Abstract
Humanin is a short endogenous peptide, which can provide protection from cell death through its association with various receptors, including the pro-apoptotic Bcl-2 family proteins Bid, Bim, and Bax. By using NMR chemical shift mapping experiments, we demonstrate that the interaction between Humanin-derived peptides and Bid is specific, and we localize the binding site to a region on the surface of Bid, which includes residues from the conserved helical BH3 domain of the protein. The BH3 domain mediates the association of Bid with other Bcl-2 family members and is essential for the protein's cytotoxic activity. The data suggest that Humanin exerts its cytoprotective activity by engaging the Bid BH3 domain; this would hinder the association of Bid with other Bcl-2 family proteins, thereby mitigating its toxicity. The identification of a Humanin-specific binding site on the surface of Bid reinforces its importance as a direct modulator of programmed cell death, and suggests a strategy for the design of cytoprotective peptide inhibitors of Bid.
Keywords: apoptosis, Bcl-2, BH3, Bid, chemical shift mapping, cytoprotection, Humanin, nuclear magnetic resonance, peptide
The Bcl-2 family proteins regulate a major pathway for apoptosis that involves intermolecular associations with each other and with a variety of other partners, leading to the release of cytotoxic molecules from the mitochondria (1). The proteins share sequence homology across four evolutionarily conserved Bcl-2 homology (BH1–BH4) domains, of which the BH3 domain is highly conserved and essential for both the cell killing activity of the pro-apoptotic family members, and for mediating protein–protein interactions among the various Bcl-2 proteins. Several proteins also contain a 20-residue hydrophobic C-terminus, believed to be responsible for their association with intracellular membranes. The six human antiapoptotic members of the family possess all four BH domains, while the pro-apoptotic family members have more diverse sequences. The latter set is subdivided into multi-domain proteins such as Bax and Bak, containing BH1, BH2, and BH3 domains, and BH3-only proteins such as Bid and Bim, containing only the BH3 domain. These BH3-only proteins are activated by upstream death signals, which trigger their transcriptional induction or post-translational modification, providing a key link between extrinsic stress cues, transmitted by the death receptors, and the mitochondrial pathway to cell death (2).
Bid is a cytosolic protein that is cleaved and activated by caspases, calpains, and lysosomal proteases, after engagement of the Fas or TNFR1 cell surface receptors, which link it to both apoptotic and necrotic cell death pathways in stroke, neuro-degeneration, hepatitis, and other ailments (3–12). Cleavage by caspase-8 produces the 15 kDa C-terminal fragment, tBid, which can interact with and be sequestered by the antiapoptotic proteins Bcl-2 and Bcl-XL (9), or interact with the multi-domain pro-apoptotic proteins Bax and Bak to promote mitochondrial apoptosis (12). In addition, tBid can translocate from the cytosol to mitochondria to induce mitochondrial membrane remodeling, the release of cytotoxic molecules, and ultimately, cell death (10–12). The BH3 domain of Bid is essential for binding to other antiapoptotic or pro-apoptotic Bcl-2 family members, however, it is not required for translocation to the mitochondria or for mitochondrial membrane remodeling, which occurs independently of other Bcl-2 proteins.
The functions of Bid, Bim, and Bax can also be regulated by Humanin (HN), a 24-residue endogenous peptide that protects neuronal cells from a variety of toxic insults (13–17). Humanin was first identified as an antiapoptotic factor capable of rescuing neuronal cells from apoptosis in familial Alzheimer's disease (13), and has been reported to bind receptors in the plasma membrane (18,19), as well as to regulate the pro-apoptotic activities of Bid, Bim, and Bax in the cytosol (20–22). Although the structures of Bid, Bim, Bax, and Humanin have been determined or characterized independently, no molecular information has been available about the interaction of Humanin with its Bcl-2 partners, and its mechanism of Bcl-2-dependent cytoprotection has been unknown.
The structures of Bid (23,24) and Bax (25) are both strikingly similar to the structures of other multi-domain pro-apoptotic and antiapoptotic Bcl-2 family proteins, despite the low level of amino acid sequence homology, while Bim is largely unfolded (26). Bid folds into eight α-helices, with the third helix coinciding almost entirely with the conserved BH3 domain. Humanin is unfolded in water but NMR shows that it adopts a helical structure in the presence of 30% trifluoroethanol, indicating a propensity for this type of secondary structure (27,28), while at high salt concentrations CD analysis shows some evidence of extended conformation (29).
In this report, we characterize the Humanin-binding site of Bid using NMR chemical shift mapping experiments. By monitoring the changes in the 1H / 15N heteronuclear single quantum correlation (HSQC) spectrum of Bid upon titration with peptides derived from Humanin we show that the interaction with Humanin affects Bid residues in the BH3 domain. The changes in 1H and 15N chemical shifts for all the titratable backbone amide signals can be modeled with a single binding equilibrium constant, demonstrating that Humanin binds specifically to Bid. The demonstration of a Humanin-specific binding site on the surface of Bid reinforces the role of this peptide as a direct modulator of Bid-dependent programmed cell death, and suggests a strategy for the development of therapeutic approaches for the many related diseases.
