq data using density profiles.. (A) GABP ChIP-Seq reads from the promoter and CpG island of the Nitric oxide synthase interacting protein gene. Hypothetical GABP binding in five cells and the corresponding DNA fragments with sequencing reads. Below, actual read data. Forward reads are displayed as small blue bands and reverse reads as small maroon bands. (B) Forward (blue) and reverse (maroon) Read Density Profiles derived from the read data contribute to the Combined Density Profile (orange). The zero x-coordinate corresponds to coordinate 54775300 of human Chromosome 19, NCBI build 36.
QuEST first constructs two separate profiles, one for forward, and one for reverse tags. These profiles are characterized by strong peaks where tags are particularly dense (Fig. 1). The distance between forward and reverse peaks is not known a priori, but it is important to account for it and to correctly combine the two separate profiles into one. Since this distance may vary considerably among experiments, QuEST estimates it from a particularly robust subset of the data. We refer to half of this distance as the “peak shift” (Methods).
Once the experiment-specific peak shift has been estimated, the forward and reverse profiles are shifted by an equal amount and added to produce the Combined Density Profile (CDP) on which all subsequent analyses are carried out (Fig. 1B; Supplementary Fig. 1; Methods). By combining the profiles in this manner, QuEST accomplishes two key aspects of ChIP-Seq analysis: first, the signals from reverse and forward reads are represented by a single classifier; second, local maxima of this classifier correspond to protein-DNA crosslinking points, providing an estimate for the location of the TFBS.
The CDP is then searched for enriched loci in a process referred to as ‘peak calling’ to identify putative transcription factor binding sites. Specifically, initial candidates for peaks are identified as positions in the reference genome corresponding to local maxima of the CDP with sufficient enrichment compared to the control data (Methods). The strongest of these are likely to be due to real binding events, whereas weaker-scoring peaks may be false positives, requiring the setting of a CDP threshold for peak calling. Since this threshold may vary considerably between experiments, the desired balance between sensitivity and specificity is achieved by a calibration procedure. Briefly, the negative control data are separated into two sets, one of which is used as a pseudo-ChIP sample in which peaks are to be predicted, and the other of which serves as a background for this sample. Any peak that is predicted in this comparison is a false positive. Hence, the false-discovery rate (FDR) estimate is given by the ratio of the number of peaks predicted in the pseudo-ChIP analysis and the number of peaks identified in the real ChIP experiment. This approach allows the user to set specific thresholds and find out the FDR, or vary the thresholds until a desired FDR is achieved (Methods; Supplementary Fig. 2).
As a final result, for each peak in the list of high-confidence peak calls, QuEST reports a score quantifying the tag enrichment at the peak and a genome coordinate that corresponds to the position of that peak. Each such coordinate is a predictor of the position of a binding event, likely an endogenous TFBS occupied by the immunoprecipitated transcription factor. The KDE-derived score QuEST reports for each peak is proportional to the frequency at which the TFBS was present in the sequenced library. Because the score reflects the amount of evidence for the peak, QuEST ranks the final peak calls accordingly.
Performance of QuEST
ChIP Datasets
To evaluate key aspects of the performance of QuEST, we generated five libraries from the human Jurkat cell line and sequenced them using the Solexa platform (Table 1). One library each was from ChIPs against the transcriptional activators GABP and SRF, two were from ChIPs against the transcriptional repressor NRSF (utilizing two different antibodies, one polyclonal and the other monoclonal), and one was a negative control library for which the immunoprecipitation step was bypassed (“reversed-crosslinks, no IP” or RX-noIP). We generated 7.9, 8.7, 8.8 and 5.4 million mapped sequence tags for GABP, SRF, NRSF polyclonal and NRSF monoclonal datasets respectively (Table 1) as well as 17.4 million mapped tags for the RX-noIP library. QuEST identified 6442 (GABP), 2429 (SRF), 2960 (NRSF polyclonal) and 2596 (NRSF monoclonal) peak positions with significant enrichment of ChIP sequencing reads. Saturation analysis indicated that these libraries were sequenced to sufficient depth to identify the majority of significant peaks (Supplementary Fig. 3).
Table 1.
