Abstract
This case study shows the application of nontraditional diagnostic methods to investigate the microbial consortia inhabiting an ancient manuscript. The manuscript was suspected to be biologically deteriorated and SEM observations showed the presence of fungal spores attached to fibers, but classic culturing methods did not succeed in isolating microbial contaminants. Therefore, molecular methods, including PCR, denaturing gradient gel electrophoresis (DGGE), and clone libraries, were used as a sensitive alternative to conventional cultivation techniques. DGGE fingerprints revealed a high biodiversity of both bacteria and fungi inhabiting the manuscript. DNA sequence analysis confirmed the existence of fungi and bacteria in manuscript samples. A number of fungal clones identified on the manuscript showed similarity to fungal species inhabiting dry or saline environments, suggesting that the manuscript environment selects for osmophilic or xerophilic fungal species. Most of the bacterial sequences retrieved from the manuscript belong to phylotypes with cellulolytic activities.
Introduction
Biodeterioration of paper and parchment in ancient books and documents represents a cause of great concern for libraries and archives all over the world. The study of the mechanisms underlying the microbiological attack of historical materials has been widely practiced and still represents one of the main focuses of those institutions and laboratories that are involved in cultural heritage conservation. Microbial investigations based on cultivation strategies are not reliable because they yield only a limited fraction of the present microbial diversity [26]. The application of molecular biology techniques on cultural heritage environments has shown that new spoiling taxa and unsuspected microbial consortia are involved in the discoloration and biodeterioration of paintings and monuments [44]. Restricted sampling from art and documental objects results in additional problems for representative floristic analyses.
Filamentous fungi colonize different organic and inorganic materials and play an important role in biodeterioration processes [30, 35, 45, 57]. They can tolerate desiccation, high salt concentrations, and heavy metal compounds that are present in inks and pigments and are thus frequent inhabitants on paper-supported objects [14, 57, 58].
The fungal and bacterial communities that can develop on a book are similar to the communities of decomposers that, in natural environments, transform nutrients bound in dead organic matter into low molecular or inorganic forms, making them available to plants. The development and maintenance of a fungal community on a shelf of a library or in a single book depends on the spores that reach the material’s surface, on the microenvironment (temperature, relative humidity, light), on the water activity of the substrate, and on the casual events that help colonization of materials (insect dispersion, human contamination, external sources of fungal diversity) [14]. A library or a single book can be compared to a virgin land that can be reached by some colonizing organisms that behave like pioneer species on a nude soil. For Wardle [53] and Mikola et al. [29], species identity and composition of decomposers have far greater impacts on ecosystem processes than species richness per se. When considering paper stored in a closed environment, its colonization and biodegradation depends on species identity and composition since only cellulolytic organisms can exploit the bulk of the substrate. Like in natural environments, the diversity-functioning relationship is driven by the presence or absence of key species [52], by niche differentiation, and species interaction. Resource partitioning or facilitative (or negative) interactions between species [50] affect the substrate exploitation process in natural environments as well as in artificial ones.
In this case study, we present the application of non-traditional diagnostic methods to investigate the microbial consortia inhabiting an ancient manuscript. Molecular methods, including polymerase chain reaction (PCR), denaturing gradient gel electrophoresis (DGGE), and the creation of clone libraries, were used as a sensitive alternative to conventional cultivation techniques.
Materials and Methods
History of the Object
A manuscript dated back to 1293 from Italy was sent to the Istituto Centrale per il Restauro e la Conservazione del Patrimonio Archivistico e Librario (ICPAL) in Rome for restoration [7]. The volume is composed of 222 sheets divided into six gatherings with a binding made of parchment. It was written on Arabic paper made of linen and was characterized by a singular and never described deterioration phenomenon that gave the paper a dramatically felted aspect, especially in the margins. The paper-supported manuscript belongs to La Spezia’s Notaries Public Archive. It was edited by a Sarzanese notary called Parente Stupio between 1293 and 1294; it contains 464 deeds recording commodities, real estate, and land transactions. The volume, utterly transformed in appearance as a result of mechanical damage, damp, and microorganisms, was no longer in a consultable condition because the lower portions of its pages were shredded (Fig. 1a and b).
Figure 1.
a, b Manuscript dated back to 1293 from Italy, which belongs to La Spezia’s Notaries Public Archive
Sampling
Sterile cotton swabs were wiped along the most damaged margins of the outer and inner pages of the manuscript to obtain samples suitable for further fungal and bacterial culturing and identification. Loose fibers, dust, and powder were also collected from the manuscript by cleaning the pages with a soft brush; these materials were mainly used for microscopic observations. Some fragments of paper (2 to 3 mm2) were also collected, mainly from the margins of the most degraded pages for microscopic examination and molecular analysis. Paper samples from the manuscript were taken by restorers after an evaluation of the areas most suitable for this purpose. Two samples of 4 mm in diameter containing ink were detached from their position: these could not be repositioned during restoration and were, therefore, used to SEM observations and specifically for ink elemental analysis.
Optical Microscope Observations
A stereoscopic microscope fitted with low temperature fiber optic lighting was used to examine the stained and deteriorated areas of the book. A Leica MZ16 dissecting microscope was used for the direct examination of the book prior to the sampling procedure. Illuminated microscopic examination of mounted slides carrying fibers and powdery materials recovered from the manuscript was performed with an Olympus AX60 microscope fitted with phase contrast and a digital camera.
Agar/Broth Cultures
The viability of fungi and bacteria was tested using agar and broth cultures. Microbial structures sampled with cotton swabs were inoculated directly on to agar plates, and swabs were then immersed in sterile Czapek and Gelatine broth [42]. Some fragments of paper were used to perform a “25 points inoculum” [14], which consists of a multiple inoculum on agar divided into a grid with 25 points made up from sub-fragments of a paper sample previously washed several times with sterile water. The aim of this procedure is to cultivate those biodeteriogens that affect cellulose fibers and to avoid the development of air-borne contaminants. When a statistically significant fraction of the 25 points develop the same fungal/bacterial species, it is reasonable to consider that strain as being a paper spoiler.
The media used to grow the inocula were malt extract agar and dichloran 18% glycerol (DG18) agar prepared according to Samson et al. [42]. All the inoculations and re-inoculations were performed in a laminar flow hood to assure sterility throughout the procedures.
