Abstract
Telomerase reverse transcriptase (TERT) and telomerase RNA (TER) function together to create a uniquely specialized polymerase. Here we have described for the first time domains of bacterially expressed Tetrahymena TERT that interacted directly with TER in the absence of assembly chaperones. We used quantitative binding assays to define TER sequence requirements for recognition by the high affinity RNA binding domain and an independent N-terminal RNA interaction domain. The TERT RNA binding domain and N-terminal RNA interaction domain had distinct, nonoverlapping requirements for TER sequence and structure that together accounted for all of the sites of TER contact inferred for full-length TERT. The TER residues important for TERT binding are only a subset of the residues required for catalytic activity. Our findings demonstrate telomerase functional specialization by an elaborate ribonucleoprotein architecture physically separable from the active site.
Eukaryotic telomeres are maintained in a dynamic balance of DNA erosion by genome replication and DNA synthesis by telomerase (1, 2). Telomerase extends chromosome 3′ ends by the addition of tandem, telomeric simple-sequence repeats. This activity was initially characterized in the ciliated protozoan Tetrahymena thermophila, in which chromosome fragmentation synchronously generates thousands of new telomere substrates (3). Telomerase activity has since been discovered in a diversity of eukaryotic species, with a tissue-specific regulation in most multicellular organisms. For example, human germ line, epithelial, hematopoietic, and cancer cells express readily detectable telomerase, whereas other cell types limit telomerase-dependent telomere length maintenance to restrict replicative capacity (4–6).
The specificity of telomeric repeat synthesis by telomerase derives from a template sequence within the RNA subunit of the ribonucleoprotein (RNP)1 complex (7). Proteins that assemble with the telomerase RNA (TER) must package it into a biologically stable RNP while leaving the template region available for substrate access. Cellular biogenesis of a telomerase holoenzyme RNP incorporates H/ACA proteins in vertebrates, Sm proteins in yeasts, and a La motif protein in ciliates (1, 8). Catalytic activation requires an additional step of RNP assembly with the telomerase reverse transcriptase protein (TERT). TERT bears the active site motifs of a reverse transcriptase (RT) with additional, unique N- and C-terminal extensions (9). The TERT N-terminal extension represents about half of the full-length protein and harbors conserved motifs shared among all TERTs or among TERTs of a particular phylogenetic group. Heterologous expression of recombinant TER and TERT can reconstitute a minimally active telomerase enzyme (10, 11).
Co-expression or combination of Tetrahymena TER and TERT in rabbit reticulocyte lysate (RRL) provides a highly amenable system for studies of TER-TERT interaction. With the use of co-immunoprecipitation assays, three regions within the 159-nucleotide T. thermophila TER (see Fig. 1A schematic) have been implicated as TERT contact sites. A predominantly single-stranded region 5′ of the template, including residues 15CAUUC19 and 37GUCA40, functions as the high affinity TERT binding/template 5′ boundary element (TBE). TER variants with TBE substitutions co-precipitate less efficiently with TERT (12) and when assembled into RNP allow aberrant copying past the normal template 5′ boundary (13, 14). Two additional regions of TER appear to contribute lower affinity interaction sites for TERT, implicated by functional complementation, sensitized co-precipitation, and site-specific cross-linking assays (15). One of these TER regions coincides with the template recognition element (TRE) 3′ of the template, proposed to position the template 3′ end in the active site (16). The second coincides with a nucleotide addition processivity element (NPE), including at least the loop of stem IV, that is required for efficient copying of the full template and for a high level of activity overall (15).