ChIP-Seq data and analysis summary.
| GABP | SRF | NRSF polycl. |
NRSF monocl. |
|
|---|---|---|---|---|
| Number of aligned ChIP reads | 7862231 | 8721730 | 8813398 | 5358147 |
| Number of peaks called by QuEST | 6442 | 2429 | 2960 | 2596 |
| FDR estimate | 1/6442 | 1/2429 | <1/2960 | 1/2595 |
| % peaks near genes (<2Kb or internal) | 83% | 72% | 53% | 53% |
Robustness and Reproducibility
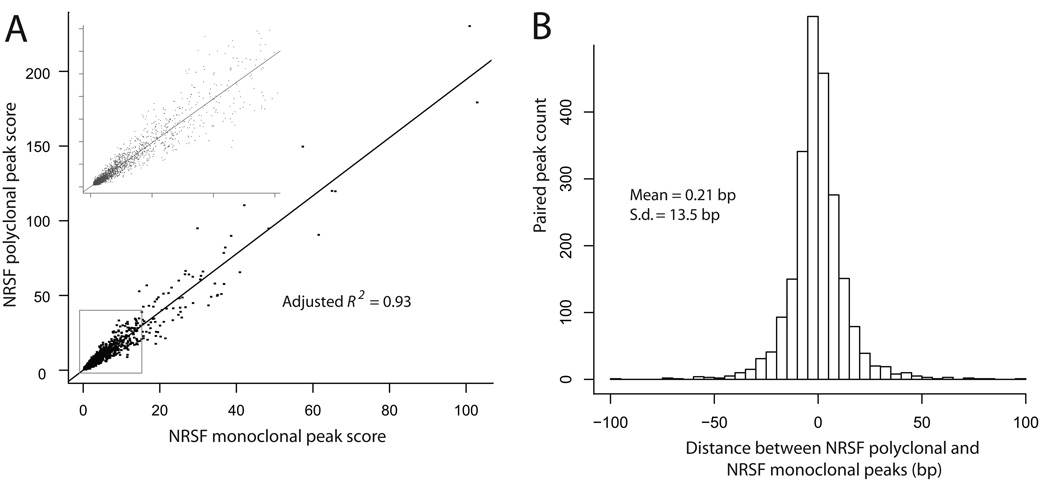
The QuEST scores of the 2320 peaks shared between the two NRSF datasets were exceptionally strongly correlated (r=0.97; Fig. 2A). The mean distance between corresponding peaks from the two data sets was 0.2 bp, with a standard deviation of 13.5 bp (Fig. 2B), demonstrating highly reproducible peak call positions. Overall, these results are evidence of QuEST’s ability to produce accurate quantification of tag enrichment that results in reproducible and robust peak calls.
Figure 2.
Reproducibility and robustness of QuEST results assessed by comparison between two independent NRSF data sets. (A) Correlation between NRSF polyclonal and NRSF monoclonal peak scores (rho = 0.97) with the inset expanding the portion near the graph origin. (B) Bar chart of the distance between NRSF polyclonal and NRSF monoclonal peak call positions.
Overlap of QuEST Peaks with Previous Studies
We identified previously described transcriptional targets of GABP, SRF, and NRSF providing some measure of validation for the peaks identified by QuEST in this study. These include GABP-regulated interleukin-16 (IL16) and cytochrome c oxidase subunits IV and Vb12 and SRF-regulated FHL221 QuEST also identified three peaks in the autoregulated SRF gene16, one in the promoter and two in one of the introns. Finally, the genes Calbidin 1, BDNF, SYT4 and NAV1 are NRSF targets in mouse embryonic stem cells20, and their orthologs were also marked by peaks in our data.