Molecular Analysis
DNA Extraction from Paper Material
DNA extraction was performed directly from the paper samples using the FastDNA SPIN Kit for Soil (Qbiogene, Illkirch, France) with modifications to the protocol as described by Michaelsen et al. [28]. To enhance DNA yields from all cells and spores on the paper surface, the samples were pretreated with lysozyme and Proteinase K as described by Schabereiter-Gurtner et al. [44]. DNA crude extracts were used directly for PCR amplification.
PCR Amplification of Extracted DNA
For the analysis of fungal sequences, fragments of about 700 bp in size corresponding to the ITS1, the ITS2 region, and the adjacent 5.8S rRNA gene were amplified with the primer pair ITS1 and ITS4 [5, 54]. For DGGE analysis, a nested PCR was performed with the PCR product of the first round as template DNA using the primers ITS1GC with a 37-base GC-clamp attached to the 5′ end [31, 32] and ITS2. All reactions were carried out as described in Michaelsen et al. [28].
For the identification of bacterial 16S rRNA sequences, DNA was amplified with the primer pair 341f/907r [31, 49]. For DGGE analysis, 200 bp fragments of the 16S rDNA were amplified with a nested PCR using the eubacterial specific primer 341f-GC with a 40-bp GC clamp added to its 5′ end [31] and the universal consensus primer 518r [33]. PCR conditions were as described by Schabereiter-Gurtner et al. [44]. All PCR products were analyzed by electrophoresis in a 2% (w/v) agarose gel.
DGGE
DGGE was performed as previously described [31] using a D-Code system (Bio-Rad) in ×0.5 TAE (20 mM Tris, 10 mM acetate, 0.5 mM Na2 EDTA; pH 7.8 with 8% (w/v) acrylamide).
For fungal fingerprints, gels contained a linear chemical gradient of 30–50% denaturants [100% denaturing solution contains 7 M urea and 40% (v/v) formamide] and were run at a constant temperature of 60°C for 14 h with a voltage of 100 V.
For bacterial fingerprints, gels contained a linear chemical gradient of 30% to 55% and were run at a constant temperature of 60°C during 3.5 h with a voltage of 200 V.
After completion of electrophoresis, gels were stained in an ethidium bromide solution and documented with a gel documentation system.
Creation of Clone Libraries and Sequence Analysis
To obtain a detailed phylogenetic identification of the microbial community members, clone libraries containing either ITS fungal regions (fungal community) or 16S rRNA fragments (bacterial community) were carried out. For fungal clone libraries, the DNA template was amplified using the primers ITS1/ITS4 [54] as mentioned above. For bacterial clone libraries, the primer pair 341f/907r was used as mentioned above. The PCR products were purified using the QIAquick PCR Purification Kit Protocol (Qiagen, Hilden, Germany) and resuspended in ddH2O water. Purified PCR products were ligated into the pGEM-T easy Vector system (Promega, Vienna, Austria) following the instructions of the manufacturer. The ligation products were transformed into Escherichia coli XL BlueTc to allow the identification of recombinants on an indicator LB medium containing ampicillin, tetracyclin, X-Gal, and IPTG as described in Sambrook et al. [41]. Clones were screened in a DGGE gel and sequenced as described by Schabereiter-Gurtner et al. [44]. Comparative sequence analysis was performed by comparing pairwise insert sequences with those available in the public online database NCBI using the BLAST search program [2].
SEM Observations and EDS Analysis
The analysis of paper samples was conducted using a variable pressure SEM instrument (EVO50, Carl-Zeiss Electron Microscopy Group) equipped with a detector for electron backscattered diffraction. In addition, a qualitative and quantitative chemical characterization of the inorganic constituents of the samples was performed by means of electronic dispersion spectroscopy (EDS), which allows for an X-ray area scanning of what is brought into focus in SEM images, thereby creating a compositional map of the paper’s surface. Only after having observed samples with SEM in Variable Pressure mode, at 20 keV, some of the samples were covered with gold with a Baltec Sputter Coater for a further analysis with SEM in High Vacuum mode. The High Vacuum allowed for a higher magnification of samples’ details.
Analysis of variance (ANOVA) was employed to investigate the differences in elemental composition of paper and ink with EDS data. In the ANOVA, each comparison is considered significant (the difference is significant) if the probability is out of the confidence interval on the basis of Tukey’s honestly significant difference (HSD) test.
Results
Optical Microscope Observations and Cultures
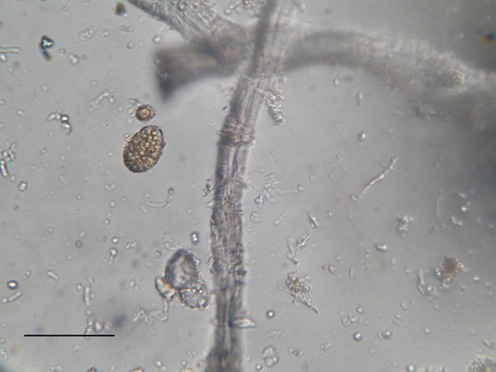
The observation with the optical microscope of some fibers showed the presence of fungal spores and pigmented cellular structures (Fig. 2). Cultures set up with fibers and powder sampled from the book and those obtained from cotton swabs resulted in the development of a few colonies of fungi (five colonies randomly distributed over 10 agar plates); distribution was not statistically significant. The colonies developed were identified as belonging to Penicillium commune Thom (two isolates) Cladosporium sphaerospermum Penzig (two isolates), Aspergillus niger van Tieghem (one isolate). None of the plates with DG18 agar for xerophilic species supported the growth of fungal colonies. The fragments of paper that were used to perform the “25 points inoculum” on agar also did not yield any positive result.
Figure 2.