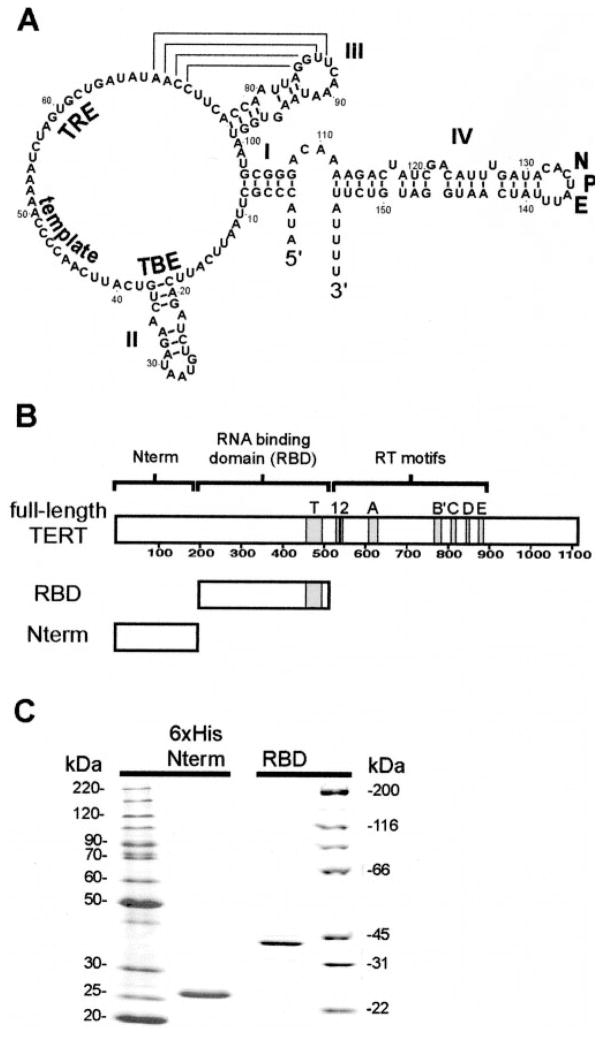
Fig. 1. Functional regions of TER and TERT.
A, schematic of T. thermophila TER with wild-type TER position numbers, functional motifs (TBE, TRE, template, NPE), and secondary structure elements (I–IV) as indicated. B, schematic of TERT and TERT domains expressed in E. coli. Motifs 1–2 and A–E are conserved among reverse transcriptases; motif T is conserved only among TERT proteins. C, SDS-PAGE analysis of N-terminally His6-tagged Nterm (left) and untagged RBD (right). Migration of molecular mass markers is indicated in kDa.
TERT requirements for interaction with TER have also been addressed using the RRL reconstitution system. Truncation analysis defined a necessary and sufficient high affinity RNA binding domain (RBD) within the Tetrahymena TERT N-terminal extension (12). Mutations in the yeast, Tetrahymena, or human TERT RBD region reduce TER interaction, suggesting evolutionary conservation of the RNA-binding function of this TERT domain (9). Indeed, the human TERT RBD region is sufficient for assembly with TER in vivo (12). For human and yeast TERTs, studies in vivo and in extract indicate that TER may also interact with TERT regions outside of the RBD (17–19). Unfortunately, current reconstitution systems are not adequate for investigating the specificity of any of these TERT-TER interaction(s) because of the limiting amount of expressed TERT protein, its inefficient assembly into RNP, and the requirement for eukaryotic accessory factors in the assembly reaction. Success in the bacterial over-expression and purification of TERT polypeptides has been reported only for a yeast TERT region including the N-terminal 160 amino acids, which demonstrates a general nucleic acid binding activity (20).
To determine the specificity and sequence requirements of Tetrahymena TERT-TER interaction with quantitative methods, we first developed a bacterial protein expression system. We over-expressed and purified two independently functional domains within the TERT N-terminal extension, each of which demonstrates direct and specific binding to TER. Together these two TERT domains, lacking any region of reverse transcriptase homology, reconstitute the entire TER interaction specificity inferred for full-length Tetrahymena TERT in RRL. No eukaryotic chaperones or cofactors were required for assembly of the purified TERT domains with TER. Curiously, the TER sequence requirements for TERT interaction represent only a subset of the TER sequence requirements for telomerase catalytic activity. This work revealed that novel protein-RNA interactions and RNA roles independent of RNP underlie the unique telomerase catalytic cycle.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The TERT RBD was expressed in untagged form using pET21. Expression was performed in BL21(DE3) pLysE cells grown in rich medium with 0.2% glucose at 37°C and induced with 1 mM isopropyl 1-thio-β-D-galactopyranoside at 21°C for 4 h. Harvested cells were resuspended in T2MG buffer (20 mM Tris-HCl, pH 8.0, 1 mM MgCl2, and 10% glycerol) with 0.2 M NaCl, 5 mM dithiothreitol, and protease inhibitors before freezing at −80°C. Thawed cells were sonicated, and extract was cleared by centrifugation. Lysate was fractionated by chromatography on Poros HS-50 using an elution gradient from 0.3 to 0.8 M NaCl. The TERT Nterm was purified in fusion with an N-terminal His6 tag using pET28, with the NdeI site encoding the first amino acid of the endogenous protein. Expression was performed in BL21(DE3) with induction and cell lysis as described for the RBD above, except that induction was performed at room temperature. For most experiments, protein was purified using a single step of chromatography on nickel-nitrilotriacetic acid-agarose with a high ratio of extract to resin.