Precision and Accuracy of Peak Calls
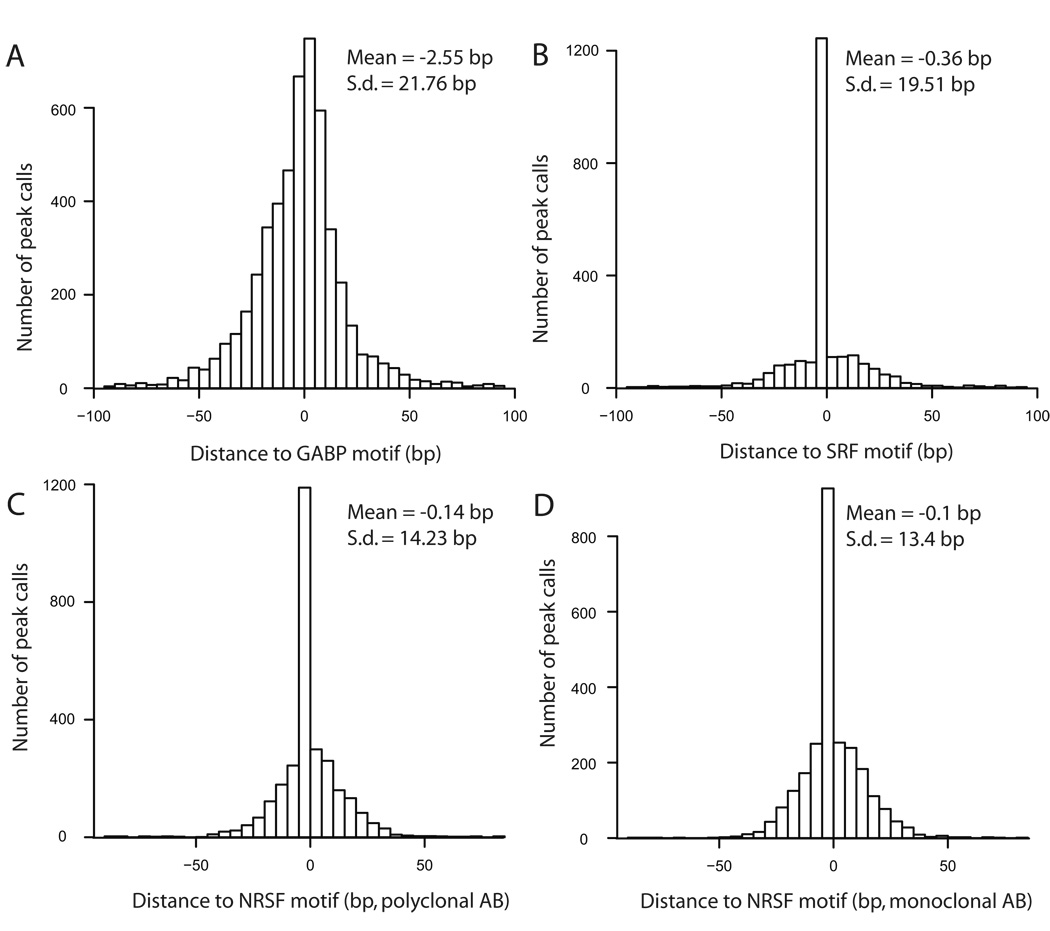
Theoretically, the genomic coordinate QuEST reports for each of its peaks should be ‘marked’ by the canonical TFBS motif. We first determined canonical motifs and their corresponding position specific scoring matrices (PSSMs) using the de novo motif finder MEME22. For each ChIP-Seq experiment, the input data into MEME was the set of 200 bp sequences from around each peak call. The resulting motifs closely corresponded to the previously established canonical recognition sites for each of the three factors12,15,23. To then determine the specific positions of motifs within the peak call regions, we asked for statistically significant matches of the PSSMs in the 200 bp around each QuEST peak, using a log-odds-ratio approach and a stringent threshold (Methods). The majority of peaks contained one or more highly significant PSSM matches, which were used to evaluate the resolution of QuEST peak calls. Remarkably, the mean distance between peak call and motif ranged from 0.1 bp in the NRSF monoclonal set to 2.55 bp for GABP, with the standard deviation ranging from 13.4 bp to 21.8 bp (Fig. 3).
Figure 3.
Resolution of QuEST as quantified by the distance between QuEST peak calls and TFBS motif centers. Histograms in each panel represent the distribution of peak distances to the nearest high-scoring motif.
