Abstract
Catalytic beacon has emerged as a general platform for sensing metal ions and organic molecules. However, few reports have taken advantage of the true potential of catalytic beacons in signal amplification through multiple enzymatic turnovers, as existing designs require either equal concentrations of substrate and DNAzyme or an excess of DNAzyme in order to maintain efficient quenching, eliminating the excess of substrate necessary for multiple turnovers. Based on the large difference in the melting temperatures between the intramolecular molecular beacon stem and intermolecular products of identical sequences, we here report a general strategy of catalytic and molecular beacon (CAMB) that combines the advantages of the molecular beacon for highly efficient quenching with the catalytic beacon for amplified sensing through enzymatic turnovers. Such a CAMB design allows detection of metal ions such as Pb2+ with a high sensitivity (LOD=600 pM). Furthermore, aptamer sequence has been introduced into DNAzyme to use the modified CAMB for amplified sensing of adenosine with similar high sensitivity. These results together demonstrate that CAMB provides a general platform for amplified detection of a wide range of targets.
A current focus of research is the development of sensors for metal ions and small organic molecules,1–5 as they play either beneficial or deleterious roles in biology or in the environment. Unlike protein and nucleic acid detections, there are few methods that can be generally applied to sensing metal ions and small organic molecules. To fill this technology gap, catalytic DNA/RNA, aptamers and aptazymes, collectively called functional nucleic acids, have been developed into sensors. As a result, catalytic DNA (called DNAzymes in this work) that are highly specific for a number of metal ions such as Pb2+,6,7 Mg2+,8 Zn2+,9 Co2+, 10,11 UO22+,12 have been isolated using in vitro selection. This high metal ion specificity makes DNAzymes an attractive and general platform to sense metal ions. Therefore, we and others have converted DNAzymes into highly sensitive and selective fluorescent,7,12,13–19 colorimetric,15,20–25, electrochemical26,27 and electrochemiluminescent28 sensors. While most of these sensors have detection limits that are below the maximum contamination levels (MCL) in water as defined by the U. S. Environmental Protection Agency (EPA), an even higher sensitivity is desired if these sensors are to be used in complex cellular or environmental samples to ensure large signal to noise ratios.
One way to improve the sensitivity is through amplified detection. An excellent example is the use of protein enzymes such as horseradish peroxidase for amplified electrochemical and colorimetric detection.29–31 Recently, a DNAzyme with peroxidase activity has been used for amplified colorimetric sensing of a number of targets.32–36 In contrast to the many amplified electrochemical and colorimetric sensors, few amplified fluorescence sensors has been reported. The fluorescent catalytic beacon is quite attractive in this regard as such a sensing system could provide in situ and real-time information for a variety of applications.7,12,13–19 In these catalytic beacon designs (Figure 1b), the DNAzyme strand acts in two roles: catalyst and quencher. Almost all reported work so far has focused on improving quenching efficiency to lower the background. For both the first-7 and the second-generation catalytic beacons, 12,13,14 the main fluorescence quenching comes from an intermolecular hybridization of a fluorophore-labeled substrate strand with a quencher-labeled DNAzyme strand. By placing a quencher at the end of the DNAzyme strand and counting on hybridization with the substrate strand to ensure effective quenching, it also limits the amount of fluorophore-labeled substrate that can be used, as quenching will primarily take place when the enzyme substrate complex is formed. With such a design it is impossible to take advantage of DNAzymes as catalysts for amplified sensing through multiple turnover reactions as only one equivalent or less of substrate is available. Therefore, despite numerous reports of catalytic beacons, to our knowledge, few reports have realized the true potential of DNAzymes as multiple turnover enzymes for signal amplification, and a new catalytic beacon design to liberate the DNAzyme strand from the role of being a quencher strand, while still maintaining low fluorescence background, is required.
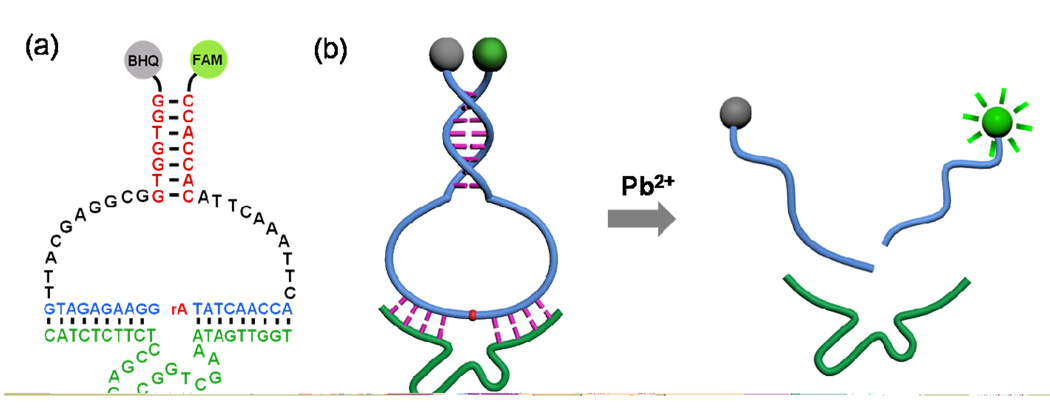
Figure 1.