Methods and Materials
Protein expression
The cDNA-encoding mouse Bid (NCBI accession number NP_031570) was inserted in the NcoI and XhoI cloning sites of a pET-23d(+) (Novagen, Madison, WI, USA) plasmid vector that had been previously modified to encode an N-terminal His tag sequence (MAHHHHHHGSPEFAV). Bid was expressed as a soluble protein in Escherichia coli BL21(DE3) cells, grown on isotopically labeled M9 minimal media. For protein expression, 5–10 μL of transformed cells from a frozen glycerol stock were transferred to 10 mL of LB media and grown for 5 h at 37 °C with vigorous shaking. Then, 1 mL of this starter culture was added to 100 mL of minimal M9 media and grown overnight. In the morning, 1 L of fresh M9 media was inoculated with the overnight culture, the cells were grown to a cell density of OD600 = 0.7, protein expression was induced by the addition of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h at 37 °C, and the cells were harvested by centrifugation, and stored at −20 °C overnight. All media contained 100 μg / mL of ampicillin. Uniformly 15N-and 13C-labeled proteins were obtained by supplying (15NH4)2SO4 and 13C-labeled glucose to the M9 salts, and selectively 15N-labeled samples were obtained by supplying individual 15N-labeled amino acids to the media (Cambridge Isotope Laboratories, Andover, MA, USA).
Protein purification
Frozen cells from 1 L of culture were lysed by French press in 30 mL of buffer A (20 mM HEPES, pH 7, 0.5 mM EDTA, 1 mM PMSF). The soluble fraction was isolated by centrifugation (48 000 × g, 4 °C, 30 min), and Bid was purified by Ni-NTA affinity chromatography (http://www.emdbiosciences.com) with 250 mM imidazole in buffer B (50 mM sodium phosphate, pH 7), followed by ion-exchange chromatography (FF-Q column, Amersham Biosciences, Piscataway, NJ, USA) with a NaCl gradient in buffer B, and size exclusion chromatography (Sephacryl S-100 column, Amersham Biosciences) in buffer C (20 mM sodium phosphate, pH 7). The His tag was not removed. The purified protein concentrations were determined by UV absorbance as described (30). NMR samples contained 0.2–1.0 mM Bid in 10% D2O, 90% H2O, 2 mM dithiothreitol (DTT), at pH 7. The truncated form of Bid (tBid) was obtained by incubation with caspase-8 as described (24). The amino acid sequence of Bid is illustrated in Figure 1A.
Figure 1.

Amino acid sequences of Bid and Humanin. Black squares represent small hydrophobic amino acids and white circles represent aromatic residues, which may be important for mediating helix–helix interactions. (A) Bid (mouse;). Helical regions are mapped above the sequence, the BH3 domain spanning helix 3 is in the boxed region, and the asterisk marks the caspase-8 cleavage site at Asp59. (B) Humanin (human). (C) Sequences of BH3 domains from mouse Bid, human Bid, human Bim, human Bax, and human Bcl-XL, with conserved hydrophobic residues in gray, and residue numbering according to mouse Bid.
Peptide synthesis
Humanin peptides, derived from the human peptide sequence (NCBI accession number AY029066), were synthesized by N-(9-fluorenylmethoxycarbonyl)-L-amino acid chemistry with a solid-phase synthesizer, and purified by HPLC. The peptide purity was confirmed by mass spectrometry. The amino acid sequence of Humanin is illustrated in Figure 1B.
NMR spectroscopy
NMR experiments were performed on Bruker AVANCE 600 and 800 MHz spectrometers (Billerica, MA, USA), at 30 °C, using a 1 second recycle delay. The chemical shifts are referenced to the 1H2O resonance, set to its expected position of 4.701 at 30 °C (31). The NMR data were processed using NMRPIPE (32), and the spectra were assigned and analyzed using SPARKY (33). For the Humanin titrations, the 1H / 15N FHSQC (fast HSQC) experiment (34) was used with 1024 points in t2 and 256 in t1. Backbone resonance assignments were obtained using standard HNCA and HNCACB experiments with constant time evolution for 15N, and by comparison with the previously determined assignments, provided to us by David Cowburn and Gerhard Wagner (23,24). Solvent suppression was accomplished with a water flip-back pulse after the original 1H-15N magnetization transfer (35–37). The spectra from some selectively 15N-labeled protein samples were necessary to resolve assignment ambiguities.
Titration of 15N-Bid with Humanin
For the Humanin titration, 1H / 15N-HSQC spectra were obtained with 0.2 mM uniformly 15N-labeled Bid and increasing amounts of unlabeled Humanin peptides. Humanin and Bid are each soluble in a variety of aqueous buffers and salts, however, the addition of full-length Humanin to Bid at pH 7 caused the precipitation of both Humanin and Bid. Sample precipitation could be avoided by working with mouse Bid instead of human Bid, shorter sequences of Humanin peptide, and reduced polypeptide concentrations in water (0.2 mM Bid, 10% D2O, 90% H2O, 2 mM DTT, pH 7), as the presence of salts promoted sample precipitation.