Leveraging QuEST Peak Calls for Biological Insight
Canonical Motifs
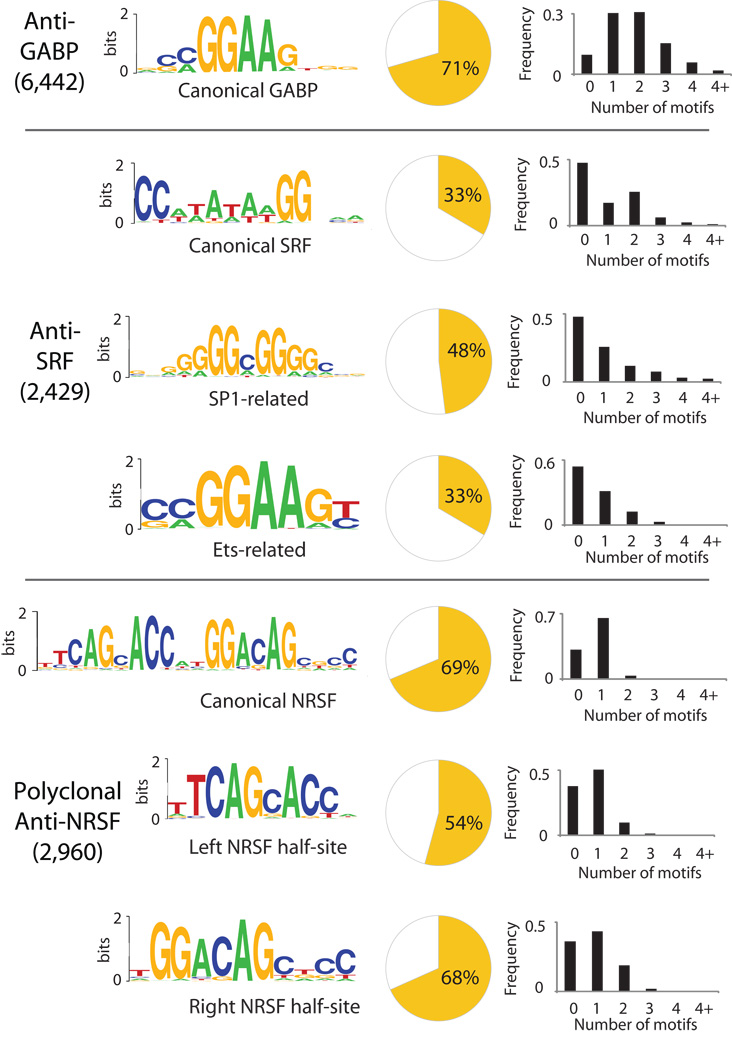
Our MEME analysis found that the canonical motifs of each transcription factor were most significantly enriched in their respective QuEST peaks (Fig. 4). Canonical motifs explain 71% (GABP), 33% (SRF) and 69% ( NRSF) of the peaks after accounting for motifs that are expected to occur by chance (Methods, Supplementary Fig. 4), illustrating QuEST’s high specificity in TFBS discovery. The comparatively low fraction of SRF peaks explained by its canonical motif is likely explained by cofactor interactions (see below). GABP and SRF, both of which assemble into a complex with a pair of DNA binding subunits12,16, most frequently contain two motifs (Fig. 4), in contrast to NRSF peaks, which typically harbor one.
Figure 4.
Motif analysis results. Each panel displays significantly overrepresented motif Weblogos24 for each of the three transcription factors. Pie-charts show the fraction of peaks with motifs in close proximity to the peak (< 100 bps). Histograms show the distribution of the motif number within 100 bps of the peak.
Interactions with other Factors
For SRF, the initial MEME analysis also yielded the SP1 motif. It explains a substantial fraction of peaks (48%), providing evidence that the previously suggested interaction25 between SP1 and SRF is common.
We also conducted a second round of MEME analyses focusing only on those peak-associated sequences that did not contain the canonical SRF motif. Such peaks may be due to indirect DNA binding of the targeted factor via a different, interacting, DNA binding protein. This analysis yielded an additional significant motif that resembled the recognition site of the Ets family of factors. This motif explained an additional 17% of the SRF peaks. The prevalence of an Ets-like motif may be due to the previously described interaction between SRF and the Ets factor ELK417,26. We note that the anti-SRF antibody has no detectable crossreactivity with other proteins on Western blots (not shown). The same strategy applied to the NRSF dataset reproduced the discovery of NRSF half-sites previously reported (Fig 4), and resulted in an additional 16% of peaks explained. No significant additional motifs were found for GABP.
