Summary
Helicobacter pylori persists deep in the human gastric mucus layer in a harsh, nutrient-poor environment. Survival under these conditions depends on the ability of this human pathogen to invoke starvation/stress responses when needed. Unlike many bacteria, H. pylori lacks starvation/stress-responding alternative sigma factors, suggesting an additional mechanism might have evolved in this bacterium. H. pylori produces polyphosphate, however, the role and target of polyphosphate during starvation/stress have not been identified. Here we show that polyphosphate accumulated during nutrient starvation directly targets transcriptional machinery by binding to the principal sigma factor in H. pylori, uncovering a novel mechanism in microbial stress response. A positively-charged-Lys-rich region at the NTD of the major sigma factor is identified as the binding region for polyphosphate (region P) in vivo and in vitro, revealing a new element in sigma 70 family proteins. This interaction is biologically significant because mutant strains defective in the interaction undergo premature cell death during starvation. We suggested that polyphosphate is a second messenger employed by H. pylori to mediate gene expression during starvation/stress. The putative “region P” is present in sigma factors of other human pathogens, suggesting that the uncovered interaction might be a general strategy employed by other pathogens.
Keywords: Helicobacter pylori, Polyphosphate, Sigma factor, Stress responses, Bacterial gene regulation
Introduction
About one half of the world population is infected with Helicobacter pylori, the causative agent of human gastritis and ulcers; and the infection is associated with gastric cancer (Marshall & Warren, 1984, Peek & Blaser, 2002, Fox & Wang, 2007). H. pylori colonizes the human gastric mucosa, where it encounters a constantly changing environment with poor nutrients and challenges from the host innate immune response (Schreiber et al., 2004, Monack et al., 2004, Allen, 2007). Thus, in order for H. pylori to persist in the host, it is critical for the bacterium to invoke a stress response when needed.
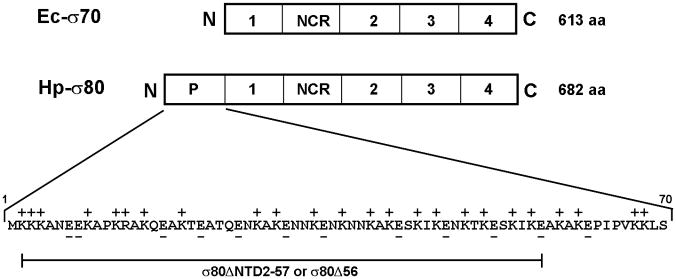
In eubacteria, typically, most transcription under regular growth conditions is initiated by RNA polymerase holoenzyme carrying a principal or housekeeping σ factor encoded by the rpoD gene. Multiple alternative σ factors control specialized regulons that are activated during different stress conditions (Gruber & Gross, 2003, Hengge, 2009, Paget & Helmann, 2003). Due to its small genome size, the H. pylori transcription machinery is simpler (Berg et al., 1997) compared to that of other bacteria such as E. coli. For example, H. pylori has only three σ factors, the primary sigma factor σ80 and two alternative σ factors, σ28 and σ54. The fact that H. pylori lacks the three starvation and stress-responding alternative sigma factors σS, σH and σE suggests that an additional stress-response mechanism might have evolved in this pathogen. Interestingly, H. pyloriσ80 (Beier et al., 1998), a member of the σ70 family proteins, has ~70 extra amino acid residues before conserved “region 1” at the N-terminal domain (NTD) compared to E. coliσ70. The role of this extra region at the NTD of σ80 is unknown.
Two small alarmone molecules, pppGpp and ppGpp [(p)ppGpp], which are associated with the stringent response (Cashel et al., 1996), are required for H. pylori survival in the stationary phase, during exposure to low pH, aerobic shock and macrophages (Mouery et al., 2006, Wells & Gaynor, 2006, Zhou et al., 2008). In H. pylori there is no RelA, and the SpoT protein is the only stringent response factor responsible for the synthesis and degradation of (p)ppGpp. Recently, we found that SpoT mediates a serum starvation response, which restricts cellular metabolic activity and limits cell growth while maintaining cell vitality for a long duration in vitro (Zhou et al., 2008). In contrast to wild type, the spoT mutant exhibits the “relaxed” growth phenotype, which confers the ability to grow to a higher density than wild type leading to premature cell death during serum starvation. Thus, H. pylori SpoT or (p)ppGpp regulates cell growth in response to nutrient starvation and stress.
Inorganic polyphosphate (polyP), a polyanion “molecular fossil” present in all living cells, is a regulator of responses to microbial stresses including nutrient deprivation (Kornberg et al., 1999, Rao et al., 2009). For example, polyP promotes ribosomal protein degradation by the Lon protease to supply amino acids needed to respond to starvation in E. coli (Kuroda et al., 2001). PolyP accumulation is regulated by (p)ppGpp, which inhibits the activity of exopolyphosphatase (PPX) in E. coli (Kuroda et al., 1997). H. pylori is able to produce polyP; but its role in the starvation/stress response and its regulation is unknown. Here we show that polyP forms stable complexes with σ80 during serum starvation in wild type H. pylori but not in the spoT mutant, and that the binding site for polyP lies in the NTD of σ80. Disruption of polyP synthesis or deletion of σ80 NTD similarly eliminates response to starvation in H. pylori leading to premature cell death. These findings reveal a novel strategy for H. pylori to respond to starvation through direct targeting of transcriptional machinery by ubiquitous polyP molecules.
Results and Discussion
σ80 is modified during serum starvation response
We recently showed that the sole stringent response factor SpoT in H. pylori mediates a serum-starvation response to protect cells from premature death (Zhou et al., 2008). In this study, we searched for differences between wild type and the spoT mutant cells in the levels of transcriptional factors, such as σ80, in response to serum starvation. H. pylori wild type strain G27 and its isogenic ΔspoT mutant were grown in a serum-free medium, and cultures were sampled at different time points. After cells were lysed by SDS, boiled cell lysates were analyzed by SDS-PAGE, followed by Western Blot with the σ80-specific polyclonal antibodies. Intriguingly, the mobility of σ80 was notably different between wild type and the spoT mutant cells, whereas the intracellular levels of σ80 appeared to be not significantly changed during the growth of both strains (Fig. 1A). The mobility of σ80 from wild type cells was significantly retarded or “up-shifted” as smeared bands from one to three days growth but became normal after day four. Under this physiological condition, wild type cells were restricted in growth and their cellular metabolic activity was limited, but they maintained viability after culturing for four days, while the ΔspoT mutant cells had a brief burst of growth for the first two days followed by rapid cell death afterward (Zhou et al., 2008). Although most of the ΔspoT mutant cells were non-cultivatable at the later time points, comparable amount of σ80 was detected during the course of the experiment, suggesting that those non-vital cells were still intact. The σ80 up-shift required the SpoT function, as σ80 was un-shifted in the ΔspoT mutant under the same growth conditions. The σ80 up-shift was in response to serum starvation, as σ80 mobility was unchanged when wild type cells were grown in a serum-supplemented medium (Fig. 1B). Thus, the σ80 up-shift is a manifestation of the SpoT-mediated serum starvation response.
Figure 1. The electrophoretic mobility of σ80 is greatly reduced during a SpoT-mediated serum starvation response.