Although the 1H and 15N chemical shifts can be sensitive to changes in sample conditions, such as pH, ionic strength, and temperature, the spectrum for each addition of Humanin was recorded at constant sample conditions, therefore the peaks that move reflect a molecular change resulting from either direct or indirect contact with Humanin. The Humanin peptides were neutralized before use, and the buffering capacity of Bid maintained the samples at pH 7 throughout the titration, as confirmed by direct pH measurements and by monitoring the 1H and 15N chemical shift frequencies of the HSQC peak from His162. This residue, located in the connecting loop between helices H6 and H7, is exposed to the aqueous environment, and has chemical shift frequencies that are highly sensitive to pH. The peak from His162 did not move throughout the Humanin titration confirming that the pH did not change (Figure 2D,G).
Figure 2.
1H / 15N heteronuclear single quantum correlation spectra of uniformly 15N-labeled Bid (0.2 mM, black) or cleaved Bid (0.2 mM, green) titrated with Humanin peptide at concentrations of (A) 0 mM, and (B–G) 1.2 mM (red). (B–G) Expanded regions of the superimposed spectra show some of the peaks that move by >0.02 p.p.m. upon addition of Humanin. (D and G) The invariant peak for H162 is labeled in bold.
The total combined change in chemical shift (Δ) for each residue was calculated by adding the changes in 1H(ΔH) and 15N(ΔN) chemical shifts, according to the equation Δ =[(ΔH)2 +(ΔN / 5)2]1 / 2, where the 15N chemical shift is scaled by 1 / 5 to account for the fivefold difference between the chemical shift dispersions of 15N and 1H (38,39).
Assuming that the change in chemical shift is due to Humanin binding, and that the titration goes to full occupancy of the Humanin-binding site, the fraction of Bid bound to Humanin can be calculated as Δ/Δmax, corresponding to the change in chemical shift at each Humanin concentration (Δ), divided by the change observed at the maximum Humanin concentration of 1.2 mM (Δmax). For each peak that moved by Δmax > 0.02 p.p.m., Δ/Δmax was calculated for each Humanin concentration of 0.0, 0.2, 0.6, 1.0, and 1.2 mM, and the dissociation constant (Kd) and maximum fraction of Humanin-bound Bid (pmax) were estimated by nonlinear curve fitting of the plots of Δ/Δmax against Humanin concentration ([HN]), using the equation for single-site binding written in terms of chemical shift change, Δ/Δmax = [HN] / (Kd + [HN]), for the binding equilibrium, [Bid] + [HN] ⇆ [Bid-HN] (38–40). The cutoff value of Δmax > 0.02 p.p.m. corresponds to chemical shift changes of more than half a line width (10 Hz), the smallest change that can be confidently measured experimentally. Data analysis and curve fitting were performed with the program PRISM (http://www.graphpad.com).
The values of Kd and pmax obtained for all the peaks with Δmax > 0.02 p.p.m. were averaged and used to calculate an average binding curve. When the average curve was used to individually fit the data for each peak, the R-square values reflecting the goodness-of-fit were all above 0.85, with an average R-square of 0.98. Overlapped peaks that could not be measured accurately were excluded from the analysis. The changes in chemical shifts were mapped onto the molecular structure of Bid and rendered with the program PYMOLa. The surface electrostatic potential of Bid was calculated with the program GRASP (41).
Results and Discussion
Changes in the NMR spectrum of Bid with addition of Humanin
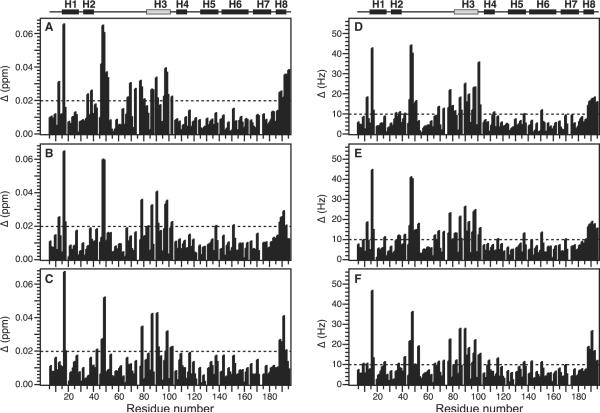
The full-length Humanin peptide binds avidly to Bid with an affinity of 10–20 nM (20), however, as the complex could not be maintained in solution at neutral pH even at low concentrations, we focused our studies on shorter versions of the peptide, spanning residues 1–12, that had been previously demonstrated to have both Bid-binding and cytoprotective activities (20,21). Several of these shorter peptides were tested, and gave chemical shift changes of similar magnitude and direction in the 1H / 15N HSQC spectrum of 15N-Bid (Figure 3) and here we present the results obtained with HN(1–12)C8A.
Figure 3.
Total change in chemical shift as a function of Bid residue number, obtained in the presence of 1.2 mM Humanin peptide. Effects are shown for three different Humanin peptides: (A and D) HN(1–12)C8A; (B and E) HN(3–12); and (C and F) HN(3–12)C8A. Total changes are shown in (A–C) p.p.m. (Δ(p.p.m.) = [(ΔH(p.p.m.))2 + (ΔN(p.p.m.) / 5)2]1 / 2), or (D–F) Hz (Δ(Hz) = [(ΔH(Hz))2+(ΔN(Hz))21 / 2). The helical regions in the NMR structure of Bid are mapped above the graph. Helix H3 with the BH3 domain is in gray.