Pigmented fungal structures, fungal spores, hyphae and conidia (Olympus AX60 microscope, light field). The black bar indicates 100 μm
SEM-EDS
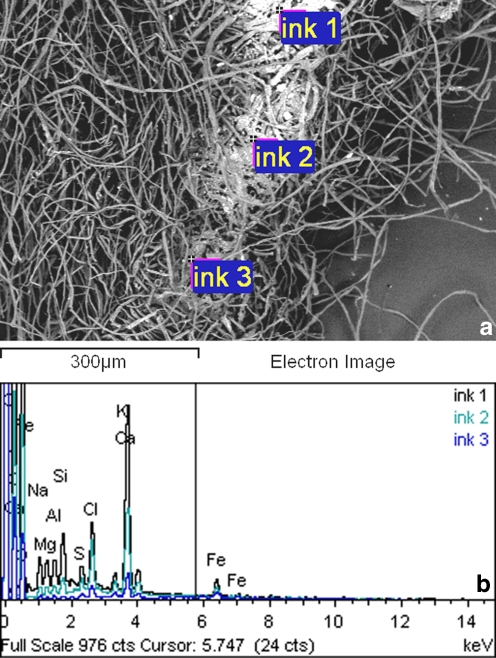
The microanalysis of the light-gray areas containing ink revealed the presence of statistically significant amounts of Fe, K, Al, Si, and Ca (Tables 1 and 2, Fig. 3a and b). Other elements present in paper but not statistically associated to inked areas were Na, Cl, Mg, S, and P. Na, Cl, and Mg relative percentages are consistent to the presence of sea salt, namely NaCl with traces of MgCl. The ink used for the writing of the manuscript can be considered an iron gall ink [7].
Table 1.
EDS microanalysis
| C | O | Na | Mg | Al | Si | P | S | Cl | K | Ca | Fe | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ink | 0.0 | 74.0 | 3.2 | 1.9 | 0.9 | 2.5 | 0.0 | 1.5 | 3.4 | 0.9 | 9.4 | 2.4 | 100.0 |
| Ink | 0.0 | 74.5 | 3.1 | 1.9 | 1.0 | 1.7 | 0.7 | 0.7 | 3.6 | 0.9 | 10.2 | 1.9 | 100.0 |
| Ink | 43.2 | 47.9 | 0.8 | 0.4 | 0.4 | 0.6 | 0.0 | 0.3 | 1.3 | 0.4 | 3.9 | 0.8 | 100.0 |
| Ink | 45.6 | 45.9 | 0.9 | 0.5 | 0.6 | 1.3 | 0.1 | 0.2 | 1.1 | 0.4 | 2.5 | 0.8 | 100.0 |
| Paper | 52.1 | 44.9 | 0.7 | 0.3 | 0.0 | 0.1 | 0.0 | 0.2 | 0.7 | 0.2 | 0.7 | 0.0 | 100.0 |
| Paper | 51.4 | 46.4 | 0.5 | 0.2 | 0.0 | 0.0 | 0.0 | 0.2 | 0.7 | 0.2 | 0.4 | 0.0 | 100.0 |
| Paper | 51.1 | 46.8 | 0.5 | 0.1 | 0.0 | 0.1 | 0.0 | 0.1 | 0.7 | 0.1 | 0.5 | 0.0 | 100.0 |
| Paper | 48.8 | 48.4 | 0.6 | 0.2 | 0.1 | 0.2 | 0.0 | 0.1 | 0.7 | 0.2 | 0.6 | 0.2 | 100.0 |
Quantitative chemical characterization of the inorganic constituents of the samples performed by means of EDS, which allows for an X-ray area scanning of what is brought into focus in SEM images, thereby creating a compositional map of the paper’s surface. The results showed were obtained analyzing surfaces with and without the graphic sign (ink/paper) as was visualized with the Backscattered detector. All elements normalized. All results in weight%
EDS electronic dispersion spectroscopy
Table 2.
Summary of all pairwise comparisons of the samples based on the average values obtained for the EDS analysis of paper and ink
| Category | Paper | Ink |
|---|---|---|
| C | 50.930 A | 22.190 A |
| O | 46.600 A | 60.560 A |
| Na | 0.549 A | 2.003 A |
| Mg | 0.202 A | 1.163 A |
| Al | 0.024 B | 0.731 A |
| Si | 0.099 B | 1.536 A |
| P | 0.000 A | 0.211 A |
| S | 0.137 A | 0.660 A |
| Cl | 0.713 A | 2.331 A |
| K | 0.159 B | 0.649 A |
| Ca | 0.570 B | 6.494 A |
| Fe | 0.038 B | 1.464 A |
α = 0.05; test used: Tukey’s, HSD; analysis of the differences between the categories with a confidence interval of 95%. Samples signed with different letters (A or B) are significantly different
Figure 3.
a, b Microanalysis of the light-gray areas containing ink revealed the presence of statistically significant amounts of Fe, K, Al, Si, and Ca
The SEM observation of the morphology of paper fibers allowed the definition of the plant origin of the cellulosic material. Some distinctive characters, like the dimension of the fibers and the presence of peculiar septa indicated that the fibers used in the manufacture of the volume’s paper were from linen [7]. Moreover, starch granules were observed with SEM analysis, and its use as sizing material was confirmed by a colorimetric test [7].
SEM analysis at high magnification performed on samples both before and after coating with gold allowed the visualization of fungal spores and hyphae attached onto fibers (Figs. 4, 5, and 6). Fungal spores appeared globose and echinated, but any attribution to a genus would be misleading because of their desiccated state, which makes any systematic description useless.
Figure 4.
Scanning electron micrograph obtained wih a backscattered electron detector (QBSD) in Variable Pressure mode
Figure 5.
Scanning electron micrograph at high magnification (magnification 10.000×) performed on samples after coating with gold. High Vacuum mode
Figure 6.
Scanning electron micrograph performed on samples after coating with gold. High Vacuum mode
Molecular Analysis
For molecular analyses, pieces of paper (approximately 2 to 3 mm2) obtained from the book were used for direct DNA extraction.
The DNA extracts were amplified by PCR with primers targeting the ITS regions of fungi, as well as the 16S rRNA gene of bacteria. The fungal and bacterial amplified fragments were further analyzed by DGGE fingerprints. DGGE analysis revealed fingerprints for both bacterial and fungal communities.
Figure 7 shows the DGGE profile derived from the fungal community colonizing paper material; DGGE profile showed a complex fungal community consisting of seven to eight dominant DGGE bands as well as some other faint bands. DGGE derived from the bacterial community showed less bands, indicating a lower biodiversity of bacteria on the paper sample (data not shown).
Figure 7.