RNA Expression and Purification
RNAs were transcribed in vitro using T7 RNA polymerase and plasmid or PCR templates by standard methods, then gel-purified, and recovered by precipitation. RNA stocks were quantified using fluorimetry and denaturing gel electrophoresis with SYBR Gold staining.
Binding Assays
Radiolabeled TER was synthesized by T7 RNA polymerase using radiolabeled UTP. The full-length transcription product was gel-purified, heated to 80°C for 3 min, and then iced for 2 min. RNA concentration was quantified by fluorimetry and competition against unlabeled RNA. Protein stocks were diluted substantially into binding reactions containing T2MG with 0.1 M NaCl, 5 mM dithiothreitol, bromphenol blue, and xylene cyanol. To each sample was added 5 μg of bovine serum albumin, 0.25 μl of RNasin, 50–250 ng of Escherichia coli tRNA as nonspecific competitor, and finally ~0.1 nM radiolabeled TER. Reactions were incubated at 30°C for 20 min before electrophoresis on a 5% acrylamide native gel (37.5:1 acrylamide:bis acrylamide, 4% glycerol, 0.5× Tris borate-EDTA) run at 200 V for 3 h at 4°C. Quantification was performed by PhosphorImager analysis. Time courses of incubation before electrophoresis revealed that binding was at equilibrium.
Activity Assays
Full-length TERT was expressed, assembled with TER in RRL, and assayed in reactions with radiolabeled dGTP as described previously (15). Activity assay reactions contained a 500 nM concentration of the primer (TG)8T2G3 and were incubated at 30°C for 30 min. Product DNA was analyzed by denaturing gel electrophoresis.
RESULTS
TER Interaction with the TERT RBD
In previous studies with RRL for protein expression, all Tetrahymena TERT polypeptides that included the RBD could efficiently co-immunoprecipitate TER (12). TER TBE substitutions inhibited TER recovery with full-length TERT or the TERT RBD, indicating that the smaller polypeptide retained determinants of full-length TERT-TER interaction specificity. We used E. coli to express the previously defined Tetrahymena TERT RBD encompassing amino acids 195–516 (Fig. 1B). This polypeptide could be purified to apparent homogeneity without an epitope tag (Fig. 1C, right). When evaluated by gel filtration, the purified RBD fractionated homogeneously at the predicted monomer molecular mass (data not shown).
We investigated the direct interaction of the RBD with TER using an electrophoretic mobility shift assay (EMSA). Each sample contained internally radiolabeled full-length TER, bearing the wild-type TBE motif 5′ of the template (Fig. 2A). A fixed, limiting concentration of radiolabeled TER was supplemented with purified RBD, incubated briefly to reach binding equilibrium, and then analyzed by native gel electrophoresis. The RBD could shift all of the radiolabeled TER to a new complex (Fig. 2B) that was not detected in assays of a mock protein purification from control extract (data not shown). An interaction affinity of ~3 nM (Fig. 2B) was observed reproducibly with independent preparations of RBD and varying concentrations of radiolabeled TER. Nonspecific competitors including tRNA, total yeast RNA, or 5 S RNA did not compete for the mobility shift of TER even when added in vast excess (see “Experimental Procedures”) (additional data not shown).
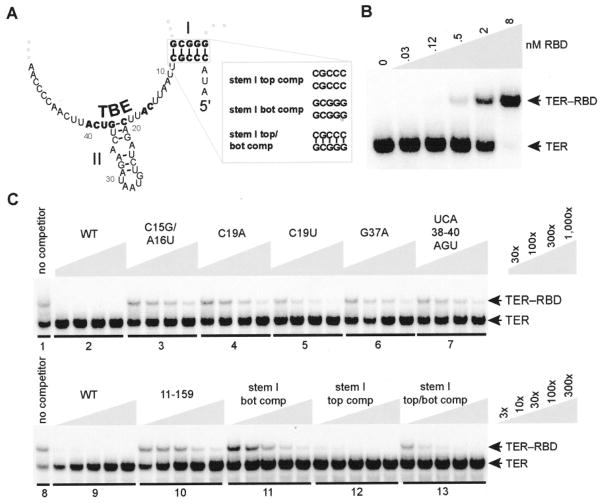
Fig. 2. TER interaction with the TERT RBD.