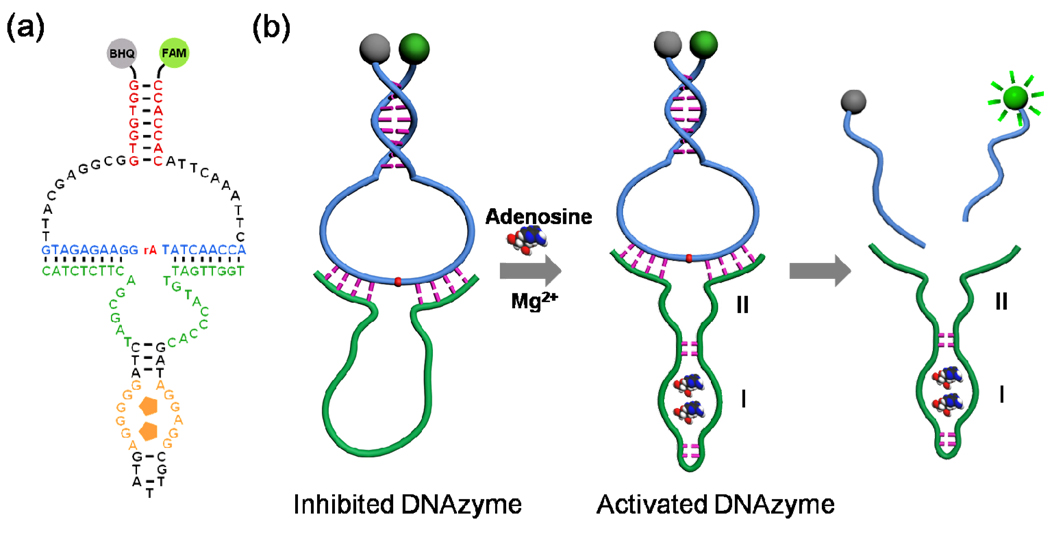
(a) Secondary structure of the CAMB on the 8–17 DNAzyme; (b) Schematic of the CAMB system for lead detection.
To realize the true potential of DNAzymes in the new catalytic beacon design, we turn our attention to molecular beacons (MBs) that have been widely used in DNA/RNA detections and some protein assays.37–39 MBs are single-stranded oligonucleotides with a stem-loop structure that brings the quencher and fluorophore into close proximity, thereby fluorescence is quenched effectively (see Scheme 1a). A unique feature of MBs is that the melting temperature of an intramolecular MB is considerably higher than that of the corresponding intermolecular MB formed from two separate DNA molecules with identical sequences.37 However, this feature is rarely used to design other fluorogenic probe systems.40 In this work, by taking advantage of this unique feature and the high quenching efficiency of the MBs, and by combining catalytic beacon’s multiple enzymatic turnover properties, we report a novel catalytic and molecular beacon (CAMB) (see Scheme 1c) for amplified sensing of Pb2+ and adenosine, with sensitivity superior to that of previously reported sensors. The use of the molecular beacon (MB) structured substrate provides several significant advantages over the linear substrate. First, the CAMB system has lower background fluorescence signal, improved signal to noise ratio and thus higher sensitivity of the sensors compared to linear substrate sensor system. Second, the efficient quenching also enables the use of excess substrates strands over the DNAzyme strand to achieve signal amplification by utilizing the multiple catalytic turnovers of DNAzyme. Third, the DNAzyme strand is liberated from the role of being a quencher, and can be readily adapted to sense a broad range of targets including metal ions, organic molecules, and DNA, as demonstrated in this manuscript. Given the above mentioned benefits the CAMB system has brought, this system can be used as a general platform for amplified detection of a number of targets with higher sensitivity than previously reported methods.
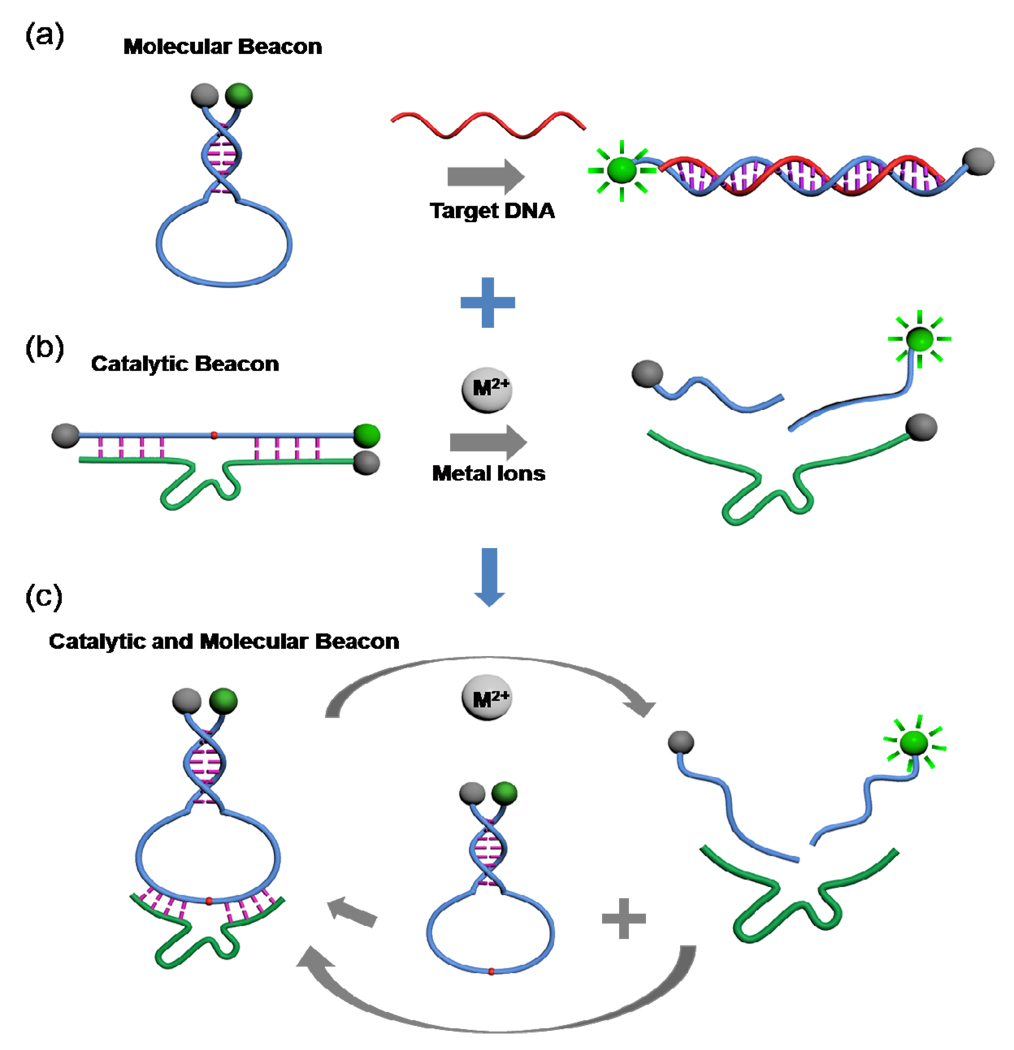
Scheme 1.