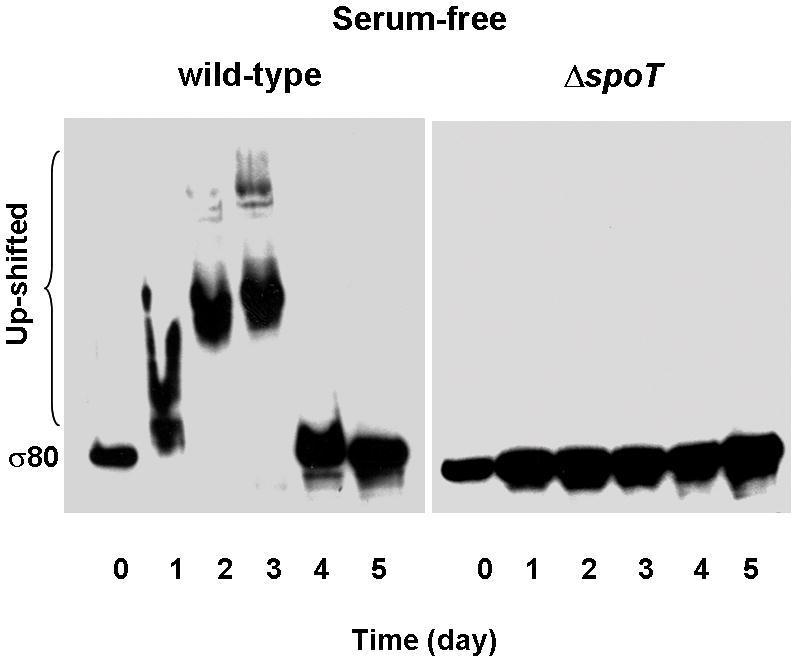
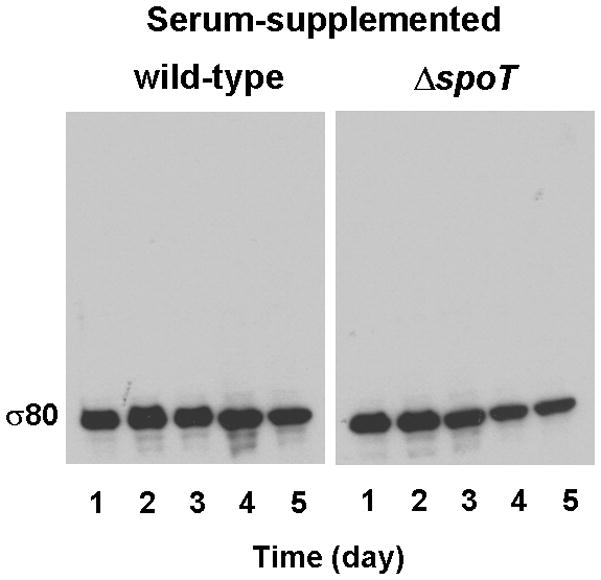
Cells of H. pylori wild type strain G27 and its isogenic ΔspoT mutant were grown in BLBB medium, which was either serum-free (A) or supplemented with 10% FBS (B). Cells were sampled at the indicated times and lysed. The σ80 protein in cell lysates was separated by 4–12% SDS-PAGE, followed by protein immunoblotting with the σ80-specific polyclonal antibodies. The positions of σ80 with normal mobility or with reduced electrophoretic mobilities (up-shifted) are indicated. The up-shifted σ80 appeared to be smeared bands, which is likely due to polyP being heterogeneous in size (see text).
Binding of polyphosphate modifies σ80
The σ80 up-shift suggested a tight association of σ80 with a putative agent, covalent modifications, or a major conformational change, resistant to SDS and boiling treatments in the protein. A cell lysate from which σ80 was up-shifted was hereafter named a shifted lysate; and a cell lysate from which σ80 was un-shifted an un-shifted lysate. The addition of increasing amounts of a shifted lysate to a fixed amount of an un-shifted lysate caused reduction of the mobility of σ80 in the latter (Fig. 2A, lanes 3–6), gradually reaching the mobility ofσ80 in the shifted lysate (lane 1). These results demonstrated that a shifted lysate contained an agent that quantitatively modified σ80 in trans. We purified the agent from shifted lysates by this assay.
Figure 2. The agent that caused σ80 up-shift is polyP.
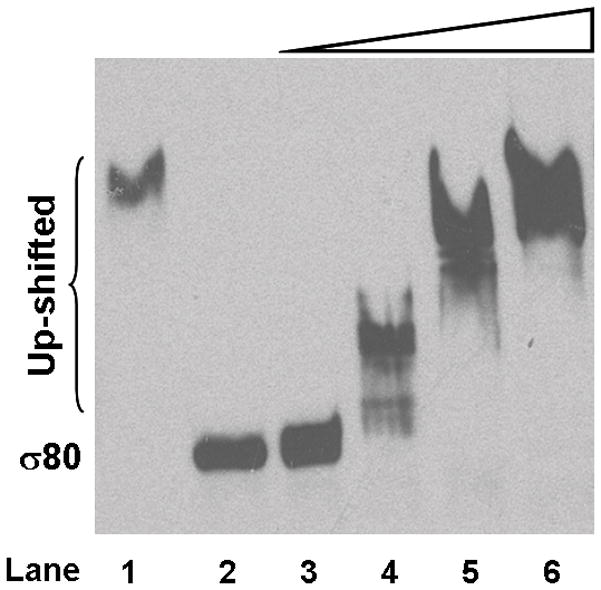
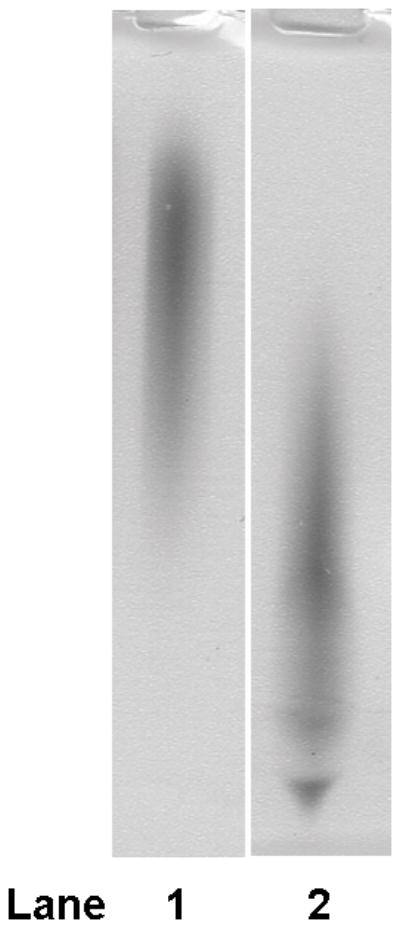
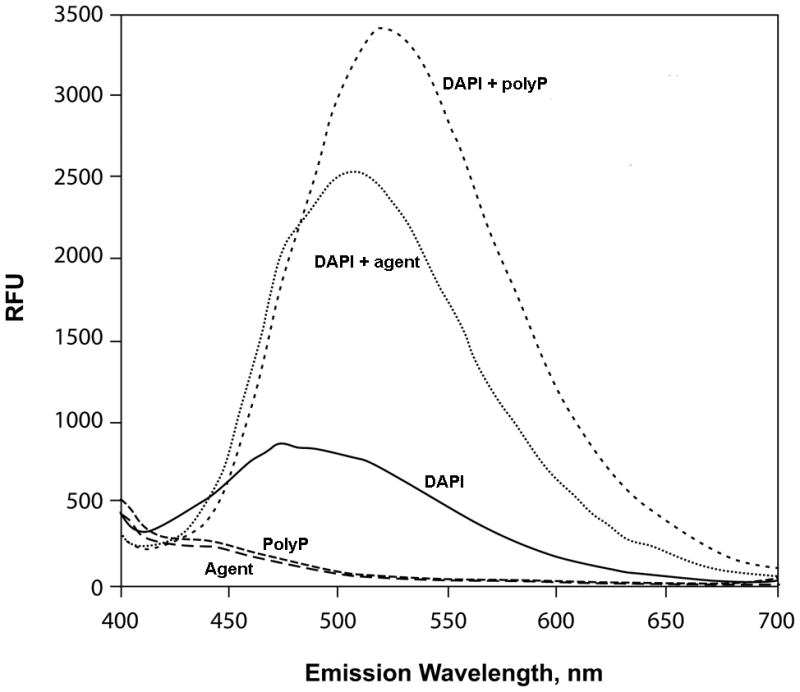
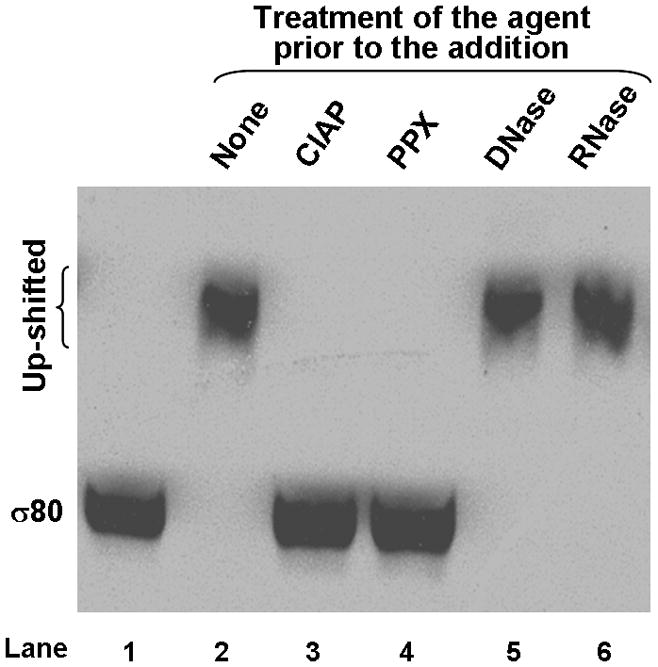
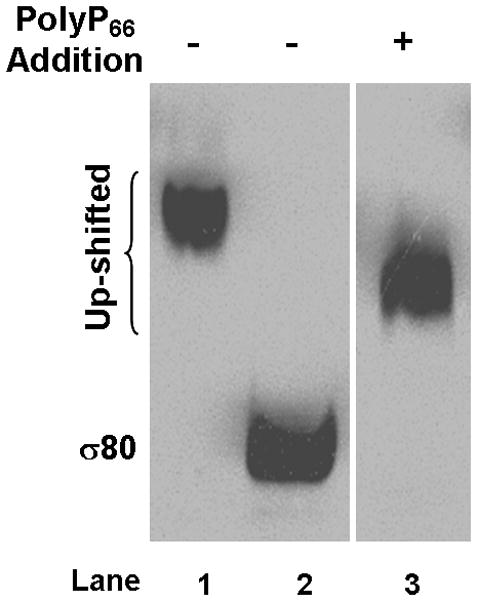
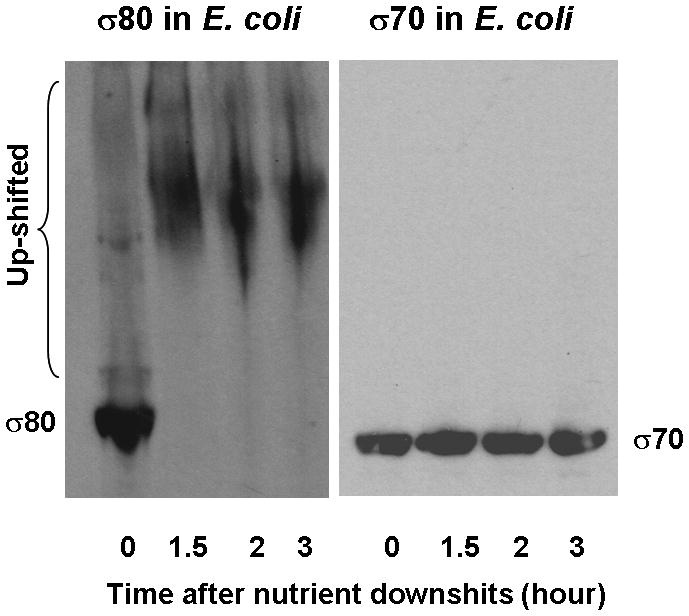
