Abstract
The proteins PLM (phospholemman), CHIF (channel inducing factor), and Mat8 (mammary tumor protein 8 kDa) are members of the FXYD family of ion transport regulatory membrane proteins. Here we describe their cloning and expression in Escherichia coli, and their purification for NMR structural studies in lipid micelles and lipid bilayers. The molecular masses of the purified recombinant FXYD proteins, determined from SDS-PAGE and from MALDI TOF mass spectrometry, reflect monomeric species. The solution NMR and CD spectra in SDS micelles show that they adopt helical conformations. The solid-state NMR spectra in lipid bilayers give the first view of their transmembrane architecture.
Keywords: Membrane protein, FXYD, Mat8, PLM, CHIF, NMR
1. Introduction
The FXYD family proteins are expressed abundantly in tissues that perform fluid and solute transport (breast/mammary gland, kidney, colon, pancreas, prostate, liver, lung and placenta), or that are electrically excitable (muscle, nervous system), where they function to regulate the flux of transmembrane ions, osmolytes and fluids [1]. While abundant in mammals, with seven members identified in humans, several FXYD family members have been identified in Zebra fish (Danio rerio), indicating that these protein sequences are conserved through evolution. A recent study, which analyzed more than 1000 related ESTs, led to a complete data base of cDNA sequences, protein signature sequences and expression patterns for the FXYD genes, and to the identification of additional family members [1].
PLM (phospholemman), one of the best characterized members of this family, is the major substrate of hormone-stimulated phosphorylation by cAMP-dependent protein kinase A and C in the heart [2], and has been recently identified as a regulator of the Na,K-ATPase [3]. PLM induces ionic currents when expressed in Xenopus oocytes, and also forms ion channels in phospholipid bilayers [4]. These channels are selective for the amino acid taurine, suggesting a physiological role in osmolyte transport and cell volume regulation. CHIF (channel inducing factor) is upregulated by aldosterone and corticosteroids in mammalian kidney and intestinal tracks, where it associates with the Na,K-ATPase pump to regulate Na+ and K+ homeostasis [5,6]. Mat8 (mammary tumor protein 8 kDa) is expressed in breast, prostate, lung, stomach, and colon, as well as in human breast tumors, breast tumor cell lines, and prostate cancer cell lines, after malignant transformation by oncogenes [7,8]. Likewise, the expression of mouse RIC (related to ion channel), and of its human homologue dysadherin, is induced by oncogenic transformation [9,10]. Dysadherin, a cancer-associated membrane glycoprotein, down-regulates E-cadherin and promotes metastasis [10]. Like PLM, both Mat8 and CHIF induce ionic currents in Xenopus oocytes, although these channels have not been characterized in lipid bilayers. Gamma (the γ subunit of the Na,K-ATPase) modulates the activity of the pump in kidney tubules, and its expression is necessary for blastocyst cavitation in mouse embryonic development [11,12]. Finally, Php (phosphohippolin) is expressed in developing mouse cerebellum and kidney [13,14], and FXYD7 is a brain and isozyme-specific regulator of the Na,K-ATPase [15]. The identification of four FXYD family members (PLM, CHIF, Gamma, FXYD7) as regulators of the Na,K-ATPase, points to a mechanism for regulation that involves the expression of an auxiliary subunit of the pump.
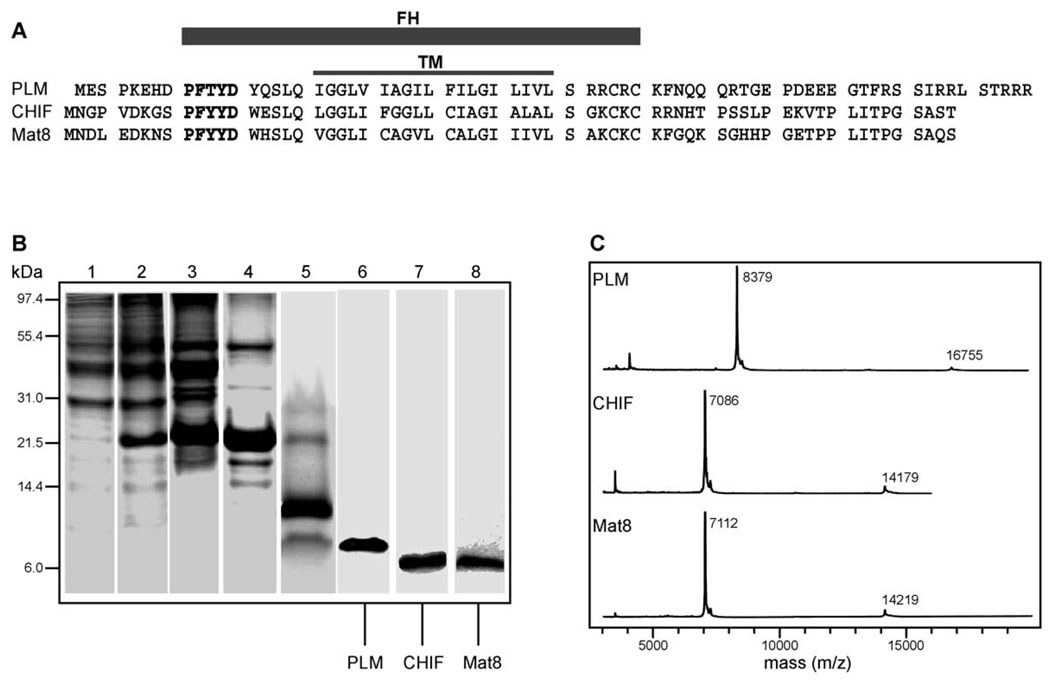
The FXYD proteins are characterized by a 35-amino acid FXYD homology (FH) domain, which includes the transmembrane domain (TM) (Fig. 1A). The short motif PFXYD (Pro, Phe, X, Tyr, Asp), before the transmembrane domain, is invariant in all known mammalian examples, and identical in other vertebrates, except for the proline. X is usually Tyr, but can also be Thr, Glu, or His. In all proteins, conserved basic residues flank the transmembrane domain, the extracellular N-termini are acidic, and the cytoplasmic C-termini are basic. The cytoplasmic domain of PLM contains consensus phosphorylation sites for both cAMP-dependent protein kinases A and C [4], and while the cytoplasmic domains of Mat8 and CHIF contain no such consensus sites, Ser49 next to Lys50, in the Mat8 sequence, may be a substrate for protein kinase C.
