Abstract
The FXYD membrane proteins constitute a family of conserved auxiliary subunits of the Na,K-ATPase, and have been the focus of recent attention due to their ability to finely regulate the activity of the enzyme complex in various physiological settings. In this review we describe the structures of the proteins, as well as their dynamics and their associations with the lipid bilayer membrane, which we have recently determined by NMR spectroscopy. Although the proteins are relatively small, their genes contain as many as six to nine small exons, and the coincidence of structured protein segments with their genetic elements suggests assembly from discrete structural modules through exon shuffling. The three-dimensional structures and backbone dynamics provide the foundation for understanding their intra-membrane association with the Na,K-ATPase α subunit, and the structure of FXYD1 suggests a mechanism whereby the phosphorylation of conserved Ser residues, by protein kinases A and C, could induce a conformational change in the cytoplasmic domain of the protein, to modulate its interaction with the α subunit.
Keywords: FXYD; Na,K-ATPase; Structure; Membrane protein; Micelles; Membrane; NMR
Amino acid sequence conservation
The FXYD family proteins are tissue-specific auxiliary subunits of the Na,K-ATPase, the principal enzyme responsible for maintaining the distribution of Na and K ion concentrations across animal cell membranes (Sweadner and Rael 2000; Cornelius and Mahmmoud 2003; Crambert and Geering 2003; Garty and Karlish 2006). Seven FXYD proteins have been identified in mammals. FXYD1 (PLM; phospholemman) is the principal substrate of hormone-stimulated phosphorylation by cAMP-dependent protein kinases A and C (PKA, PKC) in heart sarcolemma. FXYD2 (gamma), unique in the family for having two alternative splice variants (FXYD2a and FXYD2b), and FXYD4 (CHIF; channel-inducing factor; corticosteroid hormone induced factor), are each expressed in distinct, specialized segments of the kidney, with unique expression patterns that help explain the physiological differences in Na,K-ATPase activity among the nephron segments. Two family members, FXYD3 (Mat8; mammary tumor protein 8 kD) and FXYD5 (dyshaderin; RIC; resembles ion channel), are expressed in cancers and play a role in tumor progression. FXYD6 and FXYD7 are expressed exclusively in the brain, and are believed to play a role in neuron excitability during postnatal development and in the adult brain.
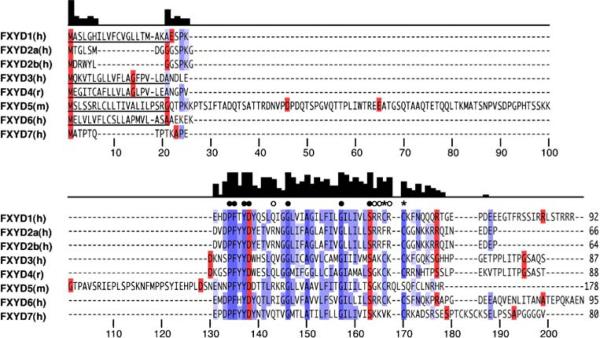
The FXYD proteins share a core homology of 35 invariant and conserved amino acids, in and around a single transmembrane segment (Fig. 1). The short signature motif PFXYD (Pro, Phe, X, Tyr, Asp), from which the family takes its name, is invariant in all seven known mammalian examples, and identical in those of other vertebrates except for Pro, and residue X is typically Tyr, but can also be substituted by Thr, Glu, or His. In all the proteins, conserved basic residues flank the transmembrane domain, the extracellular N-termini are acidic, and the cytoplasmic C-termini are basic, however, outside the signature homology motif there is little sequence conservation among the family members. The presence of two conserved Gly residues in the transmembrane helices of all FXYD proteins, suggests that it is involved in specific intramembrane helix–helix interactions.
Fig. 1.