Design strategy of the catalytic and molecular beacon (CAMB). (a) Schematic of the molecular beacon (MB) design. (b) Schematic of the catalytic beacon design. (c) Schematic of the CAMB. The DNAzyme substrate strand is incorporated in the loop of the MB, which binds to the enzyme strand to form a complex structure. Addition of metal ions cleaves the substrate and cuts the molecular beacon into two pieces, resulting in stem dehybridization and enhanced fluorescence signal. In the presence of excess MB substrate, one DNAzyme can catalyze the cleavage of multiple MB substrates and achieve an amplified fluorescence signal.
The design of the new CAMB is shown in Scheme 1c. The substrate strand of the DNAzyme can be converted into a molecular beacon by extending the original DNAzyme substrate strand at both ends. Between the original DNAzyme substrate and the MB stem, a 10-base spacer is inserted at each end of the original substrate to ensure sufficient conformational flexibility in the loop for intermolecular hybridization with the DNAzyme without disrupting the stem structure (Figure 1a). Upon the addition of the target metal ions, the DNAzyme will catalyze the cleavage of the MB substrate, converting the stable intramolecularly hybridized MB stem into two much less stable intermolecularly hybridized DNA strands. It is known that the intramolecularly hybridized MB stem is more stable than the two intermolecularly hybridized DNA strands of the same sequence by up to 20 °C (Tm) or 7.8 kcal/mol (ΔG, calculated by the Mfold program, and DINAMelt program, respectively).41 Due to this stability difference, the single-piece MB substrate that is hybridized to the DNAzyme strand can be released from the DNAzyme strand as two product pieces, causing the quenched MB fluorophore/quencher pair to be separated from each other and thus a dramatic increase of the fluorescent signal in the presence of the target metal ion. The Stojanovic group reported a catalytic molecular beacon made by linking the 10–23 DNAzyme strand with a molecular beacon, so that the activity of the DNAzyme could be modulated by the target oligonucleotide complementary to the beacon stem loop sequence.42 However, this design showed high background fluorescence and slow DNAzyme cleavage rate due to incomplete hybridization between the substrate and the DNAzyme strands. The Mao group and Wang group have reported small molecular beacons as substrates for the DNAzyme,43,44 but the hairpin structures of their substrates were opened before the catalytic cleavage reaction happened, resulting in high fluorescence backgrounds. The Willner group has developed HRP-mimicking DNAzyme-containing hairpin structures for amplified colorimetric detection of target DNA,45 adenosine monophosphate or lysozyme.46
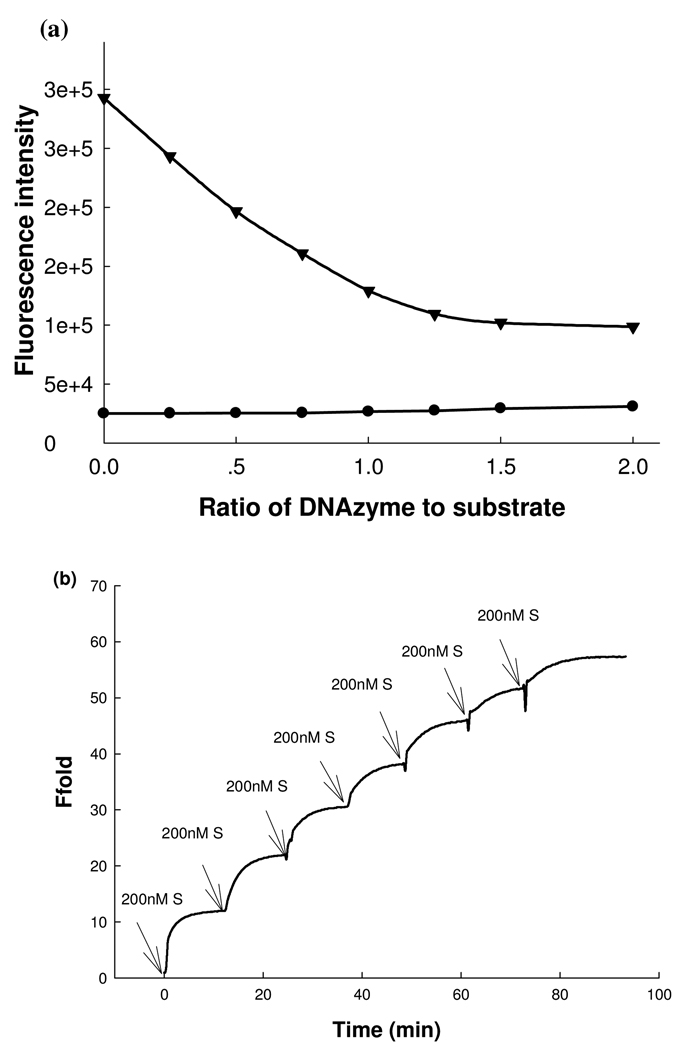
To demonstrate the use of CAMB, we chose the 8–17 DNAzyme (Figure 1a, 1b), as it has been employed to design a series of Pb2+ biosensors.7,13,15,20–22,26,27,28 In addition, its catalytic reaction mechanism has been well investigated.47,48 In the absence of DNAzyme strand, the background fluorescence of the original two-quencher catalytic beacon13 is very high (~ 2.9×105 a.u.. Figure 2a), suggesting lack of intramolecular quenching efficiency. Addition of increasing concentrations of the DNAzyme strand with a quencher resulted in more efficient quenching (Figure 2a), making it difficult to take advantage of the catalytic turnovers for signal amplification. In contrast, the background fluorescence of the new CAMB system is dramatically lower, at about 2.6×104 a.u. (Figure 2a). More importantly the background signal is not affected by the DNAzyme/substrate ratios, making it possible to use excess substrate for catalytic turnovers, generating higher fluorescent signal for the same concentration of metal ion.