We observed that a large fraction of SRF peaks (29%) occurred within 100 bp of GABP peaks, while NRSF peaks almost never coincided with either SRF or GABP peaks. The close proximity of SRF and GABP peaks might suggest that SRF not only physically interacts with the Est factor ELK417 but, in some promoters, with GABP as well.
Inference of Transcription Factor Function by Analysis of Genes with Peak Calls
QuEST analyses can be combined with orthogonal genome-wide data or resources such as GO to provide general insights into the functions of proteins targeted by ChIP-Seq experiments. For both GABP and SRF, a large majority of peak calls (83% and 72% respectively) were within 2kb of a gene. By contrast, only 53% of NRSF peak calls were within 2 kb of a gene, suggesting that NRSF’s effects on gene regulation are, on average, exerted over longer distances than those of GABP and SRF. Having obtained a set of peak-associated genes, we conducted gene expression profiling and Gene Ontology (GO) analyses to gain additional functional insight into the three DNA binding proteins of our study.
Gene expression profiling revealed that NRSF-associated genes were expressed at significantly lower relative levels than the average of all genes (Wilcoxon test, p-value < 2.2e-16, nNRSF=1274 mvs. nall=20588). This result is consistent with NRSF’s known general function as a transcriptional repressor and with previous results5. By contrast, both SRF-associated genes and GABP-associated genes were significantly higher expressed than the average gene (Wilcoxon test, both p-values <2.2e-16, nSRF=1936 and nGABP=5394 vs nall=20588, Supplementary Fig. 5), which is consistent with their activator functions 12, 15.
GO analysis27 (Supplementary Tables 1–3) revealed that NRSF-associated genes are mostly involved in neuronal function, which is consistent with previous results5. Both SRF and GABP had significant enrichment of genes that are involved in basic cellular processes, particularly those related to gene expression. These results are consistent with both GABP and SRF being fundamental regulators of basic cell biology, rather than specialized factors with specific physiological roles. GABP is the more broadly acting of the two factors, as reflected by its almost three-fold larger number of QuEST peaks and associated genes.
DISCUSSION
ChIP-Seq is rapidly becoming the approach of choice for genome-wide discovery of protein-DNA interactions, generating a need for robust and transparent analytical methodologies that leverage its inherent strengths. We developed QuEST to meet this need and utilized it as part of a work flow that is effective at producing a high-confidence list of specific and active TFBSs.
The high resolution of QuEST peak calls, evident for each of the diverse transcription factors we analyzed, is perhaps the most noteworthy methodological aspect of our results. For example, 89% of peaks that contained a significantly matching canonical TFBS motif in the NRSF polyclonal data were within 25 bp of the motif center, and 56% were within 10 bp (Fig. 3). QuEST thereby brings within reach an important goal in annotative functional genomics, which is to identify at high confidence the precise locations at which DNA binding proteins interact with the genome.
One feature that merits some discussion is the score QuEST generates for each peak, according to which the peaks are ranked. The score is directly proportional to the amount of tag enrichment in the set of DNA fragments that yielded sequences. Thus, a peak with a score of 50 is due to a TFBS that was twice as abundant in the DNA sample as a TFBS with a peak score of 25. While both scores may be above the reporting cutoff chosen (by the desired FDR), and are therefore considered real, there is twice the support for (and hence the confidence in) the stronger peak.
One potential drawback of QuEST is that it does not convert peak scores into definitive P-values. Instead, the stringency of peak calls is determined by the score threshold at which the peaks are reported, and the FDR is calculated for this threshold. Users can either use the default threshold or specify their own and assess the stringency through the FDR.
Model-free analysis as implemented in QuEST may be considered less powerful than approaches that leverage the additional power of an explicit model for the ChIP-Seq data. However, such explicit modeling will likely be elusive in the near future because of the many experimental and biological factors that influence the eventual enrichment signal that is detected by ChIP-Seq. Some part of the enrichment signal ought to reflect occupancy by the transcription factor, but confounding factors such as antibody specificity, epitope accessibility, and susceptibility of TFBS-adjacent DNA to shearing will be difficult to model explicitly. Furthermore, downstream manipulation necessary for library building, especially library amplification and sequencing, introduce additional biases into the enrichment signal. Together, these factors contribute to increased variance of signal strength across the binding sites, and complicate detection of weak binding signals. Application of QuEST or similar approaches will enhance our empirical understanding of ChIP-Seq data over the course of the next few years.