The 1H and 15N chemical shifts from protein amide sites are very sensitive to changes in protein conformation or chemical environment, and can be used to monitor the equilibrium exchange between states arising from free and ligand-bound protein (31). If the exchange rate is faster than the difference between the chemical shifts measured for the two states then the system is in fast exchange and one peak is observed at the population-weighted average chemical shift of the two states. Nuclear magnetic resonance can be used to detect weak binding or minor conformational rearrangements, and chemical shift changes as small as 0.02 p.p.m. have been reported for minor local effects on protein structure resulting from binding of small molecules or modifications, while much larger changes (>1 p.p.m.) can reflect major conformational rearrangements (38,39,42,43).
The addition of Humanin caused some of the Bid peaks to move significantly with increasing amounts of peptide up to a Humanin / Bid molar ratio of 6 / 1, while many other peaks from residues throughout the Bid amino acid sequence did not move indicating that the Humanin-bound protein does not undergo a major conformational change. Examples of the peak movements are shown in the expanded regions of the HSQC spectra in Figure 2, for free (black) and Humanin-bound (red) Bid. Several peaks moved by a total combined 1H and 15N frequency >0.02 p.p.m., including residues in helix H1 (I16), in helix H2 (L35, and L38), in the disordered loop (A46, W48, A50, L52, Q62, Q69, E77, and T78), in helix H3 (I82, I86, A87, L90, A91 E96, and D98), and in helix H8 (L189, and R191). These effects are highlighted in a plot of the total chemical shift change against Bid amino acid sequence (Figure 3).
No broadening or splitting of peaks was observed, indicating that free and Humanin-bound Bid are in fast exchange on the time-scale of the chemical shift interactions, and the moving peaks could be assigned by tracking their linear movement from their starting assigned positions in the spectrum of free Bid, to their end-points in the spectrum obtained at the maximum Humanin concentration. The chemical shift changes were complete at a Humanin / Bid molar ratio of 6 / 1. The linear peak movement indicates that Humanin binding results in a single concerted change in the molecular environment of Bid, and since the largest observed frequency difference was approximately 40 Hz, for the amide 1H chemical shift of W48, the dissociation rate for the Bid–Humanin complex must be at least 250 / second (k >> 2π40). As illustrated in Figure 3, the three different peptides examined in this study have very similar chemical shift mapping profiles, suggesting that residues 3–12 of Humanin are sufficient to cause the spectral changes. whether they are also sufficient for cytoprotection remains to be determined.
Treatment with caspase-8 cleaves Bid at residue Asp59 within the flexible loop, but was previously shown to maintain the protein conformation and the association of N- and C-terminal fragments intact (24). This is confirmed by the close similarity of the spectra of Bid and cleaved Bid in Figure 2A, where only small changes are seen near the caspase-8 cleavage site. The addition of Humanin peptide to cleaved Bid produced additional small changes in the HSQC spectrum, similar in magnitude to those observed for intact Bid, indicating that Humanin binding does not induce either the separation of the N- and C-terminal fragments or a conformational change in the cleaved protein. Examples of these peak changes are shown in the expanded regions of the HSQC spectra in Figure 2, for free (green) and Humanin-bound (red) cleaved Bid.
Mapping the Humanin-binding site of Bid
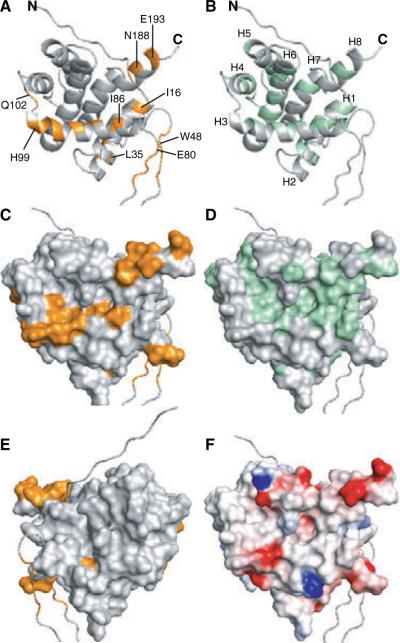
To further examine the Humanin-binding site, the chemical shift changes for residues affected by the addition of Humanin were mapped onto the three-dimensional structure of Bid previously determined by Cowburn, Wagner, and co-workers (23,24). In the structure, six amphipathic α-helices (H1–H5, and H8) fold around two more hydrophobic helices (H6–H7) at the protein core (Figure 4). Helix H3 is connected to the first two helices by a long flexible loop which includes the caspase-8 cleavage site, corresponding to Asp59 in the sequence of mouse Bid, or Asp60 in that of human Bid (Figure 1A).
Figure 4.
Molecular backbone and surface representations of Bid. The co-ordinates of mouse Bid were obtained from the Protein Data Bank (PDB ID 1DDB; 23). (A, C, and E) Amino acids with 1H / 15N heteronuclear single quantum correlation peaks that move by a total chemical shift >0.02 p.p.m., are colored gold. (A) Specific residues are labeled. (B) Helices and termini are labeled. (E) Molecular surface of Bid viewed 180° from the back with respect to all the other representations. (B and D) Surface-exposed hydrophobic residues with Kyte-Doolittle hydrophobicity >0 (52) are colored green. (F) The molecular surface is color coded with regions of electrostatic potential less than −8kBT in red, and regions of electro-static potential greater than +8kBT in blue, where kB is the Boltzmann constant and T is the temperature.