A, schematic of the TER region required for RBD binding. Bold font indicates residues analyzed by sequence substitution. Inset, stem I top comp, complement sequence in the top strand of stem I; stem I bot comp, complement sequence in the bottom strand of stem I; stem I top/bot comp, complement sequences in the top and bottom strands of stem I. B, EMSA analysis of radiolabeled TER with the indicated concentrations of RBD (0–8 nM). C, EMSA competition with unlabeled wild-type TER (WT) and TER variants at concentrations indicated by the keys at right, in which x = ~0.1 nM radiolabeled TER. Competitor RNA was added to RBD (1 nM) before addition of radiolabeled wild-type TER. Samples in each panel were from the same gel.
To investigate the sequence specificity of RBD-TER interaction, we tested nonradiolabeled variants of TER as mobility shift competitors. In samples with a fixed amount of radiolabeled TER and a fixed concentration of RBD, the addition of unlabeled wild-type TER competed the mobility shift as expected (Fig. 2C, lane sets 1–2 and 8–9; note the -fold excess of competitor indicated at right). We next tested a panel of TBE sequence variants analyzed previously for assembly with full-length TERT in RRL (12, 13). In the EMSA, TER variants with TBE substitutions did not compete as effectively as wild-type TER for RBD binding (Fig. 2C, lane sets 3–7; substituted residues are indicated in bold in Fig. 2A). In direct parallel to previous findings, the substitution C15G/A16U was strongly inhibitory, whereas the substitution C19U had the least impact. These results indicate that the bacterially expressed, purified Tetrahymena TERT RBD has a sequence-specific TER binding activity fully consistent with that of Tetrahymena TERT expressed in RRL.
The EMSA allows quantitative dissection of protein-RNA interaction specificity in a manner more rigorous than possible using co-immunoprecipitation from RRL. To investigate the contribution of TER motifs apart from the TBE in RBD-TER interaction, we tested numerous TER sequence variants for EMSA competition. Many TER variants demonstrated little difference from wild-type TER in competition for RBD binding, even using fine titrations of unlabeled TER. A notable exception was discovered in the analysis of stem I variants (substituted residues are indicated in bold in Fig. 2A). Stem I bottom strand deletion or bottom strand sequence substitution to its complement reduced competition for RBD binding by ~300-fold (Fig. 2C, lane sets 10 and 11; note the -fold excess of competitor indicated at right). Curiously, substitution of the top strand sequence had less impact (Fig. 2C, lane set 12), and substitution of both top and bottom strands to restore base-pairing potential had an intermediate effect (lane set 13). These experiments revealed that RBD interaction with TER depends on the TBE as shown previously (12) and also on the sequence and structure of stem I.
TER Interaction with the TERT N Terminus
RNA oligonucleotides containing the template and TRE (positions 43–63) or the distal end of stem-loop IV with the NPE (positions 128–142; see Fig. 1A) retain function when physically unlinked from the remainder of the TER molecule (15, 16). These regions of TER also form site-specific cross-links to TERT at short range (15). Our EMSA competition analysis did not reveal an RBD-TER interaction dependent on wild-type TRE or NPE sequence (data not shown). This observation suggested that regions of Tetrahymena TERT beyond the RBD might provide additional sites for TER interaction. Of the many regions of Tetrahymena TERT tested, we were able to over-express and purify a polypeptide encompassing amino acids 1–195 (Fig. 1B) indicated as the Nterm domain. This polypeptide could be purified to apparent homogeneity in fusion with an N-terminal His6 tag (Fig. 1C, left). When evaluated by gel filtration, purified Nterm fractionated homogeneously at the predicted monomer molecular mass (data not shown).
The purified TERT Nterm shifted radiolabeled full-length TER to a distinct complex (Fig. 3A) that was not detected in assays of a mock protein purification from control extract (data not shown). A titration of Nterm could shift all of the radiolabeled TER (Fig. 3A; additional data not shown). An interaction affinity of ~500 nM was reproducibly observed with independent preparations of protein and varying concentrations of radiolabeled TER. Notably, this affinity is ~150-fold less than we measured under the same EMSA conditions for the RBD-TER interaction. Nonspecific RNAs did not compete with TER binding by Nterm even when added in vast excess (see “Experimental Procedures”; additional data not shown). DNA oligonucleotides also failed to compete with the TER-Nterm interaction (data not shown), in contrast to the competition observed in assays of TER binding to an N-terminal domain of recombinant yeast TERT (20).