Multiple sequence alignment of the seven mammalian FXYD family proteins. Signal peptides are underlined. The intensity of blue color reflects percent amino acid identity. Amino acids in red mark the starts of coding exons. The bar graph shows sequence conservation per residue. Filled circles mark invariant residues, open circles mark conserved Arg or Lys, and asterisks mark conserved Cys. FXYD2 has two alternative splice variants (FXYD2a, FXYD2b). Amino acid numbers below the alignment serve as markers but do not reflect the actual residue numbers in the sequences. The NCBI gene accession numbers are: NM_005031 (human FXYD1); NM_001680 (human FXYD2a); NM_021603 (human FXYD2b); NM_005971 (human FXYD3); NM_022388 (rat FXYD4); NM_008761 (mouse FXYD5); NM_022003 (human FXYD6); NM_022006 (human FXYD7)
Correlation of gene and protein structures
Although the FXYD proteins are relatively small, ranging from about 60 to 160 amino acids, they are all encoded by genes with six to nine small exons. Interestingly, their secondary structures, determined by NMR in lipid micelles, mirror the intron-exon structures of their corresponding genes, suggesting that the proteins are assembled from discrete structured domains, which may serve to confer different functional properties in various physiological settings (Franzin et al. 2005).
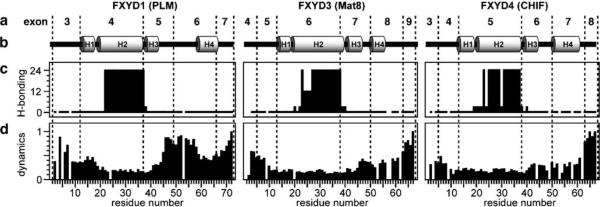
In lipid micelles, the FXYD proteins adopt unique helical structures, except for the long N-terminal extension of FXYD5 which is unstructured both in the presence and absence of lipids. The transmembrane helix is preceded by a short helical stretch of approximately five residues, beginning at the Asp residue of the FXYD signature sequence (Fig. 2). This helix-break-helix motif is observed in both lipid micelles and lipid bilayers (Franzin et al. 2007). It spans exactly the entire length of exon 4 in FXYD1, exon 6 in FXYD3, and exon 5 in FXYD4, and is followed by a third short helix, spanning the stop-transfer signal sequence of each protein, and encoded by the next exon (exon 5 in FXYD1; exon 7 in FXYD3; exon 6 in FXYD4). In all the three proteins, these three helices, plus the preceding PFXYD signature motif, constitute a common core module, which spans the entire 35-residue FXYD protein homology region, over three exons. Within the core modules, all residues have similar local backbone dynamics, and the three helices are rigidly connected, while additional backbone motions, are present in residues near the N- and C-termini, and at the boundaries of the core modules, marked by the junction between exons 5 and 6 in FXYD1, exons 7 and 8 in FXYD3, and exons 6 and 7 in FXYD4.
Fig. 2.
Correlation between gene and protein structure for FXYD1, FXYD3, and FXYD4. Residue numbering begins at 1 after the signal sequence. a Exon numbers are shown above the structures, and dashed lines mark the intron–exon junctions. b Secondary helical structures of the proteins. c The amide hydrogen exchange rates (classified as rapid <1 h, short <3 h, medium <12 h, long >24 h), highlight the strong hydrogen bonds in the transmembrane helices. d The profiles of backbone dynamics (classifed from static ~0, to mobile ~1) identify flexible linkers between helices H3 and H4, particularly prominent for FXYD1
FXYD1 has a fourth well-defined helix from residues 60 to 68, which contains the Ser63 and Ser68 consensus sites for phosphorylation by PKA and PKC, and is separated from the core module by a flexible linker. The position of the flexible linker coincides with the junction between exons 5 and 6, and the fourth helix ends at the end of exon 6. Both helices 3 and 4 of FXYD1 are tightly associated with the lipid micelle, in agreement with the NMR data for FXYD1 in lipid bilayers, which indicate the presence of a helical segment associated with the membrane surface (Franzin et al. 2005).