Figure 2.
(a) Comparison of fluorescence backgrounds of the original catalytic beacon system (17DS-FD and 17E-Dy) and CAMB at varying ratios of DNAzyme to substrate (MB2): (filled triangles) original 8–17 DNAzyme catalytic beacon system; (filled circles) new designed catalytic and molecular beacon system. The substrate concentration was fixed at 200 nM. (b) Multiple-turnover catalytic activity of CAMB system by using the 8–17 DNAzyme and MB substrate (MB2). The kinetics of fluorescence enhancement were recorded in a 200 nM of 8–17 DNAzyme solution in the presence of 10 µM Pb2+ as a cofactor. The buffer contained 25 mM HEPES (pH 7.0) and 50 mM NaCl.
For the CAMB system in Figure 1b to work well, a three-step process has to take place to generate a fluorescence signal increase: cleavage of the substrate, release of the cleaved substrate strand from the enzyme strand and dissociation of the cleaved MB stem for signal generation. The large difference in melting temperature between the intramolecular MBs hybrid and identical hybrid formed from separate molecules after cleaving by the DNAzyme forms the basis for our proposed fluorescence sensing system. An ideal substrate molecular beacon should not only provide a low fluorescence background by forming a stable stem duplex, but also enable the prompt dissociation of the stem duplex upon substrate cleavage. Therefore, the appropriate stem stability is the key factor in optimal sensor design. To optimize the sensing performance, we first investigated the effect of stem length on the sensing results. When a substrate called MB1 containing an 8 base pair stem (6 G-C base pairs and 2 A–T base pair, see Table S1 in Supporting Information) was used to test fluorescence enhancement after the cleavage reaction catalyzed by the DNAzyme in the presence of 10 µM Pb2+, the fluorescence intensity increase slowly and reached about 5 fold enhancement after 20 min (see Figure S1 and Figure S2 in Supporting Information). As the DNAzyme catalyzed cleavage reaction is highly efficient and normally completes in less than 5 min 12 at Pb2+ concentration higher than 2 µM, we reason that the dissociation of the stem of the cleaved molecular beacon (i.e., step 3) might be the rate limiting step. To test this hypothesis, we removed a G-C base pair from the MB1 and modified the stem sequence (designated as MB2) to lower the stem stability and facilitate stem dissociation upon substrate cleavage. As shown in Figure S1 and Figure S2 (see Supporting Information), MB2 showed a much improved fluorescence increase rate, reaching saturation in 5 min, and 13 fold fluorescence enhancement. Control experiments for the time-dependent fluorescence change of MB1 and MB2 in the absence of DNAzyme were also carried out and no obvious fluorescence change was observed, indicating little background variation for our MB substrates (see Figure S1 in Supporting Information). Molecular beacon substrate with an even shorter stem length was not tested as it has a calculated melting temperature less than 38.9 °C in 50 mM salt concentration buffer, which is not enough to form stable hybridization and thus allow efficient quenching. Therefore, we chose MB2 as our optimized substrate molecular beacon design and used it for further investigation.
To investigate the multiple turnover capability of the 8–17 DNAzyme towards MB2 and thus signal amplification, we kept the 8–17 DNAzyme concentration at 200 nM and then monitored the kinetics of the fluorescence enhancement of the DNAzyme solution after gradual addition of MB2 in 200 nM increments. As shown in Figure 2b, the fluorescence signal increased notably with each incremental addition of MB2 substrate until the ratio of MB2 to DNAzyme reached 5. This result indicates that the active DNAzyme molecule could be regenerated after catalyzing the cleavage reaction so that one DNAzyme molecule could cleave several MBs substrate molecules. To provide further support for multiple-turnover capability in the DNAzyme system, gel electrophoresis was used to measure the cleavage activity of 1 µM DNAzyme over 1, 3 and 6 µM of MB2 substrate in the presence of 20 µM Pb2+. As shown in Figure S3 (see Supporting Information), most of the MB substrates were cleaved in all the three lanes. These results indicated that the 8–17 DNAzyme performed multiple-turnover reactions towards the MB based substrate as designed.
Having demonstrated the superior performance of CAMB over a catalytic beacon alone, we then explored the application of the system toward Pb2+ detection. To achieve the best sensing performance, the arm lengths of the DNAzyme strand was first optimized. An increase of the arm length of the DNAzyme from 7 bases on each site of the cleavage site to 8 and then to 9, resulted in faster increase of fluorescence signal (see Figure S4), suggesting that the longer arm length could facilitate the hybridization of the substrate with the DNAzyme and enhance the enzyme catalyzed cleavage reaction. However, if the arm length is too long, it provides overly strong hybridization between the DNAzyme and cleaved product, resulting in slow product releasing kinetics and affecting sensing performance. The optimal length was determined to be 9 bases on each side of the cleavage site (17E-2(9+9)). In addition, fluorescence kinetic rates increased when the buffer pH was increased from 6.0 to 8.0, while the end-point fluorescence enhancement ratio increased when buffer pH was increased from 6.0 to 7.0 and then decreased when pH is 7.5 or 8.0. Since it is known that fluorescence intensity of FAM increases significantly with increasing pH, resulting in a higher fluorescence background and thus decreased relative ratio of fluorescence enhancement, the decrease of end-point fluorescence enhancement ratio at 7.5 and 8.0 might be due to the high pH dependence of the FAM fluorophore (see Figure S5). Finally, sensing performance increased when the salt concentration was increased from 50 mM to 100 mM, but then decreased when the salt concentration increased to 150 mM (see Figure S6). Based on these results, a DNAzyme with 9 bases on each arm (17E-2(9+9)) in a pH 7.0 buffer solution containing 100 mM NaCl was chosen for further Pb2+ sensing experiments.