The residues with HSQC peaks that undergo total movements of more than 0.02 p.p.m. (Figure 4, gold color) map to one face of the Bid structure (Figure 4C), whereas the reverse face (Figure 4E) is not affected by the interaction with Humanin, indicating that Humanin binding is site-specific. The most prominent structural feature in the binding site is defined primarily by helix H3, and to some extent by helices H1, H2, H6, and H8, which pack against it, and it overlaps with the BH3 domain and with a central hydrophobic area on the surface of the protein (Figure 4D,F).
Significant chemical shift changes (>0.015 p.p.m.) are also observed for I101 at the end of helix H3, and for L151 and V159 in helix H6. These residues line a hydrophobic depression that forms on the surface of Bid at the site where helices H3 and H6 pack against each other. In addition, some residues at the end of helix H8 are affected. This is probably due to their close proximity to helix H3, since the direct interaction of a 12-residue helical Humanin peptide with the entire length defined by helices H3–H8 (Figure 4C) is unlikely. Although, we cannot exclude the possibility that Humanin associates with Bid in an extended conformation, this would be counter to the propensity for helical structure of the peptide determined by NMR analysis (27,28).
Several peaks from residues in the disordered loop of Bid were also affected by the addition of Humanin. These residues are not randomly distributed throughout the loop but grouped in two regions, one proximal to helix H2 and the other to helix H3, therefore, it is possible that the chemical shift perturbations experienced by the corresponding NMR peaks are indirect effects, relayed by Humanin binding to the helical regions of the protein. However, the possibility of a direct involvement of the Bid loop in Humanin binding is also interesting, as it could indicate a previously unknown function for this disordered region of Bid, as observed for the unstructured loops of Bcl-XL and Bcl-2 (44,45).
Affinity of the Bid–Humanin interaction
Plots of the total chemical shift change (Δ) against Humanin peptide concentration are illustrated in Figure 5, for selected amino acids that displayed changes in chemical shifts >0.02 p.p.m. Although the changes vary in magnitude, the titration data for each residue fit to a single-site binding curve (Figure 5A). Furthermore, when the normalized chemical shift change (Δ/Δmax) is plotted against Humanin concentration, the data points for each residue with peak changes >0.02 p.p.m. fall on the single curve that is obtained by nonlinear fitting of the data with an average value for the dissociation constant of 220 ± 60 μM (Figure 5B). Figure 5 also shows that complete saturation of the Bid-binding site was not obtained, however, saturating the binding site at the observed binding affinity and stoichiometries may not be possible due to the limited sample solubility at high polypeptide concentrations.
Figure 5.
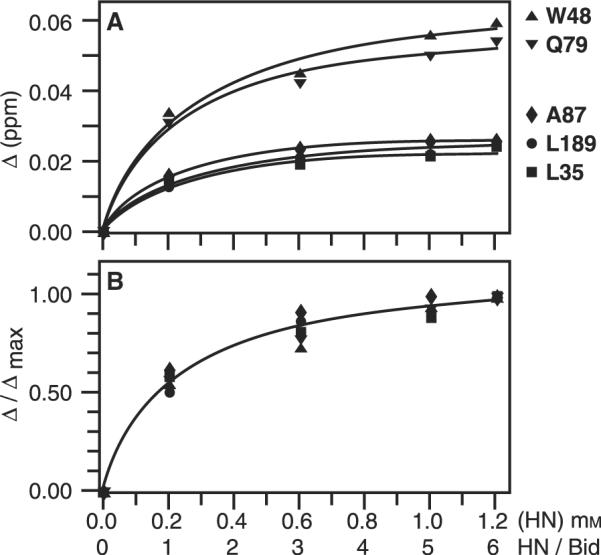
Change in chemical shift as a function of Humanin concentration ([HN]) and molar ratio of Humanin to Bid (HN / Bid), for representative backbone amide sites of Bid. (A) Total chemical shift change (Δ) and nonlinear curve fits for individual peaks. (B) Normalized total chemical shift change (Δ/Δmax). This is equivalent to the fraction of Bid bound to Humanin and was used to estimate the dissociation constant (Kd) and the maximum fraction of Humanin-bound Bid (pmax) for each peak. The binding curve was calculated using the average values of Kd (220 ± 60 μM) and pmax (1.15 ± 0.08), obtained for all the peaks in the spectrum with Δmax > 0.02. (∎) L35 in helix H2; (▴) W48 in the disordered loop; (▾) Q79 in the disordered loop near the BH3 domain; (◆) A87 in helix H3 and the BH3 domain; (●) L189 in helix H8.