FXYD3 also appears to form a fourth helix at the start of exon 8, although it is very short and not as well defined by the NMR data. A short and somewhat more flexible stretch of amino acids is also evident at the junction between exons 7 and 8 connecting helices 3 and 4. FXYD4, on the other hand, has a very well defined fourth helix (Franzin et al. 2007), beginning at the start of exon 7 and spanning its entire length before reaching the mobile C-terminus, which coincides with exon 8.
The presence of conserved modules in different FXYD family members suggests that they are derived from a common ancestor gene, from which FXYD5 appears to have been the first to diverge (Garty and Karlish 2006). The coincidence of intron–exon junctions with helical structures and flexible connecting segments, further supports this hypothesis, and indicates that the proteins in this family may have been assembled from discrete structural modules through exon shuffling. The multiple exon organization of the FXYD genes could serve to confer high structural and functional diversity among the family members.
Structure and dynamics of FXYD1
The structure of the Na,K-ATPase complex has been determined at 8 Å resolution by cryo-electron microscopy analysis of two-dimensional crystalline native kidney membranes (Hebert et al. 2001; Purhonen et al. 2006). In this structure, the electron density map clearly identifies the arrangement of the ten transmembrane helices of the α subunit, as well as distinct regions assigned to the transmembrane domains of the β and FXYD subunits. The map indicates that the FXYD protein associates with the α subunit in a groove assigned to transmembrane helices M2, M4, M6, and M9.
In heart and skeletal muscle sarcolemma, FXYD1 is an integral part of the Na,K-ATPase enzyme complex, and its activity in different physiological settings is regulated by PKA and PKC phosphorylation. In addition, FXYD1, as well as at least three other FXYD family members, can induce ionic currents in Xenopus oocytes and in phospholipid bilayers, however, the direct formation of ion channels has not been demonstrated in vivo, and recent evidence indicates that the role of FXYD proteins in ion transport regulation is solely related to their association with the Na, K-ATPase and other ion transporters.
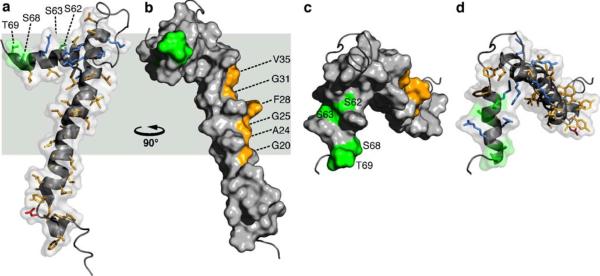
The three-dimensional NMR structure of FXYD1 is shown in Fig. 3 (Teriete et al. 2007). The FXYD motif, which may be required to stabilize the interaction with the α subunit, forms a disordered segment with some propensity for helical structure, preceding the short helix H1 (Fig. 3a). In the transmembrane helix of FXYD1, the Phe28 aromatic ring forms a significant protrusion out of a groove lined by small residues (Gly20, Ala24, Gly25, Gly31, Val35) along one side of the helix length (Fig. 3b). The transmembrane residues Gly20 and Gly31 are fully conserved in the sequences of all FXYD proteins across species (Fig. 1), and positions 24, 25, and 35 typically take small side-chains (Ala, Gly, Val or Leu). Position 28, on the other hand, displays more variability in the type and size of amino acid that it can accommodate, and is occupied by Phe in FXYD1, FXYD2 and FXYD5–7, or by the smaller residues Cys and Ala in FXYD3 and FXYD4 (human sequences). A recent study showed that a Cys residue placed at position 28 (FXYD1 numbering) in the transmembrane helices of FXYD1, FXYD2, and FXYD4, allows cross linking to Cys145 in transmembrane helix M2 of the α subunit (Garty and Karlish 2006). Interestingly, the proximity of Phe28 to this site also places it near Phe146 in M2, raising the possibility that aromatic ring stacking could stabilize the FXYD-α interaction in the membrane.