DGGE profile derived from the fungal community colonizing paper material
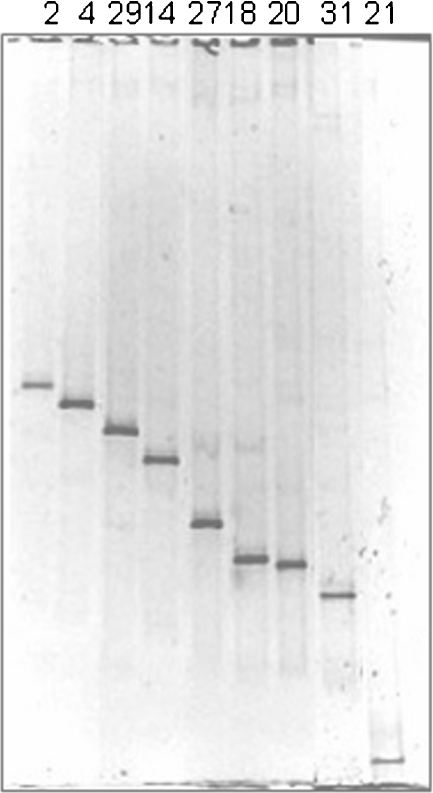
For phylogenetic identification of the individual members of the fungal and bacterial communities, two clone libraries containing either the fungal ITS regions and the 5.8S rRNA gene or the bacterial 16S rRNA gene were carried out. The resulting fungal and bacterial clones were further screened by DGGE. The obtained sequences were compared with ITS regions and 16S rRNA gene sequences of known fungi and bacteria, respectively, listed in the EMBL database. Figure 8 shows an example of the profiles derived from bacterial clones screened on DGGE. Tables 3 and 4 show the phylogenetic affiliations of the fungal and bacterial clones obtained in this study, respectively.
Figure 8.
Example of the profiles derived from bacterial clones screened on DGGE
Table 3.
Phylogenetic affinities of partial 16S rRNA coding sequences derived from bacterial clones libraries performed with paper samples obtained from the book written by Parente Stupio between 1293 and 1294
| Clones | Length | Closest identified phylogenetic relatives [EMBL accession numbers] | Similarity (%) | Accession no. |
|---|---|---|---|---|
| K2 | [589] | Bacillus sp. [DQ993298] | 98.0 | FN394538 |
| Caryophanon latum [X70319] | 98.0 | |||
| Uncultured bacterium clones [EU771735; EU466988] | 98.0 | |||
| K4 | [589] | Acinetobacter sp. [FJ544340] 16S rDNA of the microorganism resources for herbaceous fibers extracting. | 99.0 | FN394539 |
| Acinetobacter sp. [FM164629; FJ587508; FJ608713] | 99.0 | |||
| K14 | [589] | Uncultured bacterium clone zd3-48 [EU527183] | 99.0 | FN394540 |
| Bacillus sp. MB-7 and MB-1 [AF326364, AF326359] manganese(II) oxidative species. | 97.0 | |||
| Virgibacillus picturae [AJ276808] isolated from biodeteriorated wall paintings. | 97.0 | |||
| K18 | [588] | Uncultured Bacilli bacterium clones [EF664900; EF075265] | 99.0 | FN394541 |
| Sulfobacillus disulfidooxidans [AJ871255] | 97.0 | |||
| K20 | [589] | Bacillus sp. [AF548878; AF548879] Strains able of Specific Ureolytic Microbial Calcium Carbonate Precipitation | 99.0 | FN394542 |
| Sporosarcina sp. [EU182901, DQ073393; DQ993301; AB245381] | 99.0 | |||
| K21 | [571] | Kocuria sp. I_GA_W_11_16 [FJ267551] an airborne bacteria in indoor air | 99.0 | FN394543 |
| Kocuria sp. [FJ357623; FJ237398; EU073079] | 99.0 | |||
| Uncultured bacterium isolate [AM697331; AM696378] from a bacterial community in indoor environment | 99.0 | |||
| Uncultured bacterium clones [FM873202; FM873503; FM872761; FM873229 ] occupant as a source of house dust bacteria | 99.0 | |||
| K27 | [589] | Stenotrophomonas maltophilia [FJ405363; AY837728; AY841799; EU034540; EU294137] | 99.0 | FN394544 |
| K29 | [563] | Clostridium colinum type strain DSM 6011T [X76748] | 94.0 | FN394545 |
| K31 | [571] | Uncultured bacterium clones [EU466486;EU778711 ] gut microbes | 99.0 | FN394546 |
| Bacillus sp. [AB112729; EU249555; AJ717382; EF422411] | 97.0 |
Table 4.
Phylogenetic identification of fungal sequences derived from fungal clones libraries performed with paper samples obtained from the book written by Parente Stupio between 1293 and 1294
| Phylum | Order | Clone | Length (bp) | Phylogenetic identification | Similarity (%) | Accession number |
|---|---|---|---|---|---|---|
| Ascomycota | Capnodiales | F25 | 203 | Cladosporium sp. [DQ780355; DQ780357; DQ092512; DQ299303] | 98 | FN394518 |
| Eurotiales | F3 | 250 | Aspergillus penicillioides strain ATCC 34946 [AY373861] | 98 | FN394514 | |
| F37 | 205 | Aspergillus sp. [FJ196620; EU862194; DQ865103] | 99 | FN394520 | ||
| K5F | 603 | uncultured Aspergillus [FM165464] detected in a 16th-century book | 96 | FN394522 | ||
| Aspergillus versicolor [EF652480] | 96 | |||||
| nF2 | 598 | uncultured Aspergillus [FM165464] detected in a 16th-century book | 99 | FN394526 | ||
| Aspergillus versicolor [EF652480] | 99 | |||||
| nF7 | 622 | uncultured Aspergillus [FM165464] detected in a 16th-century book | 99 | FN394528 | ||
| Aspergillus versicolor [EF652480] | 99 | |||||
| nF13 | 603 | uncultured Aspergillus [FM165464] detected in a 16th-century book | 99 | FN394530 | ||
| Aspergillus versicolor [EF652480] | 99 | |||||
| nF19 | 602 | uncultured Aspergillus [FM165464] detected in a 16th-century book | 99 | FN394533 | ||
| Aspergillus versicolor [EF652480] | 99 | |||||
| nF24 | 613 | uncultured Aspergillus [FM165464] detected in a 16th-century book | 95 | FN394536 | ||
| Aspergillus versicolor [EF652480] | 95 | |||||
| nF28 | 523 | Aspergillus terreus [FJ011536; EU515150; AY360402] | 98 | FN394537 | ||
| K31F | 627 | Eurotium halophilicum isolate NRRL 2739 [EF652088] | 95 | FN394525 | ||
| nF6 | 711 | Eurotium halophilicum isolate NRRL 2739 [EF652088] | 99 | FN394527 | ||
| nF14 | 692 | Eurotium halophilicum isolate NRRL 2739 [EF652088] | 99 | FN394531 | ||
| nF21 | 701 | Eurotium halophilicum isolate NRRL 2739 [EF652088] | 99 | FN394535 | ||
| nF12 | 604 | Penicillium sp. OY12007 [FJ571473] | 100 | FN394529 | ||
| nF18 | 626 | Penicillium chrysogenum [EF200090; AY373902; AF033465] | 99 | FN394532 | ||
| K23F | 645 | Uncultured Penicillium clone 1F33 [FM165470] detected in a 16th-century book | 98 | FN394524 | ||
| Penicillium pinophilum [AB369480; AF176660; AB194281] | 98 | |||||
| Dothideales | nF20 | 634 | Uncultured Penicillium clone 1F33 [FM165470] detected in a 16th-century book | 99 | FN394534 | |
| Penicillium pinophilum [AB369480; AF176660; AB194281] | 99 | |||||
| F8 | 227 | Aureobasidium pullulans [FJ515165] | 100 | FN394516 | ||
| Pleosporales | F42 | 209 | Alternaria sp. [FJ467349; FJ545250; FJ618522] | 99 | FN394521 | |
| Basidiomycota | Wallemiales | F12 | 160 | Wallemia sebi strain UAMH 7897 [AY625073; AY302517] | 100 | FN394517 |