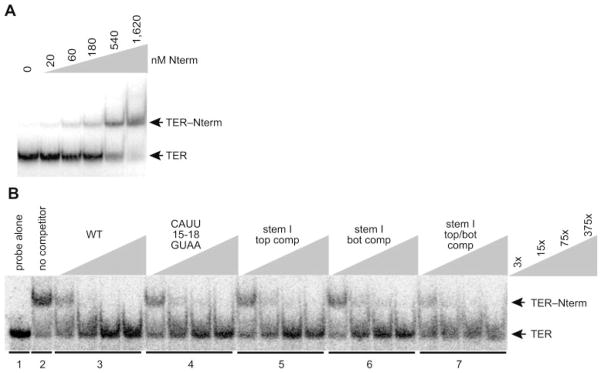
Fig. 3. TER interaction with the TERT Nterm.
A, EMSA analysis of radiolabeled TER with the indicated concentrations of Nterm (0–1,620 nM). B, EMSA competition with competitor RNA added to Nterm (500 nM) before the addition of radiolabeled wild-type TER. Unlabeled wild-type TER (WT) and TER variants described in the text were added at concentrations indicated by the key at right, in which x = ~0.1 nM radiolabeled TER. Stem I top comp, complement sequence in the top strand of stem I; stem I bot comp, complement sequence in the bottom strand of stem I; stem I top/bot comp, complement sequences in the top and bottom strands of stem I.
To investigate the sequence specificity of Nterm-TER interaction, we again used mobility shift competition. In contrast to RBD-TER interaction, Nterm-TER interaction was efficiently competed by TER variants bearing TBE or stem I substitutions (Fig. 3B). This result indicates that the TER binding specificities of the RBD and Nterm are distinct. We next tested Nterm binding competition using TER variants with substitutions and deletions throughout the full-length TER sequence (see Fig. 4A schematic). These assays revealed an influence of the NPE (Fig. 4B) and TRE (Fig. 4C) on Nterm-TER interaction.
Fig. 4. Specificity of TER interaction with the TERT Nterm.
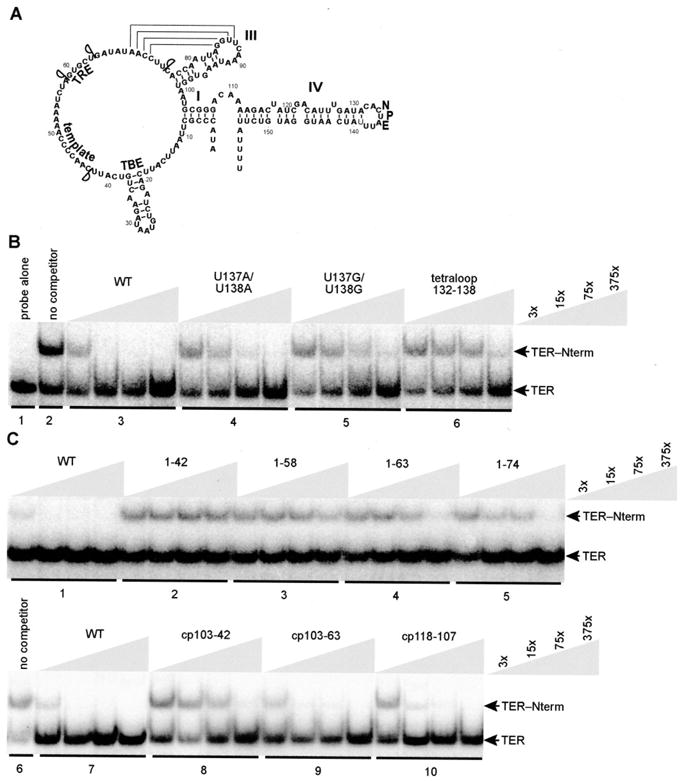
A, schematic of TER. End points of 3′ truncations are indicated. B and C, EMSA competition with unlabeled wild-type TER (WT) and TER variants at concentrations indicated by the keys at right, in which x = ~0.1 nM radiolabeled TER. Competitor RNA was added to Nterm (500 nM) before addition of radiolabeled wild-type TER. Samples in each panel were from the same gel.