Fig. 3.
Molecular backbone and surface representations of FXYD1 (PDB accession code: 2JO1). In the helical regions, basic side-chains are shown in blue, acidic side-chains are red, and apolar side-chains are yellow. The three Ser residues (S62, S63, S68) and Thr69 in the cytoplasmic helix are in green. Residues in the transmembrane helix (G20, A24, G25, F28, G31, V35), predicted to interact with the Na,K-ATPase α subunit are shown in yellow. Side views. The structures are viewed (a, d) from the membrane side, or (c, d) down the membrane surface from the cytoplasm
The specific activities of the FXYD proteins are related, in part, to subtle differences in their transmembrane sequences, and the very high conservation of amino acids in the transmembrane helices of all FXYD proteins suggests their involvement in specific intramembrane helix–helix interactions with the Na,K-ATPase, and possibly other partners. For example, while FXYD2 (Phe at position 28) reduces the Na affinity of the Na,K-ATPase, FXYD4 (Ala at position 28) increases it, and their opposing activities and unique expression patterns in distinct segments of the kidney help define the physiological differences in Na,K-ATPase activity among the kidney nephron segments (Sweadner and Rael 2000; Crambert and Geering 2003; Garty and Karlish 2006). Furthermore, in kidney membranes the transmembrane helix of FXYD2 associates with the transmembrane domain of the α subunit, while the mutation G41R (corresponding to position 31 in FXYD1) causes misrouting of the protein by inhibiting its association with the Na, K-ATPase, and is linked with renal or intestinal familial hypomagnesemia (Meij et al. 2000). The data, therefore, indicate that the conserved Gly residue at this position plays a role in mediating helix–helix interactions between the FXYD transmembrane helix and the Na,K–ATPase. The structure of FXYD1 shows that Gly31 and the other conserved transmembrane residues line one strip along helix H2 that is unencumbered by the cytoplasmic helix H4, and free to interact with the α subunit.
Helix H4 associates with the membrane surface, and is buried in the membrane to a similar extent as residues 39 to 45 at the end of the transmembrane segment. The Arg and Lys side-chains from residues 38, 39, 41, 43 in H3, and residues 61, 65, 66 in H4, are sufficiently long to reach the membrane surface with their positively charged groups, thus anchoring these segments to the cytoplasmic lipid-water interface. The cytoplasmic Ser residues that are phosphorylated by PKA (S68) or PKC (S63, S68), point to opposite directions of the helix (Fig. 3c, d), and are exposed to the surface and available for interaction with the kinases. The other two potential phosphorylation sites, Ser62 and Thr69, are also surface exposed.
The cytoplasmic helix of FXYD1 is highly basic, which helps explain its propensity to associate with the membrane surface. However, it is connected to the rest of the protein by a relatively long flexible linker, and therefore would be capable of undergoing reorientation to interact with the cytoplasmic domain of the α subunit. Helix reorientation from the membrane surface could be triggered by phosphorylation, which introduces repulsive negative charges at Ser63 and Ser68. Indeed, the NMR data indicate that Ser68 phosphorylation by PKA increases the backbone motions in helix H4.
Interestingly, the binding of both PKC and its substrate polypeptides to acidic phospholipids is important for phosphorylation, and may serve both to co-localize the enzyme and the substrate, and to stabilize a specific substrate conformation that can be recognized by PKC (Vinton et al. 1998; Newton 2003). Thus, the membrane surface association of H4 could be important for FXYD1 substrate recognition by PKC, while the addition of negatively charged phosphate groups could trigger helix reorientation, thereby providing a mechanism for modulating the interaction of FXYD1 with the α subunit.