| Erythrobasidiales | F28 | 192 | Rhodotorula aurantiaca [AB093528] | 100 | FN394519 | |
| Non-clasified | K14F | 562 | Fungal sp. [AY843071] melanized fungi from rock formations in the central mountain system of Spain | 91 | FN394523 | |
| F4 | 210 | Uncultured basidiomycete isolate dfmo0688_100 [AY969394] | 99 | FN394515 |
Microflora Detected on Paper Material
Bacteria
Table 3 shows the phylogenetic affiliations of bacterial clones obtained in this study. Five bacterial sequences (clones K2, K14, K18, K20, and K31) showed high score similarities, between 97% and 99% similarity, with Bacillus-related sp. as well as with some uncultured bacterial clones. Clone K4 showed a high score similarity with Acinetobacter sp. and clone K21 showed a high score similarity with Kocuria sp. as well as with some uncultured bacterium inhabiting indoor environment and house dust.
Clone K27 showed high score similarity with Stenotrophomonas maltophilia and clone K29 showed high score similarity with Clostridium colinum.
Fungi
Table 4 shows the phylogenetic affiliations of fungal clones obtained from the manuscript. Clone nF28 showed high similarity (98%) with Aspergillus terreus Thom. One sequence (clone F8) showed maximum score similarity (100%) with Aureobasidium pullulans (de Bary) G. Arnaud. Two sequences (clones K23F, nF20) showed a high score similarity with Penicillium pinophilum Thom.
Six fungal sequences (K5F, nF2, nF7, nF13, nF19, nF24) showed high score similarities, between 95% and 99%, with Aspergillus versicolor (Vuill.) Tirab.
Four fungal sequences (clones K31F, nF6, nF14, nF21) showed high score similarity with Eurotium halophilicum Chr., Papav. & Benj. Among the fungal DNA sequences obtained from the manuscript, clone F3 presented a high similarity with A. penicilloides, and clone F12 totally corresponded to Wallemia sebi (Fr.) Arx (100% similarity). Clone F28 gave a 100% similarity with a xerophilic fungal species: Rhodotorula aurantiaca (Saito) Lodder is an anamorphic basidiomycetous yeast species [12].
Four clones (F42, nF12, F25, F37) showed high similarity to sequences addressed to defined fungal genera but could not be identified at the species level [55]. These are all fungi that can be both considered dust inhabitants and paper spoilers, namely Cladosporium sp., Alternaria sp., Penicillium sp., and Aspergillus sp.
Discussion
The poor conservation condition of the manuscript presented in this case study did appear to be the result of biological activity, with SEM observations proofing several fungal spores attached to cellulose fibers.
Agar-based cultivation methods showed the presence of living fungal spores of few species that can be considered surface contaminants and dust inhabitants but not in a statistically significant number to be considered responsible for paper biodegradation. The conditions leading to a transformation from a surface contaminant into a paper spoiler are the micro-environmental conditions, the characteristics of the substrate, and the physiological attitudes of the organism itself. From the EDS and chemical characterization of the manuscript, we knew that its paper did not only consist of cellulose but also a complex mixture of starch, cellulose fibers, iron-containing ink, and other inorganic elements that could have represented, as a whole, a substrate for microbes. DNA sequence analysis confirmed the existence of fungi and bacteria on paper material that could not be cultivated with traditional methods.
Potential Deteriorative Actions by the Detected Microflora
Bacteria
Several bacteria have been already isolated from paper materials showing foxing deterioration [15, 25, 30], but to our knowledge, only few studies concerning their taxonomical identification or paper degrading activity has been published [11]. From the ecological point of view, the requirements of prokaryotes are very similar to the environmental needs of fungi and yeasts. One essential condition for bacterial life is a high level of humidity in the environment; thus, bacteria have to withstand drying to exist, as the sporogenic and osmophilic species discovered on this Italian manuscript do.
Facultative anaerobic or microaerophilic bacteria, as Bacillus and Clostridium strains have been detected in the book investigated in this study. They are cellulolytic bacteria and seem to play an active role in the deterioration processes. Clostridium sp. is forming a dominant group of cellulolytic bacteria in municipal solid waste [6, 10]. Bacillus sp. has been already isolated from paper affected by foxing [11] as well as from wooden art objects in museum environments [37]. Furthermore, Bacillus and related species have been shown to be the most commonly detected bacteria (up to about 20%) among the variety of microbial species isolated from the pulp and paper mill environment [8, 36, 46, 47, 51]. In addition, Bacilli have been found as the predominant cellulolytic group of bacteria in landfill, where cellulose accounts for 40% to 50% of the municipal solid waste [38], and they form a significant proportion of the intestinal microbial community of soil invertebrates, especially among cellulose degraders [23].
Acinetobacter sp. and Kocuria sp. are osmophilic bacteria described as food spoilers [19, 20]. These species has also been detected as part of the microflora of the gut of termites and other invertebrates, and they are also involved in the degradation of polymeric material, as cellulose and hemicellulose, under oxygen limitation [22]. It can be hypothesized that these bacterial strains colonized cellulose fibers due to an occasional wetting event that raised water content in the manuscript to high values and that favored bacterial dispersal and growth.