TER variants with a UUCG tetraloop in substitution of portions of stem-loop IV demonstrated ~50-fold reduced competition for Nterm binding, even when only the stem IV loop residues were replaced (Fig. 4B, lane sets 1–3 and 6). Within the loop, substitution of the conserved residues 137UU138 reduced competition almost as much as replacement of the entire loop (Fig. 4B, lane sets 4–6). To investigate Nterm interaction requirements for TER regions beyond distal stem IV, we tested a panel of 3′ truncations progressively removing stem IV, the top strand of stem I, the pseudoknot, the TRE, and the template (3′ truncation end points are indicated in Fig. 4A). Truncations up to position 63 reduced competition only ~50-fold (Fig. 4C, lane sets 1 and 4 and 5), a defect no greater than that observed with substitution or deletion of the stem IV loop alone.
Subsequent truncations that removed TRE and template residues ultimately reduced competition to an undetectable level (Fig. 4C, lane sets 2 and 3). To verify the significance of the TRE and template in the presence of an intact stem IV loop, we tested EMSA competition by circular permutation (cp) TER variants with joined wild-type 5′ and 3′ ends (16) and new 5′ and 3′ ends throughout TER to form an internal deletion (cpTER deletion variants are annotated using new 5′-3′ ends). A TER variant with the cpTER backbone and internal deletion of the pseudoknot competed for Nterm binding as well as wild-type TER (Fig. 4C, lane sets 6 and 7 and 9; cp 103–63). Additional removal of the TRE and template reduced competition for Nterm binding by ~50-fold (Fig. 4C, lane set 8; cp 103–42). As a control, a cpTER with internal deletion of part of the top strand of stem IV competed for Nterm binding nearly as well as wild-type TER (Fig. 4C, lane set 10; cp 118–107), despite a predicted increase in TER conformational flexibility. These EMSA competition assays suggested that the NPE and TRE are specific, independent determinants of Nterm interaction affinity.
Sequence Requirements for Nterm Binding Versus Catalytic Activity
TER sequence substitutions in the NPE or TRE affect the telomerase product profile as well as the overall level of activity (12, 15, 21, 22). NPE substitutions can reduce nucleotide addition processivity, limiting the amount of complete repeat synthesis, whereas TRE substitutions can affect repeat addition processivity, decreasing the number of repeats added to any given substrate. To determine whether the sequence requirements for Nterm-NPE or Nterm-TRE interaction correlate with the NPE or TRE sequence requirements for catalytic activity, we tested TER variants in parallel with both an EMSA competition for Nterm binding (Figs. 4B and 5, A and B) and a catalytic activity assay with full-length TERT in RRL (Fig. 5C).
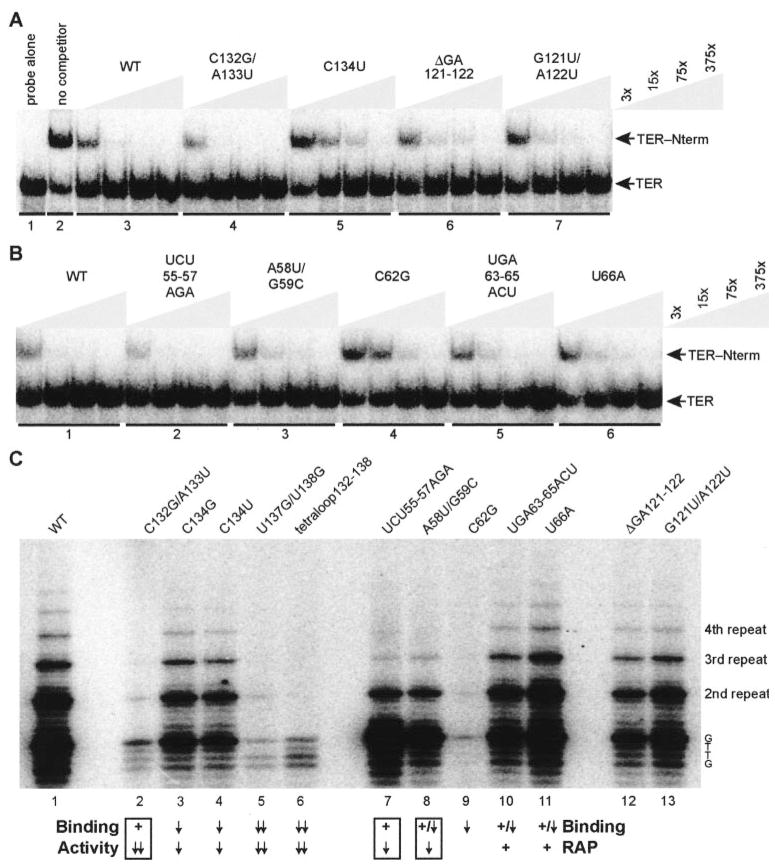
Fig. 5. TER sequence requirements for Nterm binding and catalytic activity.