(A) Addition of a shifted lysate reduced the mobility of σ80 from an un-shifted lysate. A shifted lysate (as 1X amount) (lane 1); an un-shifted lysate (2X amount) either was alone (lane 2) or mixed with increasing amounts of the shifted lysate of approximate 0.5X (lane 3), 1X (lane 4), 2X (lane 5), and 4X quantities (lane 6). (B) Both the agent (lane 1) and polyP66 (lane 2) were analyzed by 6% urea-PAGE. They stained pink by Toluidine blue O. (C) The fluorescence spectrum of DAPI shifted by the addition of the purified agent, a result similar to the addition of polyP (polyP66). Excitation wavelength: 372 nm; and the emission maximum of DAPI alone was ~ 475 nm. RFU: relative fluorescence unit. (D) Theσ80 up-shifting activity of the agent was sensitive to the treatments of CIAP and PPX (lanes 3–4) and resistant to DNase and RNase treatments (lanes 5–6). An un-shifted lysate (lane 1); the un-shifted lysate was mixed with the purified agent, which was untreated (lane 2); or pre-treated with an indicated enzyme (lanes 3–6). (E) A polyP addition up-shifted σ80 from an un-shifted lysate. A shifted lysate as a control (lane 1); and an un-shifted lysate was either alone (lane 2) or mixed with polyP66 (lane 3). (F) The H. pyloriσ80 protein expressed in nutrient downshifted E. coli cells was up-shifted while the mobility of the hostσ70 protein was unchanged under the same conditions. At time 0, the cultures were nutrient downshifted from LB to a minimal medium containing a low concentration of Pi and no amino acids for three hours. The samples in (D to F) were analyzed as described in the legend of Figure 1, and the E. coliσ70 protein in (F) was detected by σ70 specific polyclonal antibodies. The up-shifted σ80 appeared to be smeared bands, which is likely due to polyP being heterogeneous in size (see text).