METHODS
Density profiles
Individual density profiles for forward and reverse reads at any position i of the genome are given by , where h is the kernel density bandwidth (we used h = 30 bases), is the Gaussian kernel density function, and C+/−(j) gives the number of 5’ read ends at position j for forward and reverse reads respectively. In contrast to the original kernel density estimator11, our density profiles represent un-normalized density estimates in which the sum is limited to sample points proximal to any given position (within 3 KDE bandwidths). These modifications were done for computational convenience (Supplemental Methods).
The CDP used in actual peak calling is calculated according to the formula H (i) = H+ (i−λ) + H− (i+λ), where λ is a peak shift parameter estimate, and H+ and H− are the positive and negative strand density profiles as defined above.
Peak shift estimation
For regions in which the number of tags exceeded 600 in a window of 300 bp, we calculated forward and reverse profiles and recorded local maxima. Regions for which the highest scoring local maximum was 20-fold or greater than the next scoring maximum, for both negative and positive strands, and for which the enrichment in the ChIP sample was at least 20-fold over that for the RX-noIP sample, were selected. The peak shift parameter value was calculated as half of the average distance between peaks on the negative and positive strand. This estimate is robust across all 4 ChIP datasets (Supplementary Fig. 6) and highly concordant for the two NRSF datasets.
Peak calling
Candidate peaks were identified where the QuEST score profile achieved a local maximum within a 21 bp window, provided their QuEST score was above the ChIP-threshold, which is determined in conjunction with the FDR procedure described below. Within each region, local peaks were identified. A peak was eliminated when the lowest point between it and the adjacent higher peak was greater than 0.9 times the CDP value of the higher peak. The remaining peaks were reported as “calls” if (1) the value of background CDP was lower than the background CDP threshold or (2) the ratio of the of ChIP CDP to the background CDP exceeded a specified threshold (referred to as “the rescue ratio”).
False Discovery Rate Estimate for the Number of Peaks
For each experiment, the RX-noIP data were split into two data sets, one of which served as a pseudo-ChIP data set (and matched the ChIP data in the number of reads) and the other served as the background set. Then, CDPs were calculated for ChIP, pseudo ChIP and background datasets. Using the same score thresholds and rescue ratios, peaks were called in the ChIP and pseudo-ChIP datasets independently by comparison to the background data. The number of called peaks in the pseudo-ChIP data is the false discovery number (FDN), and the FDR is simply the FDN divided by the number of peaks called in the ChIP-Seq experiment. For identification of peaks that were used in subsequent MEME analyses, a rescue ratio of 10 was used for all data sets, and for each dataset the score threshold was set such that the FDN was 1.
MEME analyses
For motif identification, we extracted, for each dataset separately, “peak-associated sequences” comprised of the set of 200 bp sequences surrounding each peak call. MEME was then applied with all default parameters to yield significantly overrepresented motifs in each dataset. To identify alternative motifs in the SRF and NRSF data, a log-of-odds threshold of 3.0 was used to remove the peaks containing canonical motifs in the 200 bp window around the peak, after which MEME was applied again. See Supplementary Methods.
MAST analyses
The number of peaks explained by a particular motif was generated by taking the maximum of the difference between the total number of peaks containing a motif, and the number that could be explained by chance, at a range of stringencies (E-values) using the MEME tool MAST. A description of E-value estimation can be found in Supplementary Methods. For MAST curves, see Supplementary Figure 4.
Additional Methods
ChIP-Seq library construction and sequencing, gene expression analysis and in further detail, density profile generation, peak calling, and MEME-based motif discovery are described in Supplementary Methods.
QuEST software is freely available for nonprofit use at http://mendel.stanford.edu/sidowlab/downloads/quest/. All data presented in this study (Rx-noIP and ChIP-Seq data, and peak call coordinates) can be found at the same website.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH Grants 5 U01 HG003162 and 1 U54-HG004576 to R.M.M., and by funds from the Stanford Pathology/Genetics Ultra-High Throughput Sequencing Initiative. We thank Larisa Tsavaler for performing the Illumina expression analysis, Wing Hung Wong, Ken McCue and members of Sidow lab for valuable discussions and suggestions.