A and B, EMSA competition with unlabeled wild-type TER (WT) and TER variants at concentrations indicated by the keys at right, in which x = ~0.1 nM radiolabeled TER. Competitor RNA was added to Nterm (500 nM) before addition of radiolabeled wild-type TER. Samples in each panel were from the same gel. C, full-length TERT was assembled in RRL with wild-type or variant TER and then assayed for activity in reactions with an excess of the primer (TG)8T2G3. Addition of the sequence 5′-GTTG-3′ completes the first telomeric repeat, with additional repeats in 6-nucleotide increments. A summary of NPE and TRE variants is provided to compare Nterm binding competition (Binding) versus overall activity (Activity) or repeat addition processivity (indicated as RAP), with each TER variant scored as functionally wild type (+), near wild type (+/down arrow), reduced (down arrow), or strongly reduced (down arrows). Repeat addition processivity could not be quantified for the C62G TER RNP because of low activity. Nterm binding competition for the C134G TER variant is from data not shown.
Substitution of the entire stem IV loop or the conserved loop residues 137UU138 strongly reduced both Nterm binding (described above) and catalytic activity (Fig. 5C, lanes 5 and 6). Substitution of the similarly conserved stem IV loop residues 132CA133, however, did not affect competition for Nterm binding (Fig. 5A, lane set 4) yet reduced activity severely (Fig. 5C, lane 2). Substitution of the loop residue C134 slightly reduced both Nterm binding (Fig. 5A, lane set 5) and catalytic activity (Fig. 5C, lanes 3 and 4), as did deletion or substitution of the conserved central stem IV GA bulge (Fig. 5A, lane sets 6 and 7; Fig. 5C, lanes 12 and 13). In the TRE, the C62G substitution strongly reduced both Nterm binding (Fig. 5B, lane set 4) and catalytic activity (Fig. 5C, lane 9). TRE substitutions of 55UCU57 or 58AG59, however, did not affect competition for Nterm binding (Fig. 5B, lane sets 2 and 3) yet inhibited repeat addition processivity (Fig. 5C, lanes 7 and 8) as reported previously (12, 21). Substitutions of 63UCA65 or U66 had only minor impact on either Nterm binding (Fig. 5B, lane sets 5 and 6) or Nterm activity (Fig. 5C, lanes 10 and 11). This comparison of Nterm binding competition and activity indicates that the TER sequence requirements for catalytic cycle function extend beyond the requirements for Nterm interaction.
DISCUSSION
Tetrahymena
TERT-TER interactions investigated previously in RRL were reconstituted here using purified, bacterially expressed domains of TERT. Over-expression of TERT domains in E. coli eliminates any potential influence of eukaryotic modifying enzymes, chaperones, or cofactors in the RNP assembly reaction. The sequence specificity of TER interaction with the RBD and Nterm suggests that each domain can fold autonomously into its functional conformation. Why then is full-length TERT assembly with TER stimulated by RRL? Full-length TERT interaction with TER could be constrained by a misfolding of recombinant TERT regions beyond the RBD and Nterm or by the missing influence of factors involved in the physiological telomerase holoenzyme assembly pathway. In isolation, the Tetrahymena TERT RBD and Nterm recapitulate all of the TERT-TER interaction specificity inferred from previous physical and functional studies of full-length TERT. This suggests that the reverse transcriptase homology region of TERT may not harbor any sites of sequence-specific TER interaction. Instead, TERT appears to have gained much of its functional specialization by an appropriation of sequence-specific RNA binding domains appended to, rather than evolved within, the domain formed by the polymerase active site motifs.