The association of Bid with full-length Humanin leads to complex formation and precipitation, indicating either that these molecular partners bind very tightly, or that Humanin binding leads to the exposure of residues that promote aggregation or precipitation. Although the N-terminal Humanin peptides used in this study were previously shown to bind Bid and cleaved Bid, and to exert cytoprotection (20), fluorescence polarization anisotropy assays with FITC-conjugated Humanin showed that the full-length peptide has a much greater affinity for Bid (10–20 nM; 20) than that determined for the shorter peptides in this study, suggesting that the C-terminal part of the peptide also plays a role in Bid binding. While this numerical estimate for the truncated peptides is not expected to reflect the kinetics of the native Bid–Humanin interaction, the ability to fit the data with a unique binding curve further suggests that Humanin binds specifically to a single site on the surface of Bid.
Implications for the mechanism of Humanin cytoprotection
Previous studies have shown that Humanin can suppress the proapoptotic activities of three Bcl-2 family proteins: the BH3-only proteins Bid and Bim, and the multi-domain protein Bax (20–22). Bid, Bim, and Bax have in common that they exist as cytosolic proteins which translocate to mitochondria upon activation, inducing the release of cytotoxic molecules and initiating an apoptotic signaling cascade (12). Bid is activated by proteolytic cleavage and myristoylation, Bim is subject to several types of post-translational regulation (46), while Bax activation is believed to involve an intramolecular rearrangement that facilitates membrane insertion (25). Humanin provides two distinct mechanisms for suppressing mitochondria-dependent cell death: it binds Bid and Bim without affecting their association with mitochondria, and suppresses Bid- and Bim-dependent apoptosis in a Bax-independent manner; furthermore, it suppresses Bax-dependent apoptosis by inhibiting the association of Bax with mitochondria (20–22).
The finding that the Humanin-binding site localizes to a region of Bid which includes the BH3 domain suggests a mechanism for cyto-protection where Humanin binding engages surface-exposed hydrophobic residues in the BH3 domain, thereby rendering them inaccessible for association with its Bcl-2 targets such as Bax and Bcl-XL. This effect of Humanin appears to be effective even after proteolytic cleavage of Bid by caspase-8, as we observe that Humanin does not disrupt the association of the N- and C-terminal fragments and does not induce a major conformational change.
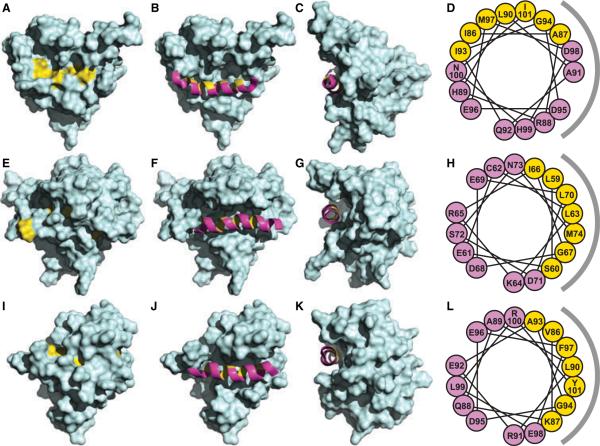
The unique arrangement of the Bid BH3 domain in the protein structure may provide an explanation for its specific interaction with Humanin (Figure 6). Although this highly conserved signature sequence is present in all pro-apoptotic and antiapoptotic Bcl-2 family proteins, the three-dimensional structures of Bid, Bax, and Bcl-XL show that while the Bid BH3 domain is surface accessible (Figure 6A–C; 23,24), those of Bax (Figure 6E–G; 25), and Bcl-XL (Figure 6I–K; 47) are buried somewhat deeper into the molecular mass, and hence, are protected from intermolecular interactions. Furthermore, helix H3 of Bid is rotated so that several of its conserved hydrophobic residues are surface-exposed and available to interact with Humanin, while the same residues in Bax and Bcl-XL are oriented toward the protein interior (Figure 6D,H,L). A recent NMR study showed that although Bim is largely unstructured in the absence of its binding partners, its BH3 domain adopts a helical structure when Bim binds pro-survival proteins (26). This suggests that the Bim BH3 domain would also be available for Humanin binding in a manner analogous to Bid.
Figure 6.
Molecular surface representations of (A–C) Bid (23), (E–G) Bax (25), and (I–K) Bcl-XL (48), with conserved hydrophobic residues in the BH3 domains shown in yellow. The (B, F and J) front and (C, G and K) side views show the BH3 domain helix as a ribbon in magenta, with hydrophobic residues in yellow. The helical wheel representations on the right show the rotations of the BH3 helices in (D) Bid, (H) Bax, and (L) Bcl-XL, colored and oriented as seen in (C, G and K) the corresponding side views. The gray semicircles to the right of the wheels represent the side of the BH3 domain facing the molecular interior. The co-ordinates were obtained from the Protein Data Bank (Bid: 1DDB; Bax: 1F16; Bcl-xL: 1LXL).