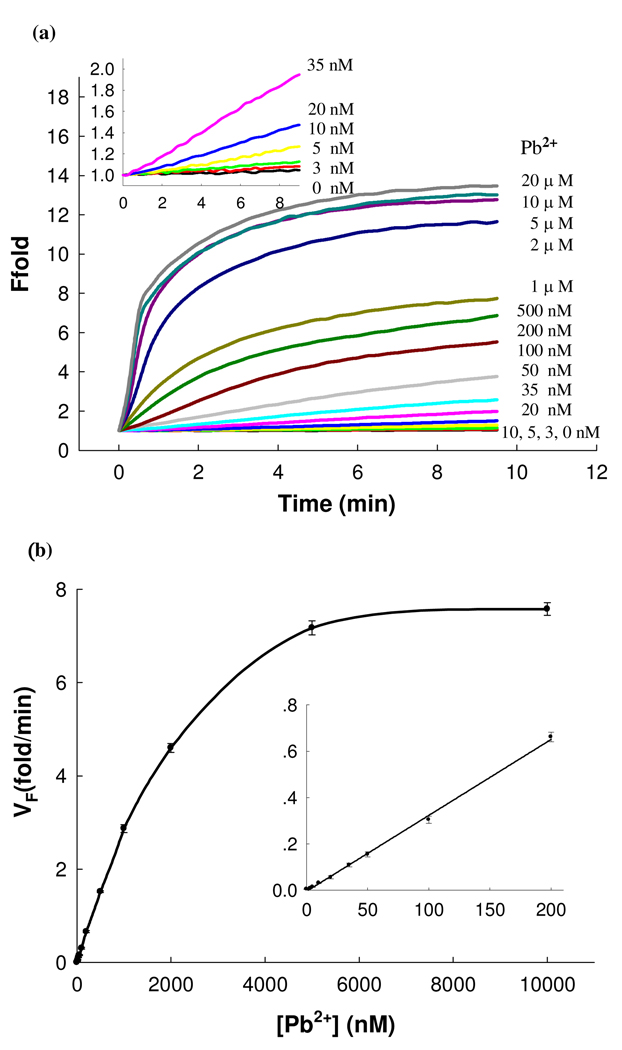
Under such conditions, a 13-fold fluorescence enhancement was observed when the Pb2+ concentration reached 5 µM (Figure 3a), which is much higher than the 4-fold enhancement of the first generation catalytic beacon with a single quencher design 7 or the 6-fold increase of the second generation catalytic beacon with a dual quencher design.13 The large fluorescence enhancement together with the multiple turnover capability of the 8–17 DNAzyme of the new CAMB system could improve the sensitivity of the catalytic beacon. Figure 3b shows the calibration curve of the MB DNAzyme sensor and it has a dynamic range from 3 nM to 5 µM and a linear range until 200 nM. This new system was very sensitive to Pb2+, with a detection limit of 600 pM determined by 3σb/slope (σb, standard deviation of the blank samples),, which is at least 10 times lower than the reported DNAzyme based lead sensors.7 As the maximum contamination level of Pb2+ in drinking water was defined by the U.S. Environmental Protection Agency (EPA) to be 15 ppb or 72 nM, this sensor is fully applicable for lead detection in drinking water.
Figure 3.
Sensitivity of the CAMB sensing system for Pb2+ detection. (a) Time-dependent fluorescence response over background fluorescence with varying concentrations of Pb2+. The concentration of DNAzyme and substrate was 200 nM, and 300 nM, respectively. The buffer contained 25 mM HEPES (pH 7.0) and 100 mM NaCl. Inset; Responses at low Pb2+ concentrations. (b) Calibration curve of the CAMB Pb2+ sensor. The curve was plotted with the initial rate of fluorescence enhancement vs. Pb2+ concentration. Inset shows the linear responses at low Pb2+ concentrations.
This ultrahigh sensitivity exemplifies the significant improvement over both catalytic beacon and molecular beacon systems reported previously. In comparison with molecular beacon, the CAMB retains the catalytic beacon’s ability to sense a much broader range of targets beyond nucleic acids and to use catalytic turnover for signal amplification. In comparison with catalytic beacons, the stable MB substrate allows much more efficient intramolecular quenching of the fluorophore in the absence of the target, making it possible to lower the background fluorescence signals, improve signal to noise ratio, and thus sensitivity of the sensor. The efficient quenching also allows the use of much more excess substrate strand over the DNAzyme strand, taking full advantage of the catalytic turnover for signal amplification, like protein enzymes.