S. maltophilia represents a rhizosphere bacterial species which has a potential agronomic importance due to its capability as biocontrol for plant diseases. Traits of S. maltophilia associated with biocontrol mechanisms include antibiotics production [18] and extracellular enzyme activities such as protease and chitinase [21, 56]. Many chitinolytic bacteria have found to produce more than one kind of chitinase. The efficient chitin degradation is assumed to be performed by the combination of these multiple chitinases. Synergistic effects on degradation of chitin or cellulose have been observed in the simultaneous action of different types of hydrolases [3, 16]. S. maltophilia could be described as a secondary colonizer of the manuscript that colonized fungal material actively growing on paper.
Fungi
Fungal cell walls are composed of chitin [12] which is a polymer containing Nitrogen. A high carbon-to-nitrogen ratio in a substrate represents a limiting factor for microbial colonization [53]. Following a fungal colonization, cellulosic material becomes more palatable for many microorganisms since it becomes enriched in nitrogen. The succession of biological “events” that could have occurred to the object is somehow recorded in the microbial and fungal dead or living material present in it. Among the fungal species that were found in the manuscript, some could be considered strongly cellulolytic and, therefore, capable of colonizing pure cellulose. It is the case of A. terreus, A. pullulans, and P. pinophilum which have all been already associated to the biodegradation of library and archival materials [39, 57, 58].
A. terreus is a fungal species used in biodegradation of lignocellulosic waste, thanks to its abilities in degrading both cellulose and phenolic compounds [13].
A. pullulans is a “black yeast” that grows on tree leaves and in salt water marshes [9]. The fungus contains multiple life forms (polymorphic) including blastospores, hyphae, chlamydospores, and swollen cells. The chlamydospores and swollen cells are considered resting forms. The fungus produces a green melanin which turns black over time.
P. pinophilum is considered an efficient producer of cellulases and hemicellulases [4].
The colonization of the manuscript by A. versicolor is probably only secondary to the growth of strong cellulolytic species, or alternatively, its growth was supported mainly by starch and gelatine used in papermaking. A. versicolor has been isolated from both paper and parchment affected by discoloration and structural damage [14, 24, 58] and as a species exhibits a high amlylolytic and gelatinolytic activity, but it is only moderately cellulolytic [17]. It is a species that can deteriorate also polymeric materials [27], grow on building materials [34], and cinematographic films [1] indicating that it has a great plasticity and physiological versatility. A. versicolor is generally xerophilic, meaning that it can grow at low water activity (<0.80). The minimum and maximum growth temperatures for A. versicolor are 4°C and 40°C with an optimum at 30°C. Its optimal water activity is 0.95 with a minimum of 0.75 [42]. The manuscript presented some portions that were effectively reduced to shreds, suggesting that paper microbial degradation occurred with detriment of the glues and the sizing materials and not of mainly cellulose fibers, thus resulting in a loss of structure but not of substance. The high number of clones obtained for A. versicolor and the enzymatic abilities of this species for the materials constituting the manuscript suggests that it had a role in the biodeterioration of some parts of the book, although the fungal material recovered from the cellulose fibers was no longer viable or culturable.
E. halophilicum is a rare species, strictly xerophilic (tonophilic), but its occurrence in the environment and on materials in dry or salted habitats may be underestimated because it commonly fails to grow on agar media. E. halophilicum has been associated to Aspergillus penicilloides by Samson and Lustgraaf [43] as inhabitants of house dust [48]. A. penicilloides and W. sebi are tonophilic fungi that can grow on substrates with very low water activity [42]. These xerophilic species can be found as biodegrading agents of salted substrata, like salted meat or vegetables [42].
The volume was utterly transformed in appearance also as a result of a soaking event, as documented by some discolorations on the margins of paper sheets. The manuscript was used by a notary to record commodities real estate and land transactions in a sea town and was probably exposed to sea air and breeze, which correlates with the high contents of NaCl and MgCl that were shown by EDS elemental analysis (Tables 1 and 2). In fact Na, Cl, and Mg were not associated to inked areas and could not be considered paper or sizing constitutive elements [40].
The presence of 3% to 4% in weight of marine salt among paper fibers (Table 1) is consistent with the presence of a considerable number of osmophilic or tonophilic fungal species. Most of the fungal clones showed, in fact, similarity with species typical for dry or salted environments, suggesting that the manuscript is characterized by a microflora with a natural selection for osmophilic or xerophilic fungal species.
Acknowledgments
The authors are grateful to Dr. Maria Luisa Riccardi (restorer at the Restoration Laboratory at the ICPL (Rome) and person in charge of the restoration of the book, for the supply of materials, and her kind collaboration). The molecular analyses included in this study, and A. Michaelsen, were financed by the Austrian Science Fund (FWF) within the framework of the project P17328-B12. G. Piñar was financed by the “Hertha-Firnberg-Nachwuchsstelle- T137” from the Austrian Science Fund (FWF).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Abrusci C, Martín-González A, Del Amo A, Catalina F, Platas G. Isolation and identification of bacteria and fungi from cinematographic films. Int Biodeterior Biodegrad. 2005;56:58–68. doi: 10.1016/j.ibiod.2005.05.004. [DOI] [Google Scholar]