The residues of TER with the most critical significance for RBD and Nterm interactions are largely single-stranded, based on evidence from phylogenetic comparison or chemical and enzymatic probing (22–24). Residues within the TBE become protected from chemical modification in the assembled RNP (24) as expected for a site of high affinity TER-RBD interaction. RBD interaction with stem I was not previously anticipated, but this finding is consistent with the conservation of both sequence and structure of this stem in the TERs of Tetrahymena species (23). Surprisingly, two TER regions distant in secondary structure each contribute to Nterm interaction. The TRE residue C62 becomes protected from chemical modification in the assembled RNP (24), and the NPE residues 137UU138 show absolute conservation among TERs of the Tetrahymena species; substitution of these residues substantially reduces Nterm binding. The Nterm domain of Tetrahymena TERT could recognize a structure formed by the NPE and TRE together, or Nterm contacts to TER could be distributed over residues in both of the motifs. Neither the RBD nor the Nterm domain of TERT harbors a recognizable amino acid consensus for nucleic acid binding, suggesting that each may utilize a novel architecture for sequence-specific recognition of RNA.
Only a subset of the TER sequence substitutions in the NPE or TRE that affect telomerase catalytic activity could be linked to a change in Nterm binding. In contrast, previous studies suggested that all of the TER sequence variants in the TBE with impact on template 5′ boundary fidelity also compromised for RBD binding (13). These results suggest a model in which Nterm-TER interaction serves to position some NPE and TRE residues for their function(s) in the catalytic cycle, whereas the RBD-TER interaction itself fulfills the function of the TBE. A high affinity of binding by the RBD suits the role of the TBE, because it must prevent residues 5′ of the template from entering the active site. The lower affinity of Nterm-TER interaction could indicate a requirement for structural rearrangement during the process of cellular RNP biogenesis or the complex cycle of telomeric repeat synthesis. The reconstitution system described here and the new insights gained into TERT-TER interaction pave the way for additional future studies of TER and TERT structure and enzyme mechanism in greater detail.
Acknowledgments
We thank Doreen Cunningham for activity assays, James Berger for help with protein characterization, and members of the Collins laboratory for critical ideas and discussion.
Footnotes
This work was supported by a predoctoral fellowship from the National Science Foundation (to C. M. O.) and National Institutes of Health Grant GM54198 (to K. C.).
The abbreviations used are: RNP, ribonucleoprotein; TER, telomerase RNA; TERT, telomerase reverse transcriptase; RRL, rabbit reticulocyte lysate; RBD, RNA binding domain; TBE, template 5′ boundary element; TRE, template recognition element; NPE, nucleotide addition processivity element; EMSA, electrophoretic mobility shift assay; Nterm, N-terminal region; RNP, ribonucleoprotein; cp, circular permutation.
References
- 1.Harrington L. Cancer Lett. 2003;194:139–154. doi: 10.1016/s0304-3835(02)00701-2. [DOI] [PubMed] [Google Scholar]
- 2.Chan SR, Blackburn EH. Philos Trans R Soc Lond B Biol Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cech TR. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 4.Collins K, Mitchell JR. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 5.Shay JW, Wright WE. Cancer Cell. 2002;2:257–265. doi: 10.1016/s1535-6108(02)00159-9. [DOI] [PubMed] [Google Scholar]
- 6.Wong JMY, Collins K. Lancet. 2003;362:983–988. doi: 10.1016/S0140-6736(03)14369-3. [DOI] [PubMed] [Google Scholar]
- 7.Chen JL, Greider CW. Trends Biochem Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Witkin KL, Collins K. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelleher C, Teixeira MT, Forstemann K, Lingner J. Trends Biochem Sci. 2002;27:572–579. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
- 10.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 11.Collins K, Gandhi L. Proc Natl Acad Sci U S A. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai CK, Mitchell JR, Collins K. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai CK, Miller MC, Collins K. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Autexier C, Greider CW. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 15.Lai CK, Miller MC, Collins K. Mol Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller MC, Collins K. Proc Natl Acad Sci U S A. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman KL, Cech TR. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beattie TL, Zhou W, Robinson MO, Harrington L. Mol Cell Biol. 2001;21:6151–6160. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriarty TJ, Huard S, Dupuis S, Autexier C. Mol Cell Biol. 2002;22:253–265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia J, Peng Y, Mian IS, Lue NF. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licht JD, Collins K. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperger JM, Cech TR. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- 23.McCormick-Graham M, Romero DP. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaug AJ, Cech TR. RNA (N Y ) 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]