We identified this agent as polyphosphate (polyP) based on several analyses. 1) The purified agent was positively stained pink with Toluidine blue O, a dye used for staining polyP (Kumble & Kornberg, 1995, Tijssen et al., 1981) as seen with synthetic polyP66 (Fig. 2B). Both the agent and polyP66 appeared heterogeneous in size, characteristic of polyP molecules (Clark & Wood, 1987), which could explain why the up-shifted σ80 appeared as smeared bands (Fig. 1A). The agent migrated slower than polyP66 in the gel, suggesting that it had a longer average chain length than polyP66. 2) Upon binding the purified agent, the fluorescent probe 4′,6-diamidino-2-phenylindole (DAPI) shifted its peak emission from 475 nm to ~ 510 nm, a known property for polyP (Allan & Miller, 1980) as seen with polyP66 (Fig. 2C). 3) The σ80 up-shifting activity of the agent was sensitive to calf intestinal alkaline phosphatase (CIAP) and E. coli exo-polyphosphatase (PPX) treatments, two enzymes known to degrade polyP in vitro (Lorenz & Schroder, 2001, Akiyama et al., 1993), whereas it was resistant to both DNase and RNase treatments (Fig. 2D). 4) Finally, addition of polyP66 to an un-shifted lysate caused the σ80 up-shift (Fig. 2E). These data positively identified the σ80 up-shifting agent as polyP molecule that tightly binds to σ80 during serum starvation.
To examine whether some other property of the H. pylori cells would be required to facilitate the interaction between polyP and σ80, we determined the behavior of H. pyloriσ80 expressed in E. coli cells containing the pQE80-rpoD (Hp). The cells were grown in LB and then down-shifted to MOPS medium with a low concentration of Pi (0.1 mM) and without amino acids. While the polyP level is minimal when E. coli cells are grown in LB, cells accumulate large amounts of polyP750 (polyP molecules with an average chain length of ~ 750 Pi residues) after the nutrient downshift (Ault-Riche et al., 1998). Before the nutrient downshift, the mobility of σ80 produced in E. coli was essentially unchanged (at time 0); however, all σ80 proteins were up-shifted after the nutrient downshift (at 1.5 to 3 hours) (Fig. 2F). In contrast, the mobility of E. coliσ70 was unchanged under the same growth conditions. These results indicated that no other element unique to H. pylori cells is required for the interaction between polyP and σ80, and that the strong interaction between polyP and the primary sigma factor is specific for H. pylori as no such interaction is detected with σ70 in E. coli.
The amino-terminal domain of σ80 is the binding site for polyP
Compared to E. coli σ70 and other σ70 family proteins, there are ~70 additional amino acid residues at the amino-terminal domain (NTD) of σ80 in the H. pylori G27 strain (Fig. 3A), which account for the difference in molecular weight of the two principal σ factors. Notably, this extra region of σ80 is highly enriched with Lys (~33%) and has twice as many positively charged residues (~34%) as those that are negatively charged (~17%). The intact σ80 protein is a basic protein with an estimated pI of 8.4. Without this extra region, the truncated σ80 would have a theoretical pI value of 5.5, thus resembling an acidic σ70 protein from E. coli.
Figure 3. PolyP binds to the Lys-rich region at the NTD of σ80.
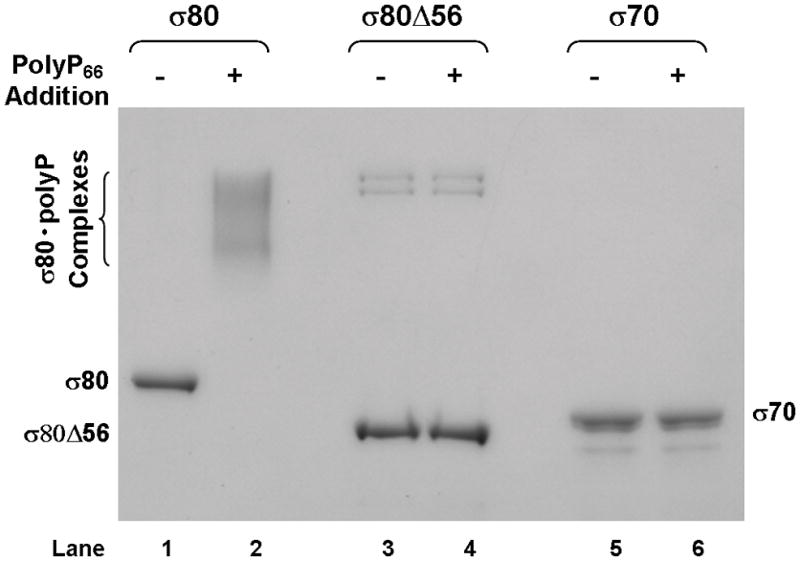
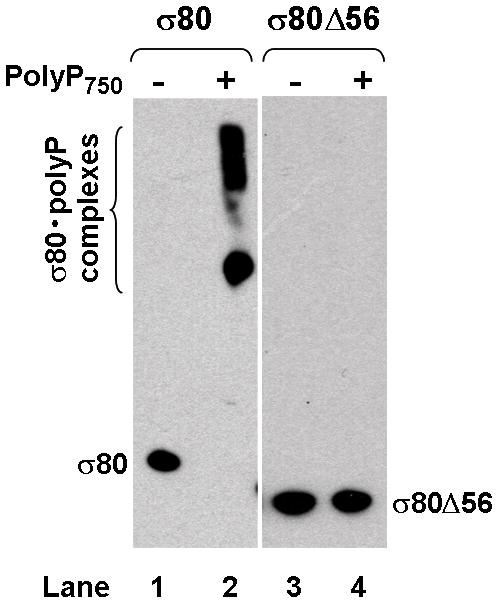
(A) There is an extra region of about 70 amino acid residues (aa) at the NTD of the H. pylori [NC_011333] σ80 compared with the E. coli [U00096] σ70. This extra region is concentrated with positively charged (+) aa including 23 Lys and one Arg, compared with 12 negatively charged (−) Glu. This extra region is named “region P” because it is a positively-charged-Lys-rich region and the target for polyP (see text). The conserved regions and non-conserved region (NCR) of members of the σ70 family proteins (Gruber & Gross, 2003) are shown. The rpoD mutation that deleted the aa 2–57 (σ80ΔNTD2-57 or σ80Δ56) is indicated. (B) A putative “region P” of the primary σ factor from Bordetella pertussis [NC_002929] or Coxiella burnetii (NC_002971). For simplicity, only the positively-charged-Lys-rich extra region (aa before conserved “region 1”) at the NTD of the σ factor indicated is shown. Positively charged (+) and negatively charged (−) aa are indicated. (C) Region P of σ80 is important for binding polyP. Purified recombinant σ80, σ80Δ56 and σ70 proteins in the absence (−) or presence (+) of polyP66 were analyzed by 4–12% SDS-PAGE. The gels were stained with QuickBlue Stain. The positions of σ80, σ80Δ56, σ70 and the σ80·polyP complexes are indicated. The other minor bands reflected impurities in protein preps. In particular, the higher molecular weight bands in lanes 3 & 4, were not related to σ80Δ56 as they were not immunostained by the σ80–specific polyclonal antibodies (data not shown). (D) PolyP is coimmunoprecipitated with the wild type σ80 protein but not with the NTD deletion under native non-denaturing condition. Purified recombinant σ80 and σ80Δ56 proteins in the absence (−) or presence (+) of polyP750 were immunoprecipitated by anti-polyHistidine antibody, and the samples were analyzed as described in the legend of Figure 1. The positions of σ80, σ80Δ56 and the σ80·polyP complexes are indicated.