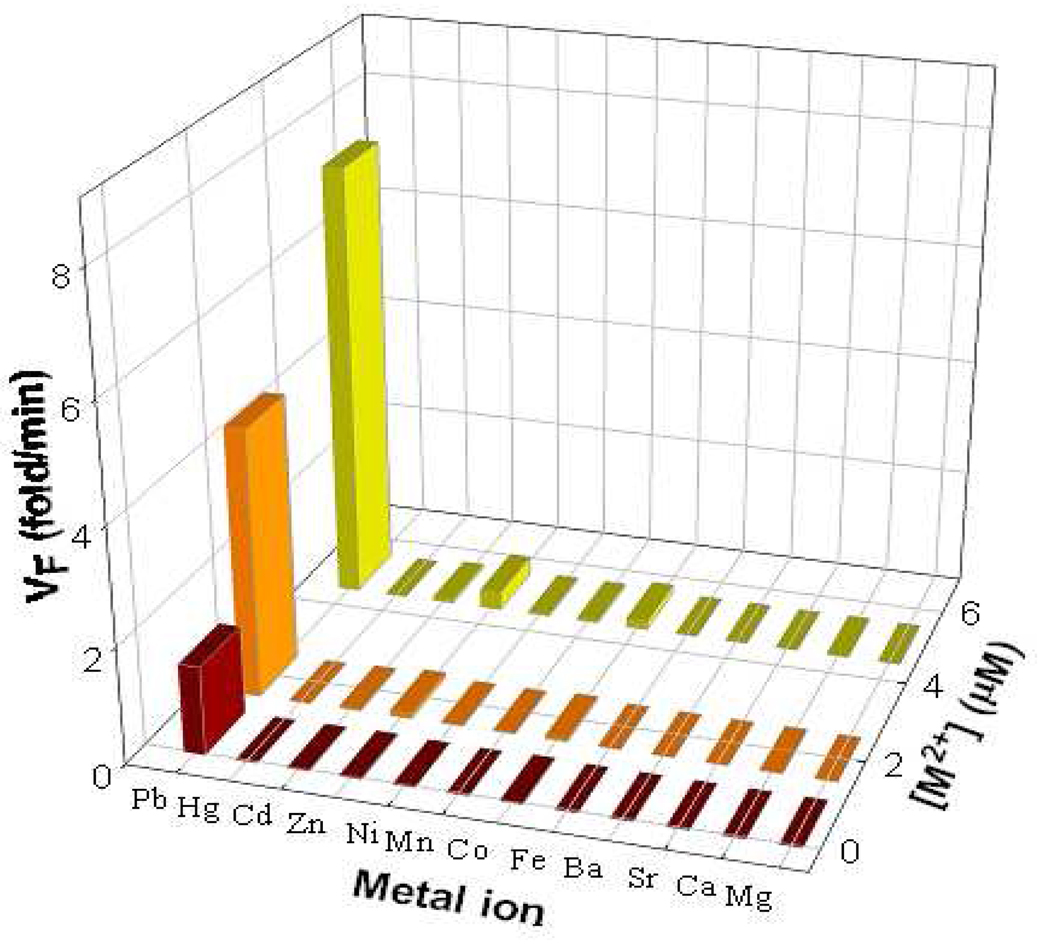
Besides sensitivity, selectivity is another important issue to assess the performance of a sensor. The original 8–17 DNAzyme-based catalytic beacon shows a good selectivity towards Pb2+ over other divalent metal ions. To investigate the effect of the MB structured substrate on the specificity of the new sensing system, its selectivity for Pb2+ over other divalent metal ions was tested by recording the sensor response to 11 competing metal ions at concentrations of 0.5, 2 and 5 µM. As shown in Figure 4, the new CAMB sensor shows good selectivity, which is consistent with the selectivity of the original 8–17 DNAzyme sensor.
Figure 4.
Selectivity of CAMB sensor for Pb2+ over other competing divalent metal ions. The initial rate of fluorescence enhancement of the sensing system induced by different metal ions at three concentrations (0.5, 2 and 5 µM) are shown. The concentration for DNAzyme and substrate was 200 nM and 300 nM, respectively. The buffer solution contained 25 mM HEPES (pH 7.0) and 100 mM NaCl.
Since our CAMB-based design can liberate the DNAzyme strand from serving the quencher role and allow true multiple turnovers, this new system could also be applied as an amplified fluorescence sensor for many other targets. However, DNAzymes are mostly known to recognize metal ions specifically. To adapt the CAMB for amplified sensing of molecules beyond metal ions, aptamers that have been shown to recognize a broad range of targets including small molecules, metal ions, proteins, virus and even cells, must be incorporated into the CAMB system.
Figure 5a and Figure 5b depict the inhibiting strategy using an allosteric DNAzyme10 for amplified fluorescent sensing of adenosine. We chose the Mg2+-dependent 10–23 DNAzyme because the sequence of the hairpin structure in its catalytic core is unimportant for its catalytic activity, and therefore can be modified without losing DNAzyme activity. Domain I corresponds to the nucleic acid sequence of the anti-adenosine aptamer,49 and domain II is the modified Mg2+-dependent DNAzyme. The sequence of the whole DNA strand was designed to avoid formation of the active secondary structure in the catalytic cores (sequences shown in Figure 5a).41 In the absence of target adenosine, the DNAzyme domain alone could not form a stable and active structure to catalyze the cleavage of the hairpin structured substrate even in the presence of Mg2+. Upon addition of adenosine, however, the aptamer region could bind to adenosine and form a compact structure, which will activate the DNAzyme by restoring the stem-loop structure of the DNAzyme. The activated DNAzyme can then catalyze the hydrolysis reaction of the MB structured substrate in the presence of Mg2+ as a cofactor, and thereby release the fluorophore-labeled DNA strand and generate a fluorescence signal enhancement.
Figure 5.
a) Secondary structure of the activated CAMB sensing system for adenosine. (b) Schematic of a fluorescence sensing CAMB system for adenosine by using CAMB as a signal amplifying label through an inhibiting strategy.