Concluding remarks
The structure of FXYD1 is the first to be determined for the FXYD family of Na,K-ATPase regulatory subunits, and highlights some key features that suggest its mode of association with the α subunit of the enzyme. A long groove formed by highly conserved amino acids runs parallel to the length of the transmembrane helix, delineating the likely binding interface of FXYD1 with the α subunit. The groove is interrupted by the aromatic ring of Phe28, which protrudes from the transmembrane helix near the middle of the hydrophobic lipid core. FXYD1 differs from FXYD2, FXYD3, and FXYD4 in the presence of a well defined cytoplasmic helix that is loosely connected to the rest of the protein by a long flexible linker. The emerging structures of FXYD2 and FXYD3 show considerably less structural order in this region, while the cytoplasmic helix of FXYD4 is more rigidly connected to the transmembrane domain. This may reflect the ability of FXYD1 to undergo helix reorientation as a means of modulating its interaction with the α subunit. Initial NMR experiments with FXYD1 phosphorylated at Ser68 by PKA indicate that phosphorylation increases the dynamics around helix H4, and it will be interesting to see whether this is accompanied by a conformational change and detachment from the lipid surface.
Acknowledgements
This research was supported by NIH grant R01-CA082864. The NMR studies utilized the NIH-supported Resource for NMR Molecular Imaging of Proteins (P41-EB002031) and the NIH-supported Burnham Institute NMR Facility (P30-CA030199).
References
- Cornelius F, Mahmmoud YA. News Physiol Sci. 2003;18:119–124. doi: 10.1152/nips.01434.2003. [DOI] [PubMed] [Google Scholar]
- Crambert G, Geering K. Sci STKE 2003. 2003:RE1. doi: 10.1126/stke.2003.166.re1. [DOI] [PubMed] [Google Scholar]
- Franzin CM, Marassi FM. NMR Structure determination of proteins in bilayer lipid membranes: the FXYD family proteins. In: Tien HT, Ottova-Leitmannova A, editors. Advances in planar lipid bilayers and liposomes. Elsevier; Amsterdam: 2005. pp. 77–93. [Google Scholar]
- Franzin CM, Yu J, Thai K, Choi J, Marassi FM. J Mol Biol. 2005;354:743–750. doi: 10.1016/j.jmb.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzin CM, Teriete P, Marassi FM. J Am Chem Soc. 2007;129:8078–8079. doi: 10.1021/ja0728371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garty H, Karlish SJ. Annu Rev Physiol. 2006;68:431–459. doi: 10.1146/annurev.physiol.68.040104.131852. [DOI] [PubMed] [Google Scholar]
- Hebert H, Purhonen P, Vorum H, Thomsen K, Maunsbach AB. J Mol Biol. 2001;314:479–494. doi: 10.1006/jmbi.2001.5137. [DOI] [PubMed] [Google Scholar]
- Meij IC, Koenderink JB, van Bokhoven H, Assink KF, Groenestege WT, de Pont JJ, Bindels RJ, Monnens LA, van den Heuvel LP, Knoers NV. Nat Genet. 2000;26:265–266. doi: 10.1038/81543. [DOI] [PubMed] [Google Scholar]
- Newton AC. Biochem J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purhonen P, Thomsen K, Maunsbach AB, Hebert H. J Membr Biol. 2006;214:139–146. doi: 10.1007/s00232-006-0056-8. [DOI] [PubMed] [Google Scholar]
- Sweadner KJ, Rael E. Genomics. 2000;68:41–56. doi: 10.1006/geno.2000.6274. [DOI] [PubMed] [Google Scholar]
- Teriete P, Franzin CM, Choi J, Marassi FM. Biochemistry. 2007;46:6774–6783. doi: 10.1021/bi700391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinton BB, Wertz SL, Jacob J, Steere J, Grisham CM, Cafiso DS, Sando JJ. Biochem J. 1998;330(Pt 3):1433–1442. doi: 10.1042/bj3301433. [DOI] [PMC free article] [PubMed] [Google Scholar]