Fig. 1.
Expression and purification of the FXYD family proteins PLM, CHIF, and Mat8. (A) Amino acid sequences of PLM, CHIF and Mat8. The FXYD homology domain (FH) encompasses the FXYD consensus sequence (in bold), and the transmembrane domain (TM). (B) Protein fractions at different stages of the purification protocol were analyzed on Coomassie stained 16% SDS-PAGE. Lane 1: Entire cell before IPTG induction. Lane 2: Entire cell after IPTG induction with the fusion protein band at 21 kDa. Lane 3: Isolated inclusion bodies enriched in fusion protein. Lane 4: Fusion protein isolated by Ni affinity chromatography. Lane 5: His9-TrpΔLE fusion partner and PLM resulting from CNBr cleavage of the fusion protein. Lane 6: Purified PLM. Lane 7: Purified CHIF. Lane 8: Purified Mat8. (C) MALDI-TOF mass spectra of purified PLM (8379 Da), CHIF (7086 Da), and Mat8 (7112 Da).
The three-dimensional structures of these proteins should provide significant insights to their roles in heart muscle regulation, carcinogenesis, and cell cycle regulation. Their small sizes make them ideal candidates for structure determination by NMR spectroscopy, enabling structural comparisons to be made for an entire family of membrane proteins involved in ion transport. NMR spectroscopy is ideally suited for the structure determination of membrane proteins in lipid environments. Solution NMR approaches to structure determination can be utilized on samples consisting of proteins dissolved in lipid micelles, while solid-state NMR approaches can be applied to samples of membrane proteins in planar lipid bilayers [16,17]. The latter technique is especially attractive because it enables structure determination within the context of lipid membranes. Furthermore, since the lipid bilayers are oriented with respect to the applied magnetic field, the samples preserve the intrinsic directional character of membrane protein structure and function.
While NMR structure determination does not require crystals of membrane proteins, it is, nevertheless, essential to obtain pure and homogeneous preparations of proteins in either micelle or lipid bilayer samples. Here, we describe the expression in Escherichia coli, purification, sample preparation and characterization of the 15N-labeled FXYD proteins PLM, CHIF, and Mat8, for NMR structure determination in lipid micelles and in oriented lipid bilayers.
2. Materials and methods
2.1. Cloning and protein expression
The human FXYD genes encoding human PLM, rat CHIF, and human Mat8 (GenBank accession numbers: NP_005022, NP_071783, and NP_005962; Fig. 1A) were cloned into the expression vector pMMHa that directs the synthesis of the fusion protein His9-TrpΔLE-FXYD [18]. The genes were adapted for E. coli codons, and constructed by synthesizing four overlapping single-stranded oligonucleotides, followed by PCR extension and amplification using Pfu (30 cycles of 1 min at 94 °C, 1 min at 40 °C, 1 min at 72 °C). The gene sequences encode the residues Asn-Gly-Met at their 5′ end, corresponding to cleavage sites for hydroxylamine (Asn-Gly) and cyanogen bromide (Met) [19,20], which provide two alternate means for cleaving each authentic FXYD from its fusion partner, and they also encode recognition sites for the restriction enzymes HindIII and BamHI. The Met residues in the CHIF (M22 and M35) and Mat8 (M31 and M36) sequences were changed to Leu in order to facilitate cyanogen bromide cleavage of the fusion proteins used for expression and purification. These are naturally occurring, conservative mutations. There are no Met residues in the amino acid sequence corresponding to PLM. The genes were digested with HindIII and BamHI, purified by agarose gel electrophoresis, and ligated using T4 DNA ligase, with the pMMHa plasmid vector that had been previously digested with the same enzymes. The resulting recombinant plasmids pTrpΔLE-FXYD were transformed into E. coli strain DH5α, positive clones were selected by restriction analysis and PCR screening, and plasmid identity was confirmed by DNA sequencing. The plasmids were finally transformed into E. coli BL21(DE3) [21], and stable transformed clones were screened for high level expression of the fusion protein in minimal M9 media (100 µg/ml of ampicillin, 7.0 g/l Na2HPO4, 3.0 g/l KH2PO4, 0.5 g/l NaCl, 11 mg/l CaCl2, 120 mg/l MgSO4, 50 mg/l thiamine, 1% v/v LB, 10 g/l d-glucose, 1 g/l (NH4)2SO4). Aliquots of each clone were stored as 15% glycerol stocks at −80 °C. For protein expression, 10 µl of the glycerol stock were used to inoculate 10 ml of LB media, and grown at 37 °C (300 rpm). After 5 h, 1 ml of this starter culture was used to inoculate 100 ml of M9 media, and the cells were grown overnight at 37 °C. In the morning, the overnight culture was used to inoculate 1 l of fresh M9 media. When the cell density reached A600 = 0.7, protein expression was induced by the addition of IPTG to a final concentration of 0.4 mM, and cell growth was continued for another 4 h at 37 °C. The cells were subsequently harvested by centrifugation and stored at −20 °C overnight. For uniformly 15N-labeled proteins, (15NH4)2SO4 (Cambridge Isotope Laboratories, Andover, MA) was supplied as the sole nitrogen source.