Prior mutagenesis studies showed that the N-terminal half of the Humanin sequence is required for association with Bid, Bim, and Bax, and that mutants with impaired binding also have impaired antiapoptotic activity, indicating that these intermolecular interactions are a critical aspect of cytoprotection by Humanin (20>–22). The first 12 amino acids of Humanin were shown to be sufficient for binding Bid and for suppressing its activity in vitro and in intact cells (20). Substituting Pro for the sole Cys8 residue in the sequence abolishes both the Bid-, Bim-, and Bax-dependent antiapoptotic activity of Humanin, and its ability to bind Bid, Bim, and Bax (20–22), while substitution of Cys8 with Ala, another small hydrophobic residue, does not affect these functions. In contrast, the Cys8Ala mutation suppresses the cytoprotective activity of Humanin on neuronal cell death induced by the amyloid precursor protein in the context of Alzheimer's disease (13), suggesting that different specific residues may be important for modulating the interactions of the peptide with different receptors.
Although a Pro residue at this position could profoundly alter the helical structure of the peptide, an Ala residue would not, suggesting that the Humanin–Bid interaction may be mediated by a Humanin helix–BH3 helix association. Interestingly, some residues in the C-terminal half of the Humanin peptide have also been shown to influence its activity. The finding that Ser14Gly is a gain of function mutation, protecting against amyloid-precursor-protein-dependent (13) as well as Bid- and Bim-dependent (20,21) cell death, and that this mutation also promotes the helical propensity of the peptide (28), particularly reinforces the idea that a specific helix–helix interaction, involving hydrophobic contacts, may be important for cytoprotection by Humanin. Glycines are abundant in the transmembrane helices of integral membrane proteins, where they often mediate helix–helix interactions through 'notch' and 'groove' interfaces (48–50), and replacing Ser14 with Gly may help stabilize helix–helix packing interactions of Humanin with its BH3-only targets (Figure 1B). Similarly, the IleGly residue pair that is exposed in the BH3 domains of Bid and Bim is also prominent in transmembrane helix–helix interactions, and may help stabilize the interaction with Humanin (Figure 1C). The presence of an Ala-X3-Ala motif (A87-X3-A91) in the sequence of the Bid BH3 domain is also notable (Figure 1C), because small-X3-small motifs, where small amino acids repeat at i and i + 4 sequential positions along one helix face, are known to stabilize hydrophobic helix–helix interactions. These considerations provide an attractive explanation for the cytoprotective effect of Humanin and its interaction with Bid and Bim, as they suggest that it might be possible to design peptide inhibitors that target BH3 domains by fine-tuning the arrangement of hydrophobic amino acids in a helical Humanin template.
Conclusions and Future Directions
The identification of a specific interaction between Humanin and Bid reinforces the role of this cytoprotective peptide as a direct modulator of Bid-dependent apoptosis. Although determining the structural details of the interaction would require the measurement of specific intermolecular contacts, with full-length Humanin, the involvement of the Bid BH3 domain, highlighted by the chemical shift mapping data in this study, provides some insight to the mechanisms which regulate the BH3-only Bcl-2 family proteins. The ability of Bid and Bim to display their BH3 domain distinguishes them from other multi-domain Bcl-2 family proteins and provides a possible explanation for the different mechanism of Humanin cytoprotection observed for Bid and Bim on one hand, and Bax on the other. Key hydrophobic residues in the BH3 domains of Bid and Bim are displayed on their molecular surfaces and would be available to interact with Humanin via hydrophobic helix–helix packing, while in the case of Bax, the molecular structure suggests that regions other than the BH3 domain may be involved. The specificity of the Bid–Humanin interaction highlighted by the NMR chemical shift mapping data, and the shared characteristic of Bid and Bim to display hydrophobic residues of their BH3 domain, suggest a strategy for the development of cytoprotective peptides, designed to target displayed BH3 domains by optimizing specific hydrophobic intermolecular contacts, as recently described for other systems (49,50).
Acknowledgments
We thank David Cowburn and Gerhard Wagner for generously providing their NMR assignments for mouse and human Bid. We thank Jinghua Yu for her assistance with NMR experiments. This research was supported by grants from the National Institutes of Health (GM065374, GM060554). The NMR studies utilized the Burnham Institute NMR Facility, supported by a grant from the National Institutes of Health (CA030199).