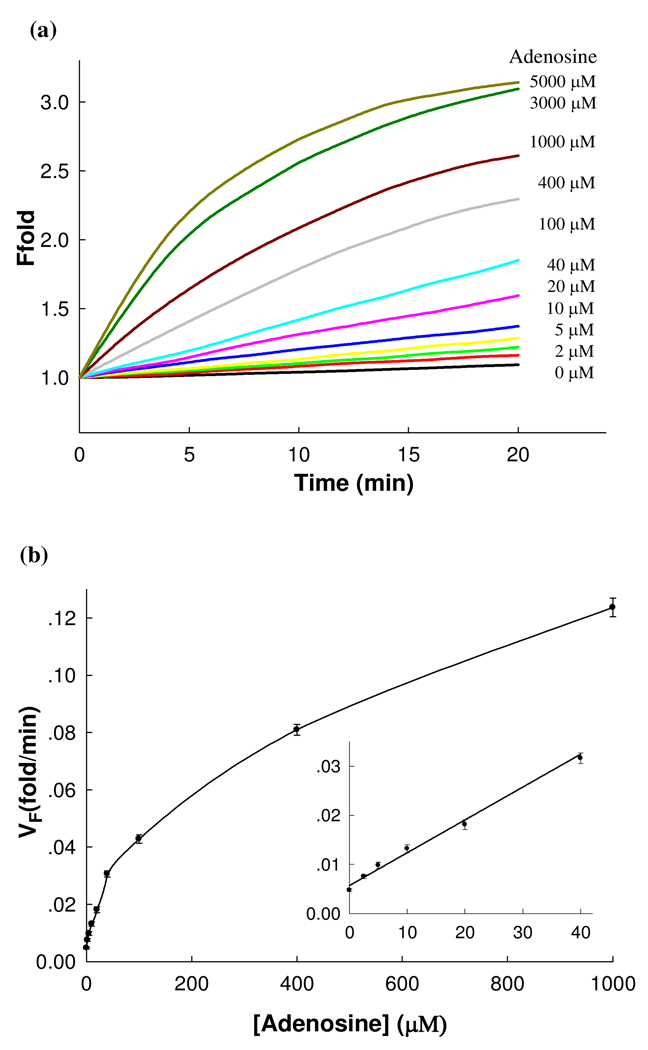
As shown in Figure 6a, in the absence of adenosine, little fluorescence enhancement was observed, indicating that the DNAzyme is inactive. In the presence of adenosine, however, a clear fluorescence increase was observed in 20 min, suggesting that the DNAzyme was activated upon the binding of adenosine. Our design affords a low background signal by effectively inhibiting the activity of the Mg2+-dependent DNAzyme in the absence of adenosine and therefore, offers a high sensitivity for the detection of adenosine. The low background of our new CAMB design at room temperature is due to the MB-based design with the substrate separated from the DNAzyme. In addition, in our design, several stable secondary structures could form (predicted by the Mfold program, see Figure S7 in Supporting Information), which could inhibit the formation of the active DNAzyme. As shown in Figure 6a, the rate of fluorescence enhancement increased with increasing adenosine concentration and reached a plateau at ~3 mM adenosine. Figure 6b depicts the calibration curve of the sensor and a linear relationship was observed until the adenosine concentration reached 40 µM (Figure 6b Inset). A low detection limit of 500 nM (3δ/slope) was obtained, which is lower than those of reported fluorescent adenosine sensors, 10 and amplified sensing methods based on a DNAzyme with peroxidase activity.34 To demonstrate the selectivity of the sensor toward adenosine, an adenosine analog, guanosine was added to the system instead. Results show that addition of 500 µM guanosine yields only minimal fluorescence changes, indicating that the sensor has good selectivity. All these experimental results indicate that the amplifying effect of the proposed fluorescence sensing system on the recognition of adenosine is realized.
Figure 6.
Sensitivity of the fluorescent CAMB system for adenosine detection. (a) Time-dependent fluorescence enhancement of the sensing system in the presence of different concentrations of adenosine with 10 mM Mg2+ as a cofactor. The concentration of substrate and the aptamer-appended DNAzyme was 300 nM and 200 nM, respectively, and the buffer solution contained 25 mM HEPES (pH 7.2) and 100 mM NaCl. (b) Calibration curve of the fluorescent CAMB sensor for adenosine. Inset shows the linear responses of sensing system to adenosine at low concentration.
In summary, we have developed a novel CAMB system by integrating a molecular beacon with a DNAzyme catalytic beacon, resulting in ultrahigh sensitivity. This performance exemplifies the significant improvement over both catalytic beacon and molecular beacon systems reported previously. In comparison with molecular beacon, the CAMB retains the catalytic beacon’s ability to sense a much broader range of targets beyond nucleic acids and to use catalytic turnover for signal amplification. In comparison with the previously reported catalytic beacon systems, this new catalytic molecular beacon system has several unique features and advantages. First, by taking advantage of the high intramolecular quenching efficiency of the molecular beacons, the MB based substrate provided a low background fluorescence, little background variation, and thereby, a large signal-to-background ratio. In addition, the ratio of the DNAzyme strand and the substrate strand is not limited to 1:1, and the signal could be amplified by cycling and regeneration of the DNAzyme, thus realizing the true potential of catalytic beacons for the first time. This large signal-to-background ratio, combined with the signal amplification effect, significantly improves the sensitivity of current DNAzyme-based sensors, resulting in a CAMB sensor for Pb2+ with a detect limit of 600 pM, much lower than those of other catalytic beacon Pb2+ sensors. By introducing aptamer into DNAzyme through an inhibition strategy, the CAMB system has been extended to sensing adenosine. Since numerous DNAzymes and aptamers have been selected to bind a wide range of targets, the CAMB system provides a new platform for sensitive detection of various targets including metal ions, small molecules and other targets, and could find wide applications in environmental and biomedical fields.
Supplementary Material
ACKNOWLEDGEMENT
We wish to thank Eric Null for proof-reading this manuscript. This work has been supported by the National Natural Science Foundation of China (Grant J0830415 and 20975034), U. S. Department of Energy (DE-FG02-08ER64568), the National Institutes of Health (ES016865), and the National Science Foundation (CTS-0120978 and DMI-0328162).
Footnotes
SUPPORTING INFORMATION AVAILABLE.