To test if the NTD is a target for polyP, we engineered a deletion of 56 amino acid residues of the NTD of σ80 including 19 Lys and 1 Arg residues (σ80Δ56). Recombinant wild type and the mutant σ80Δ56 proteins were purified and their ability to bind polyP66 in vitro was examined. PolyP and wild type σ80 protein formed stable complexes in the purified system (Fig. 3C, lane 2), demonstrating polyP is necessary and sufficient to cause σ80 up-shift in the cell lysates. In contrast, neither the recombinant σ80Δ56, nor the E. coliσ70 protein bound polyP under the same conditions, as no changes in the mobility of the two proteins were detected in the presence of polyP.
When the same samples were analyzed using a non-denaturing gel in the absence of SDS, however, the σ80 protein did not migrate into the gel (data not shown). Thus, we performed coimmunoprecipitation assay using purified components to determine the interaction between polyP and the NTD of σ80 under native conditions. PolyP and wild type σ80 protein formed stable complexes as they were coimmunoprecipitated (Fig. 3D, lane 2), demonstrating that the uncovered interaction between polyP and σ80 is biologically significant. No such complex was detected with the σ80Δ56 protein under the same condition in vitro (Fig. 3D, lane 4). As described below, the σ80Δ56 protein in the H. pylori mutant containing a chromosomal rpoD(σ80Δ56) deletion was un-shifted during serum starvation. The data unambiguously identified the NTD of σ80, which we named “region P” as a positively-charged-Lys–rich region and the binding site for polyP, a new element in σ70 family proteins.
This unique feature (a positively-charged-Lys–rich region) of the NTD of σ80 is conserved among different H. pylori strains whose genome sequences are available at this time, although there are some variations in the numbers of Lys residues within the region. For example, compared to the NTD of σ80 protein from the G27 strain, there is an 11 aa deletion including four Lys residues in the counterpart from the H. pylori 26695 strain; however, the σ80 protein in cell lysates of the 26695 strain was still up-shifted in the presence of polyP (data not shown). Our data indicated strongly that the interaction of polyP and σ80 is likely a common mechanism conserved in H. pylori. The factors affecting the interaction remain to be determined. Bioinformatic analysis of the σ70 family proteins from the database suggested that putative “region P” is present in primaryσ factors of other human pathogens including Bordetella pertussis and Coxiella burnetii (Fig. 3B). It remains to be tested whether those σ factors also bind polyP during microbial starvation/stress.
PolyP·σ80 complex is critical for bacterial persistence
Next, we evaluated the biological significance of the interaction between σ80 and polyP during serum starvation by mutational analysis. We made an isogenic H. pylori strain carrying a chromosomal rpoD deletion (σ80Δ56) missing the “region P” and another with a null ΔppK mutation rendering the cell incapable of synthesizing polyP (Tan et al., 2005). Both mutants grew as well as the wild type, as the sizes of colonies of the two mutants were comparable to that of wild type on serum-supplemented plates after incubation at 37°C for 3–4 days (data not shown). This result indicated that σ80Δ56 remained functional under regular growth conditions.
In contrast to wild type cells, σ80 was un-shifted during serum starvation in theσ80Δ56 and ΔppK mutant cells, just as in the ΔspoT mutant cells (Fig. 4A, see also Fig. 1A). These results confirmed that the region P of σ80 is the target site for polyP in vivo. Importantly, the survival rates of the σ80Δ56 and the ΔppK mutant cells grown in a serum-free medium were significantly reduced compared to wild type, as measured by colony forming unit (CFU) determination. The reduction was most pronounced at days 3–5 of the starvation approaching the lowest survival rate of the ΔspoT mutant cells (Fig. 4B). These data revealed a strong correlation between a lack of σ80·polyP complex in the starved cells and a premature death phenotype arguing that the interaction between σ80 and polyP is critical for H. pylori survival during serum starvation, and they suggested a potential new target for drug design against the pathogen. The previously reported deficiency of other ppK mutants in colonization of mice might also derive from the inability of σ80 to regulate the starvation response in the absence of polyP (Tan et al., 2005, Ayraud et al., 2005). If this is the case, one would expect the σ80Δ56 andΔspoT mutants to be defective in mouse colonization similarly to the ppK mutants because the interaction between σ80 and polyP is prevented in all of the three mutants.
Figure 4. Mutational analysis of the determinants during serum starvation response.
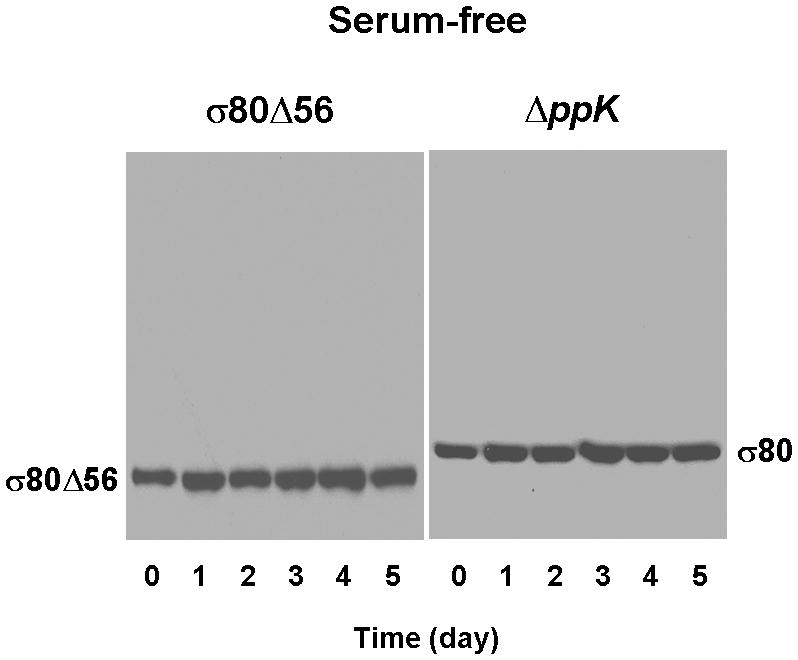
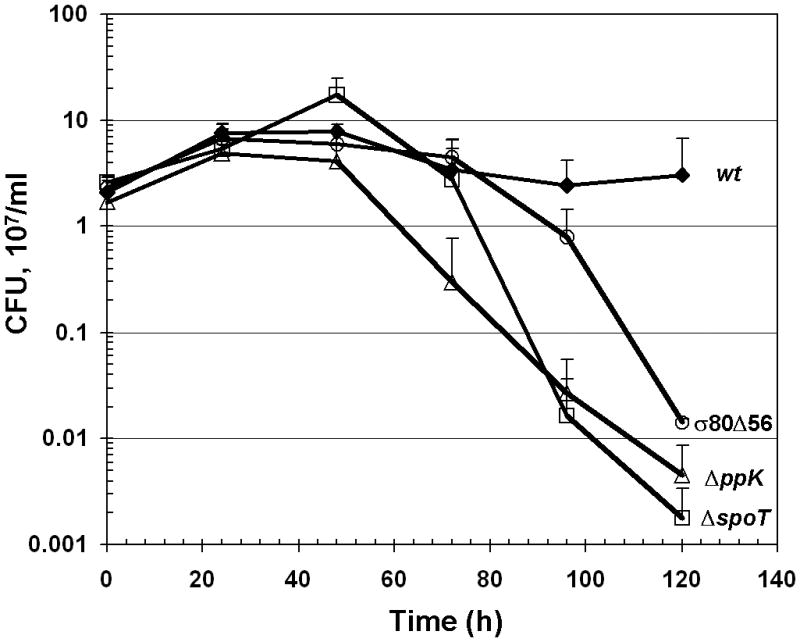
(A) Both the σ80Δ56 and ΔppK mutants were defective in forming theσ80·polyP complexes during serum starvation. σ80 was un-shifted in both the σ80Δ56 and ΔppK mutant cells grown serum-free. Cells were analyzed as described in the legend of Figure 1. (B). Both the σ80Δ56 and ΔppK mutants exhibited reduced survival during serum starvation, approaching the premature death phenotype of the ΔspoT mutant at day 5. The numbers of viable bacteria are expressed as colony-forming unit (CFU). The error bars indicate the deviations from the means, and there are no error bars indicated when the deviations were smaller than the symbol. The data are representative of three independent experiments.