Abbreviations
- BH domain
Bcl-2 homology domain
- DTT
dithiothreitol
- Fmoc
N-(9-fluorenylmethoxycarbonyl)-L-amino acid
- HN
Humanin
- HSQC
heteronuclear single quantum correlation
- FHSQC
fast HSQC
Footnotes
DeLano W.L. (2005) PyMol. Available at: http://www.pymolorg
References
- 1.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 2.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 3.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 5.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 6.Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1 / Fas death. J Biol Chem. 1999;274:1156–1163. doi: 10.1074/jbc.274.2.1156. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, He H, Zhan S, Krajewski S, Reed JC, Gottlieb RA. Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia / reperfusion. J Biol Chem. 2001;276:30724–30728. doi: 10.1074/jbc.M103701200. [DOI] [PubMed] [Google Scholar]
- 8.Cirman T, Oresic K, Mazovec GD, Turk V, Reed JC, Myers RM, et al. Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem. 2004;279:3578–3587. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- 9.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 10.Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, et al. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 11.Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 2000;7:1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Proc Natl Acad Sci USA. 2001;98:6336–6341. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, Oyama F, et al. Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochem Biophys Res Commun. 2001;283:460–468. doi: 10.1006/bbrc.2001.4765. [DOI] [PubMed] [Google Scholar]
- 15.Jung SS, Van Nostrand WE. Humanin rescues human cerebrovascular smooth muscle cells from Abeta-induced toxicity. J Neurochem. 2003;84:266–272. doi: 10.1046/j.1471-4159.2003.01524.x. [DOI] [PubMed] [Google Scholar]
- 16.Kariya S, Takahashi N, Ooba N, Kawahara M, Nakayama H, Ueno S. Humanin inhibits cell death of serum-deprived PC12h cells. Neuroreport. 2002;13:903–907. doi: 10.1097/00001756-200205070-00034. [DOI] [PubMed] [Google Scholar]
- 17.Kariya S, Takahashi N, Hirano M, Ueno S. Humanin improves impaired metabolic activity and prolongs survival of serum-deprived human lymphocytes. Mol Cell Biochem. 2003;254:83–89. doi: 10.1023/a:1027372519726. [DOI] [PubMed] [Google Scholar]
- 18.Nishimoto I, Matsuoka M, niikura T. Unravelling the role of Humanin. Trends Mol Med. 2004;10:102–105. doi: 10.1016/j.molmed.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Niikura T, Chiba T, Aiso S, Matsuoka M, Nishimoto I. Humanin: after the discovery. Mol Neurobiol. 2004;30:327–340. doi: 10.1385/MN:30:3:327. [DOI] [PubMed] [Google Scholar]
- 20.Zhai D, Luciano F, Zhu X, Guo B, Satterthwait AC, Reed JC. Humanin binds and nullifies Bid activity by blocking its activation of Bax and Bak. J Biol Chem. 2005;280:15815–15824. doi: 10.1074/jbc.M411902200. [DOI] [PubMed] [Google Scholar]
- 21.Luciano F, Zhai D, Zhu X, Bailly-Maitre B, Ricci JE, Satterthwait AC, et al. Cytoprotective peptide humanin binds and inhibits proapoptotic Bcl-2 / Bax family protein BimEL. J Biol Chem. 2005;280:15825–15835. doi: 10.1074/jbc.M413062200. [DOI] [PubMed] [Google Scholar]
- 22.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, et al. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 23.McDonnell JM, Fushman D, Milliman CL, Korsmeyer SJ, Cowburn D. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 24.Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 26.Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- 27.Benaki D, Zikos C, Evangelou A, Livaniou E, Vlassi M, Mikros E, et al. Solution structure of humanin, a peptide against Alzheimer's disease-related neurotoxicity. Biochem Biophys Res Commun. 2005;329:152–160. doi: 10.1016/j.bbrc.2005.01.100. [DOI] [PubMed] [Google Scholar]
- 28.Benaki D, Zikos C, Evangelou A, Livaniou E, Vlassi M, Mikros E, et al. Solution structure of Ser14Gly-humanin, a potent rescue factor against neuronal cell death in Alzheimer's disease. Biochem Biophys Res Commun. 2006;349:634–642. doi: 10.1016/j.bbrc.2006.08.087. [DOI] [PubMed] [Google Scholar]
- 29.Arakawa T, Niikura T, Tajima H, Kita Y. The secondary structure analysis of a potent Ser14Gly analog of anti-Alzheimer peptide, Humanin, by circular dichroism. J Pept Sci. 2006;12:639–642. doi: 10.1002/psc.773. [DOI] [PubMed] [Google Scholar]
- 30.Schendel SL, Azimov R, Pawlowski K, Godzik A, Kagan BL, Reed JC. Ion channel activity of the BH3 only Bcl-2 family member, BID. J Biol Chem. 1999;274:21932–21936. doi: 10.1074/jbc.274.31.21932. [DOI] [PubMed] [Google Scholar]
- 31.Cavanagh J. Protein NMR Spectroscopy: Principles and Practice. Academic Press; San Diego: 1996. [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2004. [Google Scholar]
- 34.Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 35.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 36.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- 37.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 38.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, et al. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman RM, Li MX, Sykes BD. The binding of w7, an inhibitor of striated muscle contraction, to cardiac troponin C. Biochemistry. 2005;44:15750–15759. doi: 10.1021/bi051583y. [DOI] [PubMed] [Google Scholar]
- 40.Evans JNS. Biomolecular NMR Spectroscopy. Oxford University Press; Oxford; New York: 1995. [Google Scholar]
- 41.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 42.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopal P, Waygood EB, Klevit RE. Structural consequences of histidine phosphorylation: NMR characterization of the phosphohistidine form of histidine-containing protein from Bacillus subtilis and Escherichia coli. Biochemistry. 1994;33:15271–15282. doi: 10.1021/bi00255a008. [DOI] [PubMed] [Google Scholar]
- 44.Chang BS, Minn AJ, Muchmore SW, Fesik SW, Thompson CB. Identification of a novel regulatory domain in Bcl-X(L) and Bcl-2. EMBO J. 1997;16:968–977. doi: 10.1093/emboj/16.5.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77 / TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 46.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 48.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 49.Sato T, Kienlen-Campard P, Ahmed M, Liu W, Li H, Elliott JI, et al. Inhibitors of amyloid toxicity based on beta-sheet packing of Abeta40 and Abeta42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin H, Slusky JS, Berger BW, Walters RS, Vilaire G, Litvinov RI, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 51.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]