2.2. Fusion protein isolation
For cell lysis, the frozen cell pellet from 1 l of cell culture was suspended in 30 ml of Buffer I (50 mM TrisCl, pH 8, 15% v/v glycerol, 50 µg/ml Lysozyme) at 23 °C for 20 min, sonicated on ice for 10 min. The soluble fraction was removed by centrifugation (48,000 × g, 4 °C, 30 min), and the pellet containing inclusion bodies was washed in 30 ml of Buffer II (50 mM TrisCl, pH 8, 1% w/v deoxycholic acid, 1% v/v IGEPAL CA-630), by sonication on ice for 10 min. The soluble fraction was removed by centrifugation (48,000 × g, 4 °C, 30 min), and the pellet dissolved in 30 ml of Buffer III (20 mM TrisCl, pH 8, 6 M GdnHCl, 0.5 M NaCl, 5 mM imidazole), transferred to a centrifuge bottle, and diluted 10-fold with water in order to precipitate the fusion protein. The precipitate was collected by centrifugation (28,000 × g, 4 °C, 1 h), dissolved in 10 ml of Buffer III, and stored at 4 °C overnight. The crude fusion protein solution from 5 l of cell culture was loaded onto a 50 ml Ni-NTA column (His Bind Resin, Novagen) and the column washed with 300 ml of Buffer III. The fusion protein was eluted with 200 ml of Buffer IV (20 mM TrisCl, pH 8, 6 M GdnHCl, 0.5 M NaCl, 500 mM imidazole), concentrated by ultrafiltration (MWCO = 10 kDa), and then precipitated by dialysis against water (MWCO = 10 kDa) to remove GdnHCl.
2.3. Protein purification
In order to cleave each FXYD protein from the TrpΔLE fusion partner, the fusion protein precipitate obtained from dialysis was collected by centrifugation, dissolved in 70% formic acid at a concentration of 10–20 mg/ml, and reacted for 2 h with a 10-fold molar excess of CNBr at 23 °C in the dark. The reaction mixture was then dialyzed against water (MWCO = 1 kDa), lyophilized to powder, dissolved in Buffer V (4 mM SDS, 10 mM sodium phosphate, pH 7.5, 12 mM DTT) and purified by size exclusion chromatography in Buffer V on Sephacryl-100 (Amersham Biosciences, Piscataway, NJ). The fractions containing pure protein were pooled, concentrated by ultrafiltration (MWCO = 10 kDa), and injected on a preparative reverse-phase HPLC column (Delta-Pak C4 column, 15 µm, 300 Å, 7.8 × 300 mm, Waters, Milford, MA) running in 90% water, 10% acetonitrile, 0.1% trifluoroacetic acid, with a linear gradient of acetonitrile. PLM, CHIF, and Mat8 eluted, respectively at 60%, 56%, and 56% acetonitrile, and were stored as lyophilized powders at −20 °C. Protein purity and identity were assessed by N-terminal amino acid sequence analysis, MALDI TOF mass spectrometry, and solution NMR spectroscopy.
2.4. SDS-PAGE
SDS PAGE was performed using the Tris–Tricine system described by Schaegger and von Jagow [22], which is well suited to membrane proteins. The gel loading buffer was supplemented with 15 mM DTT, and gels were stained with Coomassie Blue G250.
2.5. Mass spectrometry
Approximately 0.2 mg of lyophilized protein were dissolved in 10 µl of solution I (75% acetonitrile, 25% water, 0.1% TFA), and 1 µl of this protein solution were mixed with 1 µl of solution II (15 mg/ml sinnapinic acid, 300 µl acetonitrile, 200 µl methanol, 500 µl Milli-Q ultra purified water). The resulting solution was spotted onto a seed layer spot on the MALDI target. The seed layer was prepared from matrix solution (6 mg sinnapinic acid, 600 µl methanol, 390 µl acetone, 10 µl of 0.1% aqueous TFA). The MALDI-TOF mass spectrum was collected in linear mode on a Voyager DE-PRO mass spectrometer (Applied Biosystems, Foster City, CA). Typically, 250–500 laser pulses were averaged per spectrum.