The σ80 up-shift occurred in wild type cells grown in serum-free medium but not in the cells grown in serum-supplemented medium suggesting that polyP levels would be higher during nutrient starvation; and further, the absence of the σ80 up-shift in the spoT mutant suggested that (p)ppGpp would positively regulate the accumulation of polyP in H. pylori (Fig. 1A). Indeed, the polyP levels were over five-fold higher in the wild type cells starved for serum for 48 hours than that in the cells without the starvation; and as expected, the polyP levels were very low in the spoT mutant cells under the same conditions (Fig. 5A).
Figure 5. The determinants during starvation/stress response in H. pylori.
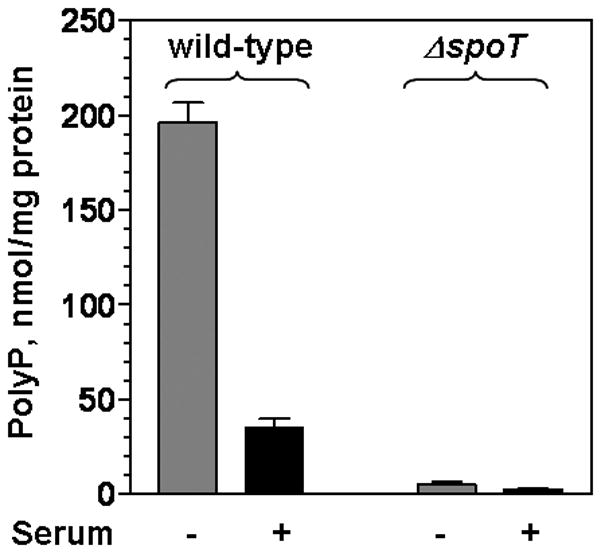
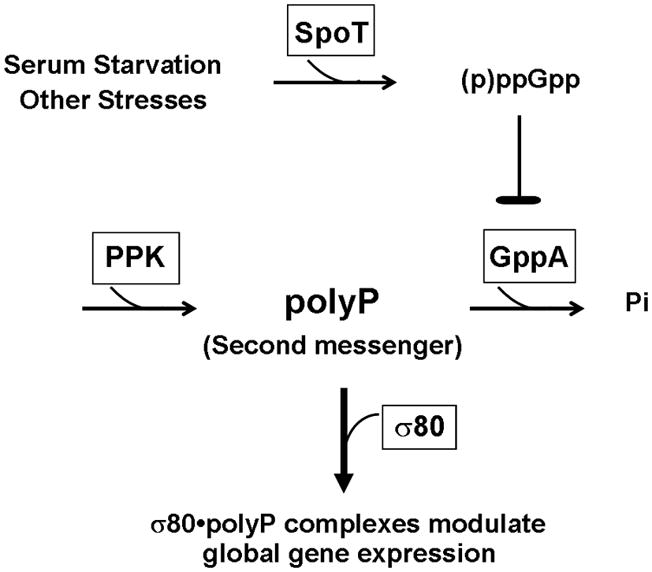
(A) Accumulations of polyP are observed in wild type cells grown serum-free and the accumulation is regulated by (p)ppGpp in H. pylori. PolyP extracted from wild type and the spoT mutant cells grown either in the absence (−) or presence of serum (+) for 48 hours were quantified. The error bars indicate the deviations from the means. The data are representative of three independent experiments. (B) This simplified scheme proposes the regulation of polyP and the role of polyP during SpoT mediated starvation/stress response in H. pylori. Serum starvation and other stresses promote SpoT to synthesize (p)ppGpp and/or to decrease the degradation of these alarmones, which can lead to accumulation of (p)ppGpp. H. pylori lacks the gene for the exopolyphosphatase PPX but contains a homologous guanosine pentaphosphate phosphohydrolase gene for GppA that is also an exopolyphosphatase (Keasling et al., 1993). Inhibition of GppA by (p)ppGpp can in turn lead to accumulation of polyP, assuming that (p)ppGpp does not inhibit polyphosphate kinase (PPK). Increased levels of polyP serve as a second messenger by forming strong association with σ80 to modify transcription machinery and global gene expression.
The determinants during starvation/stress response in H. pylori are summarized in Fig. 5B. PolyP accumulation is likely linked to the starvation/stress response, just as in E. coli. In E. coli, the level of polyP increases during amino acid starvation and by exposure to other stresses (Ault-Riche et al., 1998), mainly because accumulation of (p)ppGpp during stress response or the stringent response (Cashel et al., 1996) inhibits the activity of exo-polyphosphatase (PPX) (Kuroda et al., 1997) that breaks down polyP. H. pylori lacks the gene for PPX but contains a homologous guanosine pentaphosphate phosphohydrolase gene for GppA that is also an exopolyphosphatase (Keasling et al., 1993) similar to PPX. In H. pylori, an increased level of (p)ppGpp, which is synthesized by the SpoT enzyme upon stress and nutrient downshift (Mouery et al., 2006, Wells & Gaynor, 2006), is likely to inhibit GppA activity, but not the activity of polyphosphate kinase (PPK), the enzyme for synthesis of polyP, causing accumulation of polyP in the cell. Therefore, a lack of (p)ppGpp synthesis in the ΔspoT mutant could account for a reduced polyP level causing no change in σ80 mobility during serum starvation in the mutant strain cells (Fig. 1A). Moreover, studying the interaction of these determinants would provide a possible explanation why the mobility of σ80 returned to normal in wild type cells after day 4 during serum starvation (Fig. 1A). The effects of other stresses on the polyP production and the regulation of synthesis and/or degradation of polyP in H. pylori are currently under investigation.
The mode of action of polyP in H. pylori differs, however, from that in E. coli. PolyP stimulates Lon protease activity to degrade ribosomal proteins supplying the amino acids needed to starved E. coli cells (Kuroda et al., 2001). In H. pylori, polyP targets the principal σ factor. PolyP unlikely mediates the degradation of σ80 as levels ofσ80 appeared to be not significantly changed upon serum starvation in the wild type H. pylori cells. Our results support the hypothesis that in responding to nutrient starvation/stress, an elevated cellular level of polyP acts as a second messenger to control gene regulation and pathogenesis by directly modulating the activity of transcriptional machinery in the cell. We are currently testing this working model experimentally.