- 2.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman JD. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisset C, Fraschini C, Schülein M, Henrissat B, Chanzy H. Imaging the Enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl Environ Microbiol. 2000;66:1444–1452. doi: 10.1128/AEM.66.4.1444-1452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown JA, Collin SA, Wood TM. Enhanced enzyme production by the cellulolytic fungus Penicillium pinophilum, mutant strain NTGIII/6. Enzyme Microb Technol. 1987;9:176–180. doi: 10.1016/0141-0229(87)90074-3. [DOI] [Google Scholar]
- 5.Buchan A, Newell SY, Moreta JIL, Moran MA. Analysis of internal transcribed spacer regions of rRNA genes in fungal communities in a south eastern US salt marsh. Microb Ecol. 2002;43:329–340. doi: 10.1007/s00248-001-1062-0. [DOI] [PubMed] [Google Scholar]
- 6.Cailliez C, Benoit L, Thirion JP, Petitdemange H, Raval G. Characterization of 10 mesophilic cellulolytic Clostridia isolated from a municipal solid waste digestor. Curr Microbial. 1992;25:105–112. doi: 10.1007/BF01570968. [DOI] [Google Scholar]
- 7.Carrarini R, Casetti Brach C (2006) Libri & Carte Restauri e analisi diagnostiche a cura di Quaderni 1, Istituto Centrale di Patologia del Libro, Gangemi editore, Roma, pp 35-38
- 8.Chandra R, Singh S, Krishna Reddy MM, Patel DK, Purohit HJ, Kapley A. Isolation and characterization of bacterial strains Paenibacillus sp. and Bacillus sp. for kraft lignin decolorization from pulp paper mill waste. J Gen Appl Microbiol. 2008;54:399–407. doi: 10.2323/jgam.54.399. [DOI] [PubMed] [Google Scholar]
- 9.Cooke WB (1959) An ecological life history of Aureobasidium pullulans (de Barry) Arnaud. Mycopathologia et Mycologia Applicata pp 1-45 [DOI] [PubMed]
- 10.Cummings SP, Stewart CS. Newspaper as a substrate for cellulolytic bacteria. J Appl Bacteriol. 1994;76:196–202. [Google Scholar]
- 11.De Paolis MR, Lippi D. Use of metabolic and molecular methods for the identification of a Bacillus strain isolated from paper affected by foxing. Microbiol Res. 2008;163:121–131. doi: 10.1016/j.micres.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Domsch KH, Gams W, Anderson T-H. Compendium of soil fungi. New York: Academic; 1980. [Google Scholar]
- 13.Emtiazi G, Naghavi N, Bordbar A. Biodegradation of lignocellulosic waste by Aspergillus terreus. Biodegradation. 2001;12(4):259–263. doi: 10.1023/A:1013155621336. [DOI] [PubMed] [Google Scholar]
- 14.Gallo F. Biological factors in the deterioration of books, Technical Notes. Rome: ICCROM; 1985. [Google Scholar]
- 15.Gallo F, Pasquariello G. Foxing, ipotesi sull’origine biologica. Boll Ist Centr Pat Libro. 1989;43:136–176. [Google Scholar]
- 16.Gaudin C, Belaich A, Champ S, Belaich JP. CelE, a multidomain cellulase from clostridium cellulolyticum: a key enzyme in the cellulosome? J Bacteriol. 2000;182:1910–1915. doi: 10.1128/JB.182.7.1910-1915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gopinath Subash CB, Periasamy A, Azariah H. Extracellular enzymatic activity profiles in fungi isolated from oil-rich environments. Mycoscience. 2005;46(2):119–126. doi: 10.1007/s10267-004-0221-9. [DOI] [Google Scholar]
- 18.Jakobi M, Winkelmann G. Maltophilin: a new antifungal compound produced by Stenotrophomonas maltophilia R3089. J Antibiot. 1996;49:1101–1104. doi: 10.7164/antibiotics.49.1101. [DOI] [PubMed] [Google Scholar]
- 19.Justè A, Lievens B, Frans I, Klingeberg M, Michiels CW, Willems KA. Present knowledge of the bacterial microflora in the extreme environment of sugar thick juice. Food Microbiol. 2008;25:831–836. doi: 10.1016/j.fm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Justè A, Lievens B, Klingeberg M, Michiels CW, Marsh TL, Willems KA. Predominance of Tetragenococcus halophilus as the cause of sugar thick juice degradation. Food Microbiol. 2008;25:413–421. doi: 10.1016/j.fm.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi DY, Reedy RM, Bick JA, Oudemans PV. Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl Environ Microbiol. 2002;68:1047–1054. doi: 10.1128/AEM.68.3.1047-1054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.König H, Fröhlich J, Hertel H. Diversity and lignocellulolytic activities of cultured microorganisms. In: König H, Varma A, editors. Intestinal microorganisms of termites and other invertebrates. Heidelberg: Springer; 2005. pp. 272–302. [Google Scholar]
- 23.König H. Bacillus species in the intestine of termites and other soil invertebrates. J Appl Microbiol. 2006;101:620–627. doi: 10.1111/j.1365-2672.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- 24.Kowalik R. Microbiodeterioration of library materials. Part 2. Microbiodecomposition of basic organic library materials. Restaurator. 1980;4:135–219. doi: 10.1515/rest.1980.4.3-4.135. [DOI] [Google Scholar]
- 25.Lippi D, Osmi M, Passeti L, De Paolis MR (1995) First observations on bacterial strains isolated from papers affected by foxing. In: Proceedings of the First International Congress Science and Technology for the Safeguard of Cultural Heritage in the Mediterranean Basin, Catania, Italy, pp. 1287-1290
- 26.Lord NS, Kaplan CW, Shank P, Kitts CL, Elrod SL. Assessment of fungal diversity using terminal restriction fragment pattern analysis: comparison of 18S and ITS ribosomal regions. FEMS Microbiol Ecol. 2002;42:327–337. doi: 10.1111/j.1574-6941.2002.tb01022.x. [DOI] [PubMed] [Google Scholar]
- 27.Lugauskas A, Levinskaitė L, Peciulytė D. Micromycetes as deterioration agents of polymeric materials. Int Biodeterior Biodegrad. 2003;52(4):233–242. doi: 10.1016/S0964-8305(03)00110-0. [DOI] [Google Scholar]
- 28.Michaelsen A, Pinzari F, Ripka K, Lubitz K, Piñar G. Application of molecular techniques for the identification of fungal communities colonising paper material. Int Biodeterior Biodegrad. 2006;58:133–141. doi: 10.1016/j.ibiod.2006.06.019. [DOI] [Google Scholar]
- 29.Mikola J, Bardgett RD, Hedlund K. Biodiversity, ecosystem functioning and soil decomposer food webs. In: Loreau M, Naeem S, Inchausti P, editors. Biodiversity and ecosystem functioning. Synthesis and perspectives. Oxford: Oxford University Press; 2002. pp. 169–180. [Google Scholar]