2.6. CD spectroscopy
The samples contained 20 µM FXYD protein, 500 mM SDS, 20 mM sodium phosphate, 1 mM sodium azide, pH 5. Protein solutions were transferred to a quartz cuvette (0.1 mm path length), and far UV CD spectra were recorded at 23 °C on an Aviv model 62A-DS CD spectrometer (Aviv Instruments, Lakewood, NJ) equipped with a temperature controller. A 5 s time constant and a 1 nm band width were used during data acquisition over a wavelength range of 184–260 nm. For each protein solution, five spectra were recorded, averaged, and referenced by subtracting the average of three spectra obtained from NMR buffer alone.
2.7. NMR spectroscopy in lipid micelles
Samples were prepared by dissolving 1–2 mg of 15N-labeled protein in 250 µl of NMR buffer (500 mM SDS, 10 mM DTT, 10% D2O, 20 mM sodium citrate, pH 5.0). The solution NMR experiments were performed on a Varian Inova 500 spectrometer equipped with a triple-resonance 5 mm probe. The two-dimensional 1H/15N HSQC [23] spectra were obtained at 45 °C. The 15N and 1H chemical shifts were referenced to 0 ppm for liquid ammonia and tetramethylsilane, respectively. The NMR data were processed using NMR Pipe and rendered in NMR Draw [24] on a Dell Precision 330 MT Linux workstation (Round Rock, TX).
2.8. NMR spectroscopy in lipid bilayers
Samples were prepared by first dissolving 2 mg of 15N-labeled protein in 0.5 ml of TFE with 50 µl of β-mercaptoethanol, and then adding 100 mg of lipid (DOPC/DOPG, 4:1, mol/mol) in 1 ml of CHCl3. This solution was spread on the surface of 40 glass slides (dimensions 11 × 20 × 0.06 mm, Paul Marienfeld, Germany), and the solvents removed under vacuum overnight. The glass slides were then stacked and equilibrated for 24 h at 40 °C in a chamber containing a saturated solution of ammonium phosphate, which provides an atmosphere of 93% relative humidity. The sample was wrapped in one layer of parafilm and one of teflon tape, and then sealed in thin polyethylene film prior to insertion in the NMR probe. The solid-state NMR spectra were obtained at 23 °C on a Chemagnetics-Otsuka Electronics CMX400 (Fort Collins, CO) spectrometer, with a wide-bore 400/89 Oxford magnet (Oxford Instruments, UK), using home-built double-resonance (1H/15N or 1H/31P) probes with square rf coils wrapped directly around the samples. The 15N spectra were obtained with single contact 1 ms CPMOIST [25,26] with continuous 1H irradiation (rf field strength 63 kHz) during acquisition, in order to decouple the 1H–15N dipolar interactions. The 31P spectra were obtained with a single pulse, and continuous 1H irradiation (rf field strength 63 kHz) during acquisition to decouple the 1H–31P dipolar interactions. The 15N and 31P chemical shifts were referenced to 0 ppm for liquid ammonia and phosphoric acid. The NMR data were processed using the program FELIX (Accelerys, San Diego, CA) on a Silicon Graphics computer workstation (Mountain View, CA).
3. Results and discussion
3.1. Protein expression and characterization
Our approach to the production of PLM, CHIF, and Mat8 utilized the E. coli pMMHa fusion protein expression vector, which directs the synthesis of the fusion protein His9-TrpΔLE-FXYD. This vector has been used successfully for the production of other membrane proteins ranging in size from 80 to 150 amino acids [16]. The TrpΔLE fusion partner, from the Trp leader amino acid sequence, is very effective at forming inclusion bodies and is thus protected from proteolysis. The fusion protein is not toxic to the E. coli host cells, and is expressed at levels up to 20% of total cellular protein in E. coli strain BL21(DE3), grown on the M9 minimal media required for isotopic labeling [16,18,21]. Intact FXYD proteins are liberated from the fusion partner using CNBr, which cleaves specifically after Met residues [20]. The use of chemical cleavage eliminates the difficulties such as poor specificity and enzyme inactivation often encountered with protease treatment of membrane proteins in detergents.
The protein content of cells isolated before and after IPTG induction is shown in Fig. 1B (lanes 1 and 2). Fusion protein overexpression is marked by the appearance of an intense band near 21 kDa. After protein expression, the separation and purification of PLM, CHIF, and Mat8 were accomplished with the four-step-protocol detailed under Materials and methods. First, the inclusion bodies containing the fusion protein were separated from the E. coli lysate by a series of wash and centrifugation steps (Fig. 1B, lane 3). Second, the fusion protein was isolated by Nickel affinity chromatography (Fig. 1B, lane 4). Third, each FXYD protein was cleaved from the fusion partner using CNBr. This yields a fragment near 12 kDa, corresponding to His9-TrpΔLE [18], and a smaller fragment corresponding to either PLM at 8.4 kDa, CHIF at 7.1 kDa, or Mat8 at 7.1 kDa (Fig. 1B, lane 5). Finally, the proteins were purified with size exclusion chromatography, followed by reverse-phase chromatography (Fig. 1B, lanes 6–8). Typically, 1.5 mg of purified protein are obtained from 1 l of cell culture in 15N-labeled minimal media.