Summary remarks
Our study revealed a novel mechanism exhibited by a human pathogen in responding to stress caused by starvation. During a SpoT mediated serum starvation response in H. pylori, polyP forms a strong association with σ80. Such an interaction is critical for the bacterial persistence during nutrient depletion, a likely environment deep in human gastric mucus layer where the pathogen lives. A positively-charged-Lys-rich region at the NTD of σ80 is identified as the binding region for polyP (region P) in vivo and in vitro, revealing a new element for σ70 family proteins. Putative “region P” is present in primary sigma factors of other human pathogens including Bordetella pertussis and Coxiella burnetii, suggesting that the uncovered interaction might be a general strategy employed by other pathogens to cope with starvation/stress.
Experimental procedures
Bacterial strains and general methods
Bacterial techniques, and growth conditions for H. pylori either with a serum-free or serum-supplemented medium (BLBB) under microaerophilic environment (5% O2, 10% CO2 and 85% N2) are as described (Zhou et al., 2008). Unless otherwise mentioned, all H. pylori strains used are G27 and its derivatives, and cells were grown serum-supplemented, either in broth or solid agar plate. H. pylori strains containing the ΔppK1-cat mutation (Tan et al., 2005) were kindly provided by Douglas E. Berg, Washington University Medical School, St. Louis, Missouri. The ΔppK1-cat mutation was introduced to the G27 strain by transformation using the mutant’s genomic DNA, and chloramphenicol (8 μg/ml) resistant recombinants were selected. The G27 strain containing the ΔppK1-cat mutation was named ΔppK. The rpoD NTD deletion mutation (σ80ΔNTD2-57 or σ80Δ56) was constructed by allelic exchange after cells were transformed with a PCR DNA fragment (~ 2.2 kb), which contained 5′-DNA sequence (NC_011333.1:95106..94374) located immediately upstream of the rpoD gene, the nonpolar aphA-3 gene, a ribosome binding site and ATG, and followed by the rpoD sequence encoding the 58th to 273rd amino acid residues of σ80 (NC_011333.1:94202..93551), and kanamycin (15 μg/ml) resistant recombinants were selected. The strategy of linking several overlapped PCR products generated with appropriate primers (sequences available on request) to make this 2.2 kb PCR DNA fragment is as described (Zhou et al., 2008). The rpoD NTD deletion mutation was verified by PCR with appropriate primers and DNA sequencing.
Cloning of the H. pylori rpoD gene and its derivatives, expression and purification of the recombinant σ80 proteins in E. coli
Standard molecular biology and DNA techniques are as described (Sambrook et al., 1989). DNA fragments containing the H. pylori rpoD gene and its derivatives flanked by BamHI and PstI sites from H. pylori DNA were generated by PCR with appropriate primers (sequences available on request) and were cloned into the BamHI and PstI sites of vector pQE80L (Qiagen). The resulting wild type recombinant rpoD construct and its derivatives encoded recombinant σ80 proteins containing the additional sequence MRGSHHHHHHGS at their amino-terminus. These clones were transformed into E. coli K12 strains and their sequence was confirmed by DNA sequencing.
To overproduce the recombinant σ80 proteins from cells harboring the pQE80-rpoD (Hp) constructs, mid-log cultures (LB containing 50 μg/ml of ampicillin) were induced with IPTG (1 mM) for about 2h before cells were harvested. Most of the overproduced proteins were soluble, and they were purified by Talon superflow metal affinity resin (Clontech, Palo Alto, CA) using a procedure essentially as described for the purification of the recombinant E. coliσ70 protein (Zhi & Jin, 2003). Rabbit polyclonal antibodies against purified recombinant wild type σ80 and σ70 proteins were prepared for us by Spring Valley Laboratories, Inc. MD.
SDS-PAGE and Western Blots
After cultures were sampled (0.1–1 ml depending on optical density (OD) at a wavelength of 600 nm), cells were spun down. The cell pellets were resuspended with 0.1 ml of protein loading buffer containing SDS (Invitrogen) and heat boiled for 5–10 min to lyse cells. About 5–10 μl of the resulted cell lysates (usually from cells of OD600 0.005–0.015 unit, which corresponded to 1/20 to 1/10 of the original cultures sampled) were analyzed by 4–12% SDS NuPAGE (Invitrogen), followed by Western Blot with σ80 specific polyclonal antibodies. In each set of experiments, an equal amount of cell lysates was analyzed.
Nutrient downshift experiments in E. coli
To determine the behavior of σ70 in E. coli cells accumulating large amounts of polyP, earlier stationary phase cultures (OD600 ~ 0.9) in LB (as time 0) were nutrient downshifted to MOPS medium with a low concentration of Pi (0.1 mM) and without amino acids for three hours as described (Ault-Riche et al., 1998). Similarly, E. coli cells harboring the pQE80-rpoD (Hp) were grown in LB supplemented with amperciline (50 μg/ml) to late-log phase (OD600 ~ 0.5), and then IPTG (1 mM) was added to induce the expression of σ80 for 90 min. After samples were taken (as time 0), cultures were nutrient downshifted to the same minimal medium supplemented with amperciline (50 μg/ml) and IPTG (1 mM) for three hours. Samples were prepared and analyzed by SDS-PAGE and Western blots as described above, and the σ70 protein was detected by σ70 specific polyclonal antibodies.
Extraction and analyses of the agent that caused σ80 up-shift, and polyP assays
The agent was extracted from shifted cell lysates (from ~15 ml cultures) using a procedure that was originally developed for isolation of plasmid DNA from H. pylori (De Ungria et al., 1998), and after a final step of isopropanol precipitation, the resulting precipitates were resuspended in ~50 μl H2O. As little as 0.5 μl of the purified agent caused σ80 up-shift from un-shifted lysates as measured by the σ80 up-shift assay.
The purified agent (~10 μl) and polyP66 (~ 2 μg) [heterogeneous molecules with an average chain length of ~ 66 inorganic phosphate (Pi) residues, purchased from Sigma] in 10 mM Tris-HCl (pH 8.0) buffer were analyzed by 6% urea-PAGE until the dye bromophenol blue reached 3 cm from the bottom of the gel, followed by staining with Toluidine Blue O (Sigma), as described (Kumble & Kornberg, 1995). DAPI fluorescence in the presence of the purified agent (5 μl) or polyP66 (0.2 mg/ml) was analyzed essentially as described (Allan & Miller, 1980), except that a SpectraMax Gemini EM Fluorescence microplate reader (Molecular Devices) was used. Samples were prepared in 25 mM Tris-HCl buffer (pH. 7.0), 1 M glycerol, either with or without DAPI (20 μg/ml). Samples were excited at 372 nm and fluorescence emission spectra were recorded. To test the sensitivity of the purified agent toward different enzymes, 1 μl of the agent was treated with an indicated enzyme in a 10 μl reaction first, and then the reaction mixtures were precipitated with isopropanol. Each of the precipitates was resuspended in 8 μl of H2O and 4 μl were used for the σ80 up-shift assays. For the control without treatment, 0.5 μl of the agent was used. Calf intestinal alkaline phosphatase (New England Biolabs), DNase and RNase (Invitrogen) were used according to the manufacturer’s instructions. E. coli recombinant exo-polyphosphatase (PPX) was purified from E. coli cells harboring pQE80-ppX (Ec) essentially as described above for σ80 proteins purification, and it was used as described (Akiyama et al., 1993).