- 30.Montemartini Corte A, Ferroni A, Salvo VS. Isolation of fungal species from test samples and maps damaged by foxing, and correlation between these species and the environment. Int Biodeterior Biodegrad. 2003;51:167–173. doi: 10.1016/S0964-8305(02)00137-3. [DOI] [Google Scholar]
- 31.Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muyzer G, Ramsing NB. Molecular Methods to study the organization of microbial communities. Water Science Technologies. 1995;32:1–9. doi: 10.1016/0273-1223(96)00001-7. [DOI] [Google Scholar]
- 33.Neefs JM, Van de Peer Y, Hendriks L, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucl Acids Res. 1990;18:2237–2317. doi: 10.1093/nar/18.19.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen KF, Holm G, Uttrup LP, Nielsen PA. Mould growth on building materials under low water activities. Influence of humidity and temperature on fungal growth and secondary metabolism. Int Biodeterior Biodegrad. 2004;54:325–336. doi: 10.1016/j.ibiod.2004.05.002. [DOI] [Google Scholar]
- 35.Yu NP. The biodeterioration of papers and books. In: Garg KL, Garg N, Mukerji KG, editors. Recent advances in biodeterioration and biodegradation. Calcutta: Naya Prokash; 1994. pp. 1–88. [Google Scholar]
- 36.Oppong D, King VM, Bowen JA. Isolation and characterization of filamentous bacteria from paper mill slimes. Int Biodeterior Biodegrad. 2003;52:53–62. doi: 10.1016/S0964-8305(02)00174-9. [DOI] [Google Scholar]
- 37.Pangallo D, Šimonovičová A, Chovanová K, Ferianc P. Wooden art objects and the museum environment: identification and biodegradative characteristics of isolated microflora. Lett Appl Microbiol. 2007;45:87–94. doi: 10.1111/j.1472-765X.2007.02138.x. [DOI] [PubMed] [Google Scholar]
- 38.Pourcher A-M, Sutra L, Hébé I, Moguedet G, Bollet C, Simoneau P, Gardan L. Enumeration and characterization of cellulolytic bacteria from refuse of a landfill. FEMS Microbiol Ecol. 2001;34:229–241. doi: 10.1111/j.1574-6941.2001.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 39.Pushalkar S, Rao KK. Production of b-glucosidase by Aspergillus terreus. Current Microbiology. 1995;30:255–258. doi: 10.1007/BF00295497. [DOI] [PubMed] [Google Scholar]
- 40.Roberts JC. Neutral and alkaline sizing. In: Roberts JC, editor. Paper Chemistry. 2. Glasgow: Blacklie A&P; 1996. pp. 140–159. [Google Scholar]
- 41.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Samson RA, Hoekstra ES, Frisvad JC, Filtenborg O (eds) (2002) Introduction to food- and airborne fungi, 6th edn. Centraalbureau Voor Schimmelculture, Utrecht
- 43.Samson RA, Lustgraaf B. Aspergillus penicilloides and Eurotium halophilicum in association with house-dust mites. Mycopathologia. 1978;64(1):13–16. doi: 10.1007/BF00443082. [DOI] [PubMed] [Google Scholar]
- 44.Schabereiter-Gurtner C, Pinar G, Lubitz W, Rolleke S. An advanced molecular strategy to identify bacterial communities on art objects. J Microbiol Methods. 2001;45:77–87. doi: 10.1016/S0167-7012(01)00227-5. [DOI] [PubMed] [Google Scholar]
- 45.Sterflinger K, Prillinger H. Molecular taxonomy and biodiversity of rock fungal communities in an urban environment (Vienna, Austria) Antonie Van Leeuwenhoek. 2001;80:275–286. doi: 10.1023/A:1013060308809. [DOI] [PubMed] [Google Scholar]
- 46.Suihko ML, Sinkko H, Partanen L, Mattila-Sandholm T, Salkinoja-Salonen M, Raaska L. Description of heterotrophic bacteria occurring in paper mills and paper products. J Appl Microbiol. 2004;97:1228–1235. doi: 10.1111/j.1365-2672.2004.02416.x. [DOI] [PubMed] [Google Scholar]
- 47.Suihko ML, Stackebrandt E. Identification of aerobic mesophilic bacilli isolated from board and paper products containing recycled fibres. J Appl Microbiol. 2003;94:25–34. doi: 10.1046/j.1365-2672.2003.01803.x. [DOI] [PubMed] [Google Scholar]
- 48.Tamura M, Kawasaki H, Sugiyama J. Identity of the xerophilic species Aspergillus penicillioides: Integrated analysis of the genotypic and phenotypic characters. J Gen Appl Microbiol. 1999;45:29–37. doi: 10.2323/jgam.45.29. [DOI] [PubMed] [Google Scholar]
- 49.Teske A, Wawer C, Muyzer G, Ramsing NB. Distribution of sulphate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and DGGE of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilman D, Lehman C. Biodiversity, composition, and ecosystem processes: theory and concepts. In: Kinzig AP, Pacala SW, Tilman D, editors. The functional consequences of biodiversity. Princeton: Princeton University Press; 2001. pp. 9–41. [Google Scholar]
- 51.Vaisänen OM, Weber A, Bennasar A, Rainey FA, Busse HJ, Salkinoja-Salonen MS. Microbial communities of printing paper machines. J Appl Microbiol. 1998;84:1069–1084. doi: 10.1046/j.1365-2672.1998.00447.x. [DOI] [PubMed] [Google Scholar]
- 52.Wardle DA. Is sampling effect a problem for experiments investigating biodiversity-ecosystem function relationships? Oikos. 1999;87:403–407. doi: 10.2307/3546757. [DOI] [Google Scholar]
- 53.Wardle DA. Communities and ecosystems: linking the aboveground and belowground components. Princeton: Princeton University Press; 2002. [Google Scholar]
- 54.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. New York: Academic; 1990. pp. 315–322. [Google Scholar]
- 55.Wu Z, Tsumura Y, Blomquist G, Wang X-R. 18S rRNA gene variation among common airborne fungi, and development of specific oligonucleotide probes for the detection of fungal isolates. Appl Environ Microbiol. 2003;69:5389–5397. doi: 10.1128/AEM.69.9.5389-5397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, Yuen GY. The role of chitinase production by Stenotrophomonas maltophilia strain C3 in biological control of Bipolaris sorokiniana. Phytopatology. 2000;90:384–389. doi: 10.1094/PHYTO.2000.90.4.384. [DOI] [PubMed] [Google Scholar]
- 57.Zotti M, Ferroni A. Microfungal biodeterioration of historic paper: Preliminary FTIR and microbiological analyses. Int Biodeterior Biodegrad. 2008;62(2):186–194. doi: 10.1016/j.ibiod.2008.01.005. [DOI] [Google Scholar]
- 58.Zyska B. Fungi isolated from library materials: a review of the literature. Int Biodeterior Biodegrad. 1997;40:43–51. doi: 10.1016/S0964-8305(97)00061-9. [DOI] [Google Scholar]