The homogeneity of the purified proteins was confirmed by MALDI TOF mass spectrometry, and N-terminal amino acid sequence analyses. The mass spectra shown in Fig. 1C demonstrate the high degree of purity obtained using this procedure. The major peaks have masses that correspond exactly to authentic PLM, CHIF, and Mat8. The small peaks at half mass arise from doubly charged species, and those at double mass from a small fraction of FXYD dimer. Importantly, the spectra show no evidence of degradation or chemical modifications. Formylation of hydroxyamino acids, resulting from exposure to formic acid during CNBr cleavage, was prevented by limiting the reaction time to 2 h, which was sufficient to yield at least 90% cleavage of the fusion protein (Fig. 1B, lane 5).
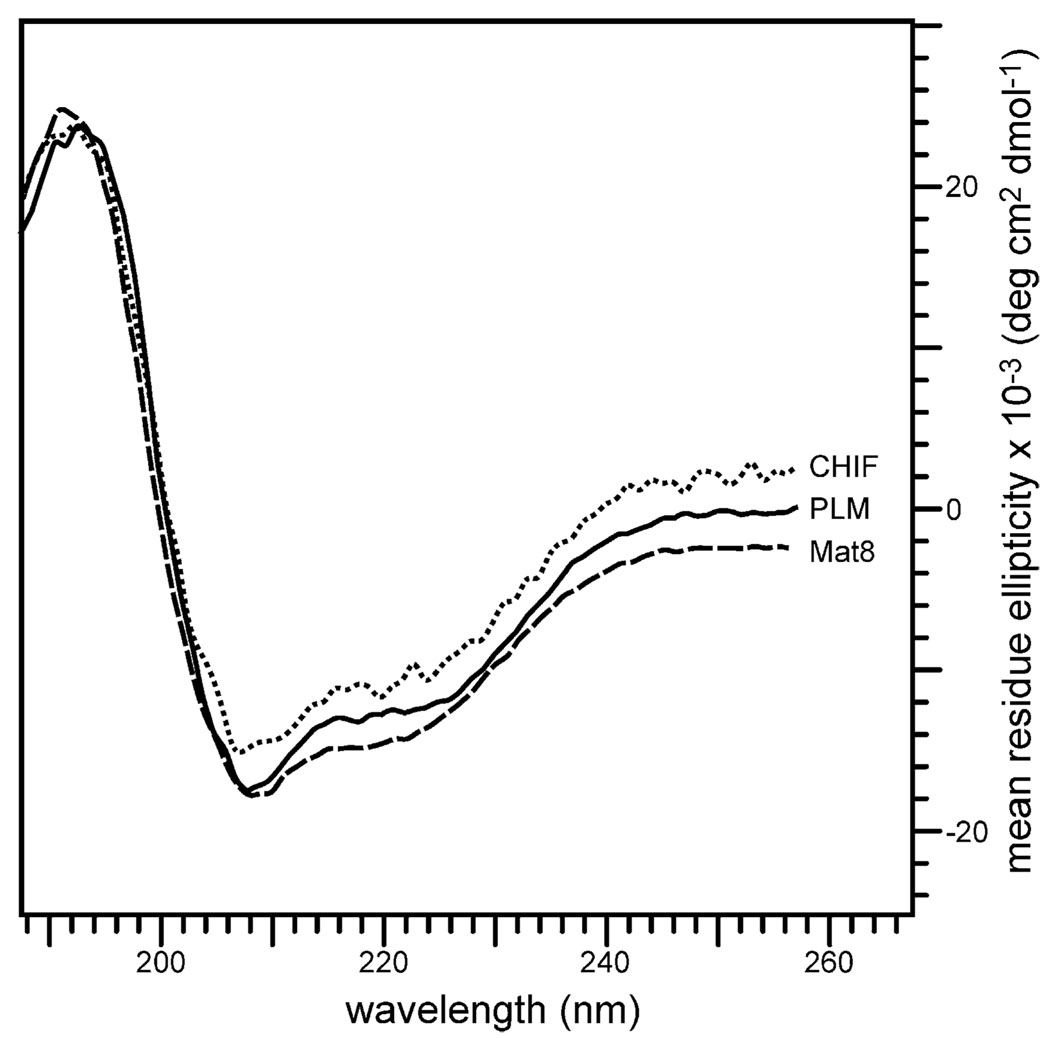
The CD spectra obtained for PLM, CHIF and Mat8 in SDS micelles are shown in Fig. 2. For all three FXYD family members, the two minima, at 208 and 222 nm, are characteristic of α-helical structures (calculated α-helicity about 40%). These purified recombinant proteins were used for NMR structural studies in lipid micelles and lipid bilayers.
Fig. 2.
CD spectra of purified PLM, CHIF, and Mat8 in SDS micelles. The samples contained 20 µM FXYD protein, 500 mM SDS, 20 mM sodium phosphate, 1 mM sodium azide pH 5.