Footnotes
AUTHOR CONTRIBUTIONS
A.V., S.B., A. Sundquist, and A. Sidow conceived the QuEST peak calling concept and developed the preliminary statistical framework. A.V. further developed and refined the statistical framework, and implemented QuEST. R.M.M. and D.S.J. devised the ChIP experiments. D.S.J., C.M., and B.A. performed the ChIP experiments. A.V. applied QuEST to the sequence data, and produced all quantitative results. A.V. and A. Sidow wrote the manuscript. A.V., D.S.J, S.B., R.M.M., and A. Sidow edited the manuscript.
EdSumm
AOP
A chromatin immunoprecipitation and sequencing (ChIP-Seq) data analysis package, QuEST, facilitates transcription factor binding site discovery at about 20 base-pair resolution.
Issue
A chromatin immunoprecipitation and sequencing (ChIP-Seq) data analysis package, QuEST, facilitates transcription factor binding site discovery at about 20 base-pair resolution.
References
- 1.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004 Feb 20;116(4):499–509. doi: 10.1016/s0092-8674(04)00127-8. 2004. [DOI] [PubMed] [Google Scholar]
- 2.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated Signal Transduction Kinases Frequently Occupy Target Genes. Science. 2006 Jul 28;Vol. 313.(no. 5786):533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 Jun 14;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieb JD. Genome-wide mapping of protein-DNA interactions by chromatin immunoprecipitation and DNA microarray hybridization. Methods Mol Biol. 2003;224:99–109. doi: 10.1385/1-59259-364-X:99. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007 Jun 8;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 6.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007 Aug;4(8):651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER. ChIP-seq: welcome to the new frontier. Nat Methods. 2007 Aug;4(8):613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 8.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007 May 8;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Wold B, Myers RM. Sequence census methods for functional genomics. Nat Methods. 2008 Jan;5(1):19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, Koch CM, Dunham I, Bieda M, Xu X, Farnham PJ, Kapranov P, Nix DA, Gingeras TR, Zhang X, Holster H, Jiang N, Green R, Song JS, McCuine SA, Anton E, Nguyen L, Trinklein ND, Ye Z, Ching K, Hawkins D, Ren B, Scacheri PC, Rozowsky J, Karpikov A, Euskirchen G, Weissman S, Gerstein M, Snyder M, Yang A, Moqtaderi Z, Hirsch H, Shulha HP, Fu Y, Weng Z, Struhl K, Myers RM, Lieb JD, Liu XS. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008 Mar;18(3):393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parzen E. On estimation of a probability density function and mode. Ann. Math. Stat. 1962 Sep;33:1065–1076. [Google Scholar]
- 12.Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol Dis. 2004 Jan-Feb;32(1):143–154. doi: 10.1016/j.bcmd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, Weng Z. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res. 2007 Jun 17;6:818–827. doi: 10.1101/gr.5623407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem. 2004 Sep 1;93(1):74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- 15.Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006 Nov;16(11):588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006 Jun 15;20(12):1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 17.Cooper SJ, Trinklein ND, Nguyen L, Myers RM. Serum Response Factor binding sites differ in three human cell types. Genome Res. 2007 Feb;17(2):136–144. doi: 10.1101/gr.5875007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins PJ, Kobayashi Y, Nguyen L, Trinklein ND, Myers RM. The ets-related transcription factor GABP directs bidirectional transcription. PLoS Genet. 2007 Nov;3(11):e208. doi: 10.1371/journal.pgen.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995 Oct;5(5):566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 20.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005 May 20;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Philippar U, Schratt1 G, Dieterich C, Mller JM, Galgczy M, Engel FB, Keating MT, Gertler F, Schle R, Vingron M, Nordheim A. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Genome Res. 2006 Feb;16(2):197–207. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 22.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park, California: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 23.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004 Jun;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madsen CS, Regan CP, Owens GK. Interaction of CArG elements and a GC-rich repressor element in transcriptional regulation of the smooth muscle myosin heavy chain gene in vascular smooth muscle cells. J Biol Chem. 1997 Nov 21;272(47):29842–29851. doi: 10.1074/jbc.272.47.29842. [DOI] [PubMed] [Google Scholar]
- 26.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004 Jan 7;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 27.Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006 Oct;16(10):1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.