Experimental details and supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference
- 1.Nolan EM, Lippard SJ. Chem. Rev. 2008;108:3443–3480. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]
- 2.Chen PR, He C. Curr. Opin. Chem. Biol. 2008;12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Xue X, Wang F, Liu X. J. Am. Chem. Soc. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 4.Qian F, Zhang C, Zhang Y, He W, Gao X, Hu P, Guo Z. J. Am. Chem. Soc. 2009;131:1460–1468. doi: 10.1021/ja806489y. [DOI] [PubMed] [Google Scholar]
- 5.McRae R, Bagchi P, Sumalekshmy S, Fahrni CJ. Chem. Rev. 2009;109:4780–4827. doi: 10.1021/cr900223a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Lu Y. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]
- 8.Breaker RR, Joyce GF. Chem. Biol. 1995;2:655–660. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 9.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF. J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 10.Mei SHJ, Liu Z, Brennan JD, Li Y. J. Am. Chem. Soc. 2003;125:412–420. doi: 10.1021/ja0281232. [DOI] [PubMed] [Google Scholar]
- 11.Bruesehoff PJ, Li J, Augustine AJ, Lu Y. Comb. Chem. Highthroughput Screening. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Lu Y. Anal. Chem. 2003;75:6666–6672. doi: 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Lu Y. J. Am. Chem. Soc. 2007;129:9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Y, Tong A, Lu Y. J. Am. Chem. Soc. 2009 DOI: 10.1021/ja905854a. [Google Scholar]
- 17.Liu Z, Mei SHJ, Brennan JD, Li Y. J. Am. Chem. Soc. 2003;125:7539–7545. doi: 10.1021/ja035208+. [DOI] [PubMed] [Google Scholar]
- 18.Chiuman W, Li Y. Nucleic Acids Res. 2007;35:401–405. doi: 10.1093/nar/gkl1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Kim Y, Liu H, Zhu Z, Bamrungsap S, Tan W. J. Am. Chem. Soc. 2009;131:8221–8226. doi: 10.1021/ja901132y. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Lu Y. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Lu Y. J. Am. Chem. Soc. 2004;126:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Lu Y. J. Am. Chem. Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, Wang Z, Liu J, Lu Y. J. Am. Chem. Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ZD, Lee JH, Lu Y. Adv Mater. 2008;20:3263–3267. [Google Scholar]
- 25.Moshe M, Elbaz J, Willner I. Nano. Lett. 2009;9:1196–1200. doi: 10.1021/nl803887y. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Rowe AA, Plaxco KW. J. Am. Chem. Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 27.Shen L, Chen Z, Li Y, He S, Xie S, Xu X, Liang Z, Meng X, Li Q, Zhu Z, Li M, Le X, Shao Y. Anal. Chem. 2008;80:6323–6328. doi: 10.1021/ac800601y. [DOI] [PubMed] [Google Scholar]
- 28.Zhu X, Lin Z, Chen L, Qiu B, Chen G. Chem. Commun. 2009:6050–6052. doi: 10.1039/b911191c. [DOI] [PubMed] [Google Scholar]
- 29.de Lumley-Woodyear T, Caruana DJ, Campbell CN, Heller A. Anal. Chem. 1999;71:394–398. doi: 10.1021/ac980770h. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Wan Y, Gau V, Zhang J, Wang L, Song S, Fan C. J. Am. Chem. Soc. 2008;130:6820–6825. doi: 10.1021/ja800554t. [DOI] [PubMed] [Google Scholar]
- 31.Rodenko B, Toebes M, Celie PHN, Perrakis A, Schumacher TNM, Ovaa H. J. Am. Chem. Soc. 2009;131:12305–12313. doi: 10.1021/ja9037565. [DOI] [PubMed] [Google Scholar]
- 32.Li T, Dong S, Wang E. Chem. Commun. 2007:4209–4211. doi: 10.1039/b712165b. [DOI] [PubMed] [Google Scholar]
- 33.Kolpashchikov DM. J. Am. Chem. Soc. 2008;130:2934–2935. doi: 10.1021/ja711192e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbaz J, Moshe M, Shlyahovsky B, Willner I. Chem. Eur. J. 2009;15:3411–3418. doi: 10.1002/chem.200802004. [DOI] [PubMed] [Google Scholar]
- 35.Deng M, Zhang D, Zhou Y, Zhou X. J. Am. Chem. Soc. 2008;130:13095–13102. doi: 10.1021/ja803507d. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama S, Sintim HO. J. Am. Chem. Soc. 2009;131:10320–10333. doi: 10.1021/ja902951b. [DOI] [PubMed] [Google Scholar]
- 37.Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 38.Venkatesan N, Seo YJ, Kim BH. Chem. Soc. Rev. 2008;37:648–663. doi: 10.1039/b705468h. [DOI] [PubMed] [Google Scholar]
- 39.Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, Colon P, Lin H, Tan W. Angew. Chem. Int. Ed. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo X, Xia F, Xiao Y, Plaxco KW. J. Am. Chem. Soc. 2010;132:1816–1818. doi: 10.1021/ja909551b. [DOI] [PubMed] [Google Scholar]
- 41.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stojanovic MN, de Prada P, Landry DW. Chembiochem. 2001;2:411–415. doi: 10.1002/1439-7633(20010601)2:6<411::AID-CBIC411>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 43.Tian Y, Mao C. Talanta. 2005;67:532–537. doi: 10.1016/j.talanta.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 44.Meng X, Wang K, Tan W, Jun Li, Tang Z, Guo Q, Huang S, Li D. Chinese Sci. Bull. 2003;48:2581–2584. [Google Scholar]
- 45.Xiao Y, Pavlov V, Niazov T, Dishon A, Kotler M. J. Am. Chem. Soc. 2004;126:7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- 46.Teller C, Shimron S, Willner I. Anal. Chem. 2009;81:9114–9119. doi: 10.1021/ac901773b. [DOI] [PubMed] [Google Scholar]
- 47.Kim HK, Rasnik I, Liu J, Ha T, Lu Y. Nature Chem. Biol. 2007;3:763–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HK, Li J, Nagraj N, Lu Y. Chem. Eur. J. 2008;14:8696–8703. doi: 10.1002/chem.200701789. [DOI] [PubMed] [Google Scholar]
- 49.Huizenga DE, Szostak JW. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.