3.2. NMR spectroscopy
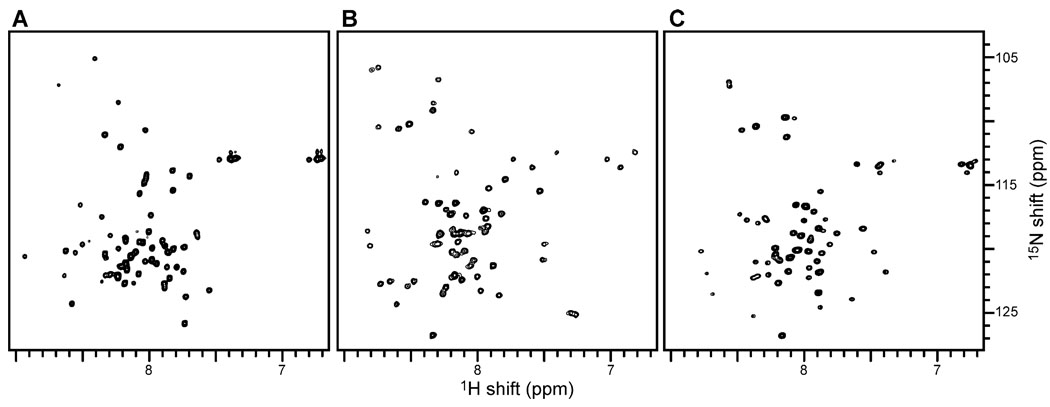
The initial solution NMR and solid-state NMR studies of PLM, CHIF, and Mat8, provide valuable information about their structure and membrane architecture. These spectra are the starting points for higher dimensional solution NMR experiments and structure determination. Solution NMR spectroscopy of the FXYD proteins in lipid micelles can be used to determine their secondary structures and three-dimensional folds, because the proteins reorient fast enough in solution to give isotropic spectra with relatively narrow line widths. The two-dimensional 1H/15N HSQC spectra of uniformly 15N-labeled FXYD proteins in aqueous SDS micelles are shown in Fig. 3. Each resonance represents a single 15N-labeled site of the protein, and is characterized by 1H and 15N chemical shift frequencies that reflect the local environment. The samples were optimized to obtain narrow resonance line widths and high resolution, and the presence of one well-defined resonance for each amide site in the protein is indicative of a high-quality micelle sample. Resonances from the Gly (15N shift = 105–110 ppm), Trp indole (1H shift = 10 ppm), and Gln and Asn sidechain (1H shift = 6.5–7.5 ppm) nitrogens are clearly resolved. The limited chemical shift dispersion is indicative of helical structures for these proteins. The measurements of as many 1H/1H NOEs as possible among resonances provide the short- and long-range distance constraints needed for structure determination, and we have acquired three-dimensional 1H/15N HSQC-NOESY spectra of uniformly 15N-labeled FXYD proteins where several well-resolved cross peaks can be identified.
Fig. 3.
Two-dimensional 1H/15N HSQC spectra of uniformly 15N labeled (A) PLM, (B) CHIF, and (C) Mat8 in SDS micelles at 40 °C. The samples contained 1–2 mM FXYD protein, 500 mM SDS, 20 mM sodium phosphate, 10 mM DTT, 1 mM sodium azide pH 5. The 15N and 1H chemical shifts are referenced to 0 ppm for liquid ammonia and tetramethylsilane.
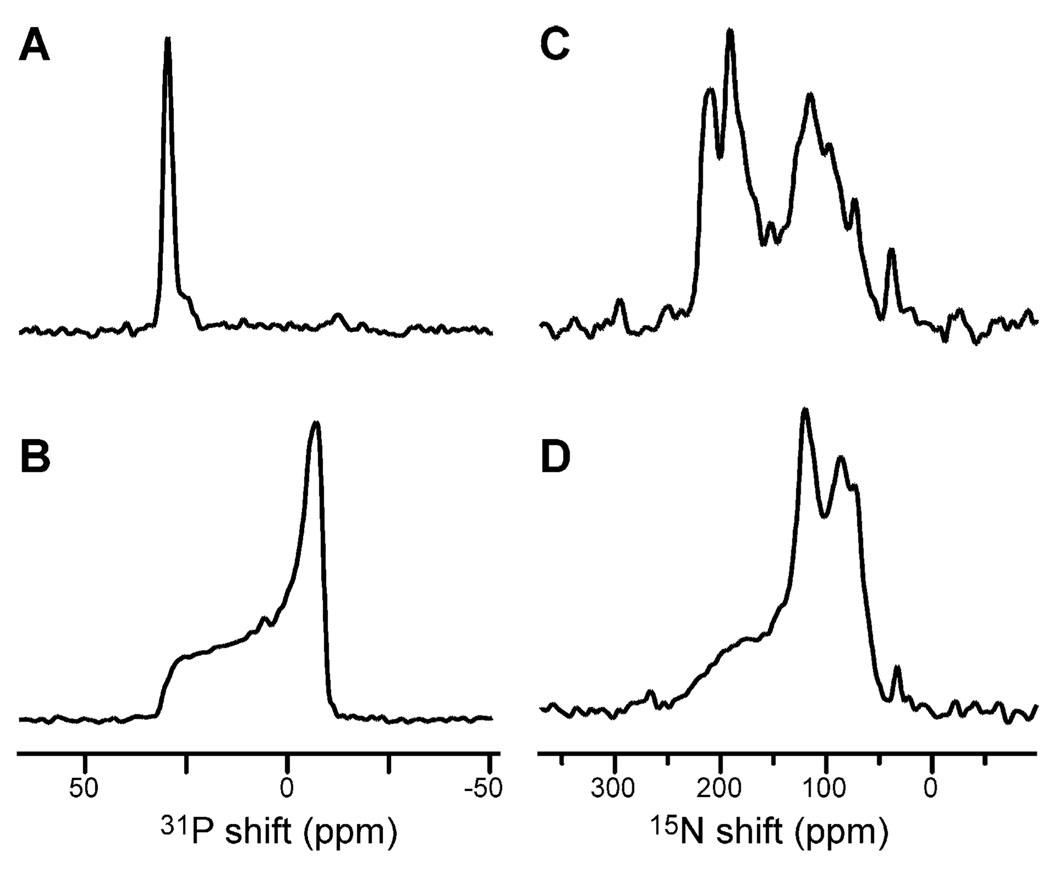
The solid-state NMR spectra of 15N-labeled CHIF in oriented and unoriented lipid bilayers are shown in Fig. 4. The degree of phospholipid bilayer alignment can be assessed with 31P NMR spectroscopy of the lipid phosphate headgroup, as shown in Fig. 4A,B. The 31P NMR spectra obtained for the CHIF samples are characteristic of a liquid-crystalline bilayer arrangement, in both oriented and unoriented samples (Fig. 4A,B). The spectrum from the oriented sample, in Fig. 4A, has a single peak near 30 ppm, as expected for highly aligned bilayers with this lipid composition (DOPC/DOPG = 4:1 molar). These spectra demonstrate that highly aligned samples of FXYD proteins in lipid bilayers can be prepared for NMR structure determination.