PolyP was extracted from cultures of H. pylori wild type and the spoT mutant cells grown either serum-free or serum-supplemented for 48 hours using Glassmilk (MP Biomedicals) as described (Ault-Riche et al., 1998). PolyP was quantified in terms of Pi residues by a two-enzyme assay as described (Ault-Riche et al., 1998). PolyP was first converted to ATP in the presence of ADP by E. coli polyphosphate kinase (PPK) (a gift from Wenqing Xu, University of Washington, Seattle, WA), and then ATP was measured by a luciferase reaction (CellTiter-Glo, Promega) as described (Zhou et al., 2008). A standard curve for ATP (1 nM to 1 μM) was used for calibration. Protein concentration in cell lysates was determined using a DC Protein Assay Reagent (Bio-Rad) using a BSA standard.
The σ80 up-shift assay, determination of the polyP-binding region of σ80 and coimmunoprecipitation assay
For the σ80 up-shift assay, unless otherwise indicated, an un-shifted lysate (typically from cells of OD600 ~0.015) was mixed with either a shifted lysate, the purified agent (0.5 μl), or polyP66 (0.25 μg) for 10 min at 37 °C. The mixtures were analyzed by SDS-PAGE and Western Blot as described above.
To detect the formation of σ80·polyP complexes and determine the polyP-binding region of σ80 using purified components, each of the purified proteins (~ 0.4 μg) was used alone or mixed with polyP66 (3 μg) in a 10 μl buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl and 1 mM MgCl) for 10 min at 37 °C. They were analyzed by 4–12% SDS NuPAGE, and the gels were stained by QuickBlue Stain (Bostonbiologicals) and dried using Gel-Dry drying solution (Invitrogen).
Coimmunoprecipitation assay was used to detect the formation of σ80·polyP complexes under non-denaturing conditions. Each of the purified recombinant σ80 proteins (~ 2.4 μg) were mixed with (~ 2.5 μg) of polyP750, which was synthesized by E. coli PPK as described (Ault-Riche et al., 1998), in 120 μl of buffer B (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 0.1 mM EDTA and 0.1 mg/ml BSA), and samples were rotated gently at room temperature (25°C) for 5 min. Then 3 μl of mouse anti-polyHistidine antibody (Sigma) was added and the mixtures were rotated in a cold room (4°C) for 1 hour, followed by the addition of 20 μl of protein A/G agarose (Santa Cruz). After incubation in the cold room overnight with rotation, the mixtures were spun down. The immunoprecipitates were washed with the buffer B (800 μl) twice before resuspended in the buffer (40 μl). Samples (4 μl) were analyzed by SDS-PAGE, followed by Western Blot with σ80 specific polyclonal antibodies as described above.
Acknowledgments
We thank D.E. Berg for strains, T. Durfee for the construction of pQE80-ppX (Ec), W. Xu for E. coli PPK, and B. Xiao and C. Cagliero for bioinformatics analysis of sigma factors. We are grateful for helpful comments for the manuscript from M. Kashlev, A. Klar, KX Jin, JL Caswell and M. Mills. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- Allan RA, Miller JJ. Influence of S-adenosylmethionine on DAPI-induced fluorescence of polyphosphate in the yeast vacuole. Can J Microbiol. 1980;26:912–920. doi: 10.1139/m80-158. [DOI] [PubMed] [Google Scholar]
- Allen LA. Phagocytosis and persistence of Helicobacter pylori. Cell Microbiol. 2007;9:817–828. doi: 10.1111/j.1462-5822.2007.00906.x. [DOI] [PubMed] [Google Scholar]
- Ault-Riche D, Fraley CD, Tzeng CM, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayraud S, Janvier B, Labigne A, Ecobichon C, Burucoa C, Fauchere JL. Polyphosphate kinase: a new colonization factor of Helicobacter pylori. FEMS Microbiol Lett. 2005;243:45–50. doi: 10.1016/j.femsle.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Beier D, Spohn G, Rappuoli R, Scarlato V. Functional analysis of the Helicobacter pylori principal sigma subunit of RNA polymerase reveals that the spacer region is important for efficient transcription. Mol Microbiol. 1998;30:121–134. doi: 10.1046/j.1365-2958.1998.01043.x. [DOI] [PubMed] [Google Scholar]
- Berg DE, Hoffman PS, Appelmelk BJ, Kusters JG. The Helicobacter pylori genome sequence: genetic factors for long life in the gastric mucosa. Trends Microbiol. 1997;5:468–474. doi: 10.1016/s0966-842x(97)01164-5. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Clark JE, Wood HG. Preparation of standards and determination of sizes of long-chain polyphosphates by gel electrophoresis. Anal Biochem. 1987;161:280–290. doi: 10.1016/0003-2697(87)90452-0. [DOI] [PubMed] [Google Scholar]
- De Ungria MC, Tillett D, Neilan BA, Cox PT, Lee A. A novel method of extracting plasmid DNA from Helicobacter species. Helicobacter. 1998;3:269–277. doi: 10.1111/j.1523-5378.1997.06085.pp.x-i1. [DOI] [PubMed] [Google Scholar]
- Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007;117:60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hengge R. Proteolysis of sigma(S) (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Keasling JD, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc Natl Acad Sci U S A. 1993;90:7029–7033. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kumble KD, Kornberg A. Inorganic polyphosphate in mammalian cells and tissues. J Biol Chem. 1995;270:5818–5822. doi: 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- Lorenz B, Schroder HC. Mammalian intestinal alkaline phosphatase acts as highly active exopolyphosphatase. Biochim Biophys Acta. 2001;1547:254–261. doi: 10.1016/s0167-4838(01)00193-5. [DOI] [PubMed] [Google Scholar]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- Mouery K, Rader BA, Gaynor EC, Guillemin K. The stringent response is required for Helicobacter pylori survival of stationary phase, exposure to acid, and aerobic shock. J Bacteriol. 2006;188:5494–5500. doi: 10.1128/JB.00366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A. 2004;101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Fraley CD, Zhang M, Dailidiene D, Kornberg A, Berg DE. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J Bacteriol. 2005;187:7687–7695. doi: 10.1128/JB.187.22.7687-7695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijssen JP, Beekes HW, Van Steveninck J. Localization of polyphosphates at the outside of the yeast cell plasma membrane. Biochim Biophys Acta. 1981;649:529–532. doi: 10.1016/0005-2736(81)90156-5. [DOI] [PubMed] [Google Scholar]
- Wells DH, Gaynor EC. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J Bacteriol. 2006;188:3726–3729. doi: 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi H, Jin DJ. Purification of highly-active and soluble Escherichia coli sigma 70 polypeptide overproduced at low temperature. Methods Enzymol. 2003;370:174–180. doi: 10.1016/S0076-6879(03)70015-9. [DOI] [PubMed] [Google Scholar]
- Zhou YN, Coleman WG, Jr, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. Regulation of cell growth during serum starvation and bacterial survival in macrophages by the bifunctional enzyme SpoT in Helicobacter pylori. J Bacteriol. 2008;190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]