Fig. 4.
One-dimensional solid-state NMR spectra of CHIF in (A,C) oriented and (B,D) unoriented lipid bilayers at 22 °C. (A,B) 31P chemical shift spectra from the phosphate headgroups of the phospholipids. (C,D) 15N chemical shift spectra of uniformly 15N-labeled CHIF. The 15N and 31P shifts are referenced to 0 ppm for external liquid ammonia and phosphoric acid.
The one-dimensional 15N chemical shift spectra of CHIF (Fig. 4C,D) provide a first view of the FXYD protein architecture in membranes. A preliminary analysis of the solid-state NMR data is possible since both CD and NMR spectroscopy in micelles show that the overall secondary structure of these proteins is α-helical. Membrane proteins in lipid bilayers are largely immobile on NMR time scales; therefore, their resonances are not motionally averaged but have frequencies that reflect the orientation of their respective sites relative to the direction of the magnetic field [16,17]. In our sample, the lipid bilayer plane is perpendicular to the magnetic field direction; therefore, each resonance frequency reflects the orientation of its corresponding protein site in the membrane.
The spectrum of CHIF in oriented bilayers (Fig. 4C) displays significant resolution with identifiable peaks at frequencies throughout the range of the 15N amide chemical shift. The resonance intensity near 200 ppm arises from backbone amide sites in the transmembrane helix that have their NH bonds nearly perpendicular to the plane of the membrane, while the intensity near 80 ppm is from sites in the N- and C-termini, with NH bonds nearly parallel to the membrane surface. The rather narrow dispersion of 15N resonances centered around 200 ppm suggests that the CHIF transmembrane helix crosses the lipid bilayer membrane with a very small tilt angle. The peak near 35 ppm results from the amino groups of the lysine sidechains and the N-terminus.
The spectrum of CHIF in unoriented bilayers is strikingly different and provides no resolution among resonances (Fig. 4D). Most of the backbone sites are structured and immobile on the time scale of the 15N chemical shift interaction (10 kHz), contributing to the characteristic amide powder pattern between about 220 and 60 ppm. Some of the CHIF backbone sites, probably near the N- and C-termini, are mobile, and give rise to the resonance band centered near 120 ppm. Therefore, while certain resonances near 120 ppm, in the spectrum of oriented CHIF, may reflect specific orientations of their corresponding sites, some others arise from mobile backbone sites.
4. Conclusions
The FXYD membrane proteins regulate ion, osmolyte, and fluid homeostasis in a variety of tissues, and are emerging as auxiliary tissue-specific and physiological state-specific subunits of the Na,K-ATPase. The cloning, expression, and purification of the recombinant FXYD family members PLM, CHIF, and Mat8 enable NMR structural studies to be performed in membrane environments, since the proteins can be isotopically labeled, and obtained in quantities that are suitable for NMR structure determination. The 1H/15N HSQC spectra in SDS micelles display excellent resolution and demonstrate the feasibility of structure determination in micelles. The HSQC and CD spectra in micelles reflect helical secondary structures for all three proteins, and the solid-state NMR spectra from oriented lipid bilayer samples provide a first view of the topological features of their transmembrane and terminal domains. The ability to produce milligram quantities of pure FXYD proteins also opens the door for functional studies that, together with structure determination, can provide important structure–activity correlations. For example, the reconstitution of Na,K-ATPase activity in the presence of FXYD proteins would be an important step in understanding the mechanism of pump regulation, while the incorporation of FXYD proteins in lipid bilayers enables ion channel activities to be characterized by measuring specific ionic currents.
Acknowledgements
This research was supported by grants from the National Institutes of Health National Cancer Institute (RO1-CA82864) and the Department of the Army (DAMD17-00-1-0506). The NMR experiments were performed at the Resource for Solid-State NMR of Proteins at the University of California San Diego, and are supported by a grant from the Biomedical Research Technology Program, National Center for Research Resources (P41RR09731). KJC is supported by a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics. 2000;68:41–56. doi: 10.1006/geno.2000.6274. [DOI] [PubMed] [Google Scholar]
- 2.Palmer CJ, Scott BT, Jones LR. Purification and complete sequence determination of the major plasma membrane substrate for cAMP-dependent protein kinase C in myocardium. J. Biol. Chem. 1991;266:11126–11130. [PubMed] [Google Scholar]
- 3.Crambert G, Fuzesi M, Garty H, Karlish S, Geering K. Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11476–11481. doi: 10.1073/pnas.182267299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moorman JR, Ackerman SJ, Kowdley GC, Griffin MP, Mounsey JP, Chen Z, Cala SE, O’Brian JJ, Szabo G, Jones LR. Unitary anion currents through phospholemman channel molecules. Nature. 1995;377:737–740. doi: 10.1038/377737a0. [DOI] [PubMed] [Google Scholar]
- 5.Attali B, Latter H, Rachamim N, Garty H. A corticosteroid-induced gene expressing an ȜIsK-likeȝ K+ channel activity in Xenopus oocytes. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6092–6096. doi: 10.1073/pnas.92.13.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beguin P, Crambert G, Guennoun S, Garty H, Horisberger JD, Geering K. CHIF, a member of the FXYD protein family, is a regulator of Na,K-ATPase distinct from the γ-subunit. EMBO J. 2001;20:3993–4002. doi: 10.1093/emboj/20.15.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison BW, Moorman JR, Kowdley GC, Kobayashi YM, Jones LR, Leder P. Mat-8, a novel phospholemman-like protein expressed in human breast tumors, induces a chloride conductance in Xenopus oocytes. J. Biol. Chem. 1995;270:2176–2182. doi: 10.1074/jbc.270.5.2176. [DOI] [PubMed] [Google Scholar]
- 8.Varaala MH, Porvari K, Kyllonen A, Vihko P. Differentially expressed genes in two LNCaP prostate cancer cell lines reflecting changes during prostate cancer progression. Lab. Invest. 2000;80:1259–1268. doi: 10.1038/labinvest.3780134. [DOI] [PubMed] [Google Scholar]
- 9.Fu X, Kamps MP. E2a-Pbx1 induces aberrant expression of tissue-specific and developmentally regulated genes when expressed in NIH 3T3 fibroblasts. Mol. Cell. Biol. 1997;17:1503–1512. doi: 10.1128/mcb.17.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ino Y, Gotoh M, Sakamoto M, Tsukagoshi K, Hirohashi S. Dysadherin, a cancer-associated cell membrane glycoprotein, down-regulates E-cadherin and promotes metastasis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:365–370. doi: 10.1073/pnas.012425299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer RW, Biemesderfer D, Bliss DP, Collins JH, Forbush B. Molecular cloning and immunological characterization of the gamma polypeptide, a small protein associated with the Na,K-ATPase. J. Cell Biol. 1993;121:579–586. doi: 10.1083/jcb.121.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones HD, Davies TC, Kidder GM. Embryonic expression of the putative g subunit of the sodium pump is required for acquisition of fluid transport capacity during blastocyst development. J. Cell Biol. 1997;139:1545–1552. doi: 10.1083/jcb.139.6.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi F, Yamaguchi K, Tai Y, Sugimoto K, Tokuda M. Molecular cloning and characterization of a novel phospholemman-like protein from rat hippocampus. Mol. Brain Res. 2001;86:189–192. doi: 10.1016/s0169-328x(00)00213-8. [DOI] [PubMed] [Google Scholar]
- 14.Saito S, Matoba R, Kato K, Matsubara K. Expression of a novel member of the ATP1G1/PLM/MAT8 family, phospholemman-like protein (PLP) gene, in the developmental process of mouse cerebellum. Gene. 2001;279:149–155. doi: 10.1016/s0378-1119(01)00745-4. [DOI] [PubMed] [Google Scholar]
- 15.Beguin P, Crambert G, Monnet-Tschudi F, Uldry M, Horisberger J-D, Garty H, Geering K. FXYD7 is a brain-specific regulator of Na,K-ATPase α1-α isozymes. EMBO J. 2002;21:3264–3273. doi: 10.1093/emboj/cdf330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opella SJ, Ma C, Marassi FM. NMR of membrane associated peptides and proteins. Methods Enzymol. 2001;339:285–313. doi: 10.1016/s0076-6879(01)39319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marassi FM. NMR of peptides and proteins in membranes. Concepts Magn. Reson. 2002;14:212–224. [Google Scholar]
- 18.Staley JP, Kim PS. Formation of a native-like subdomain in a partially folded intermediate of bovine pancreatic trypsin inhibitor. Protein Sci. 1994;3:1822–1832. doi: 10.1002/pro.5560031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bornstein P, Balian G. Cleavage at Asn–Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- 20.Gross E, Witkop B. Selective cleavage of the methionyl peptide bonds in ribonuclease with cyanogen bromide. J. Am. Chem. Soc. 1961;83:1510–1511. [Google Scholar]
- 21.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 22.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 23.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. 1995;108B:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipe. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 1973;59:569–590. [Google Scholar]
- 26.Levitt MH, Suter D, Ernst RR. Spin dynamics and thermodynamics in solid-state NMR cross-polarization. J. Chem. Phys. 1986;84:4243–4255. [Google Scholar]