Abstract
Benzo[a]pyrene (BaP) is a prototypical polycyclic aromatic hydrocarbon (PAH) found in combustion processes. Cytochrome P450 1A1 and 1B1 enzymes (CYP1A1, CYP1B1) can both detoxify PAHs and activate them to cancer-causing reactive intermediates. Following high dosage of oral BaP (125 mg/kg/day), ablation of the mouse Cyp1a1 gene causes immunosuppression and death within ~28 days, whereas Cyp1(+/+) wild-type mice remain healthy for >12 months on this regimen. In the present study, male Cyp1(+/+) wild-type, Cyp1a1(−/−) and Cyp1b1(−/−) single-knockout, and Cyp1a1/1b1(−/−) double-knockout mice received a lower dose (12.5 mg/kg/day) of oral BaP. Tissues from 16 different organs––including proximal small intestine (PSI), liver, preputial gland duct (PGD)––were evaluated; microarray cDNA expression and >30 mRNA levels were measured. Cyp1a1(−/−) mice revealed markedly increased CYP1B1 mRNA levels in the PSI, and between 8 and 12 weeks developed unique PSI adenomas and adenocarcinomas. Cyp1a1/1b1(−/−) mice showed no PSI tumors but instead developed squamous cell carcinoma of the PGD. Cyp1(+/+) and Cyp1b1(−/−) mice remained healthy with no remarkable abnormalities in any tissue examined. PSI adenocarcinomas exhibited striking up-regulation of the Xist gene, suggesting epigenetic silencing of specific genes on the Y-chromosome; the Rab30 oncogene was up-regulated; the Nr0b2 tumor suppressor gene was down-regulated; paradoxical over-expression of numerous immunoglobulin kappa and heavy chain variable genes was found––although the adenocarcinoma showed no immunohistochemical evidence of being lymphatic in origin. This oral BaP mouse paradigm represents an example of “gene-environment interactions” in which the same exposure of carcinogen results in altered target organ and tumor type, as a function of just one or two globally absent genes.
Keywords: oral benzo[a]pyrene, CYP1 enzymes, Cyp1a1 gene, Cyp1b1 gene, adenocarcinoma of proximal small intestine, immunoglobulin-secreting tumor, squamous cell carcinoma of preputial gland duct, Xist gene, Rab30 oncogene, Nr0b2 tumor suppressor gene, non-agouti locus, Olfr genes
Introduction
Carcinogens widely distributed in the environment include many types of polycyclic aromatic hydrocarbons (PAHs). Environmental PAHs occur as byproducts of cigarette smoke, gas- and diesel-engine exhaust, charcoal-grilled food, creosote railroad ties, and coal and coke distillation ovens in the petroleum industry. The most thoroughly studied prototypic PAH is benzo[a]pyrene (BaP)1–3. In many different tissues and cell types from numerous mammalian studies, metabolically-activated BaP is well known to cause cytotoxic, teratogenic, genotoxic, mutagenic and carcinogenic effects4,5. BaP and other PAHs in cigarette smokers are implicated as causative agents in lung cancer6,7 and atherosclerosis5.
The etiology of toxicity and cancer caused by PAHs is very complex. By way of the aromatic hydrocarbon receptor (AHR), PAHs induce numerous enzymes involved in both activation and detoxication of PAHs. Typically, PAHs are metabolically activated by phase I enzymes to reactive intermediates that bind covalently to nucleic acids and proteins1,2,4,5; however, PAHs are also detoxified8,9 by phase I (functionalization) as well as by phase II (conjugation) enzymes. In addition, PAHs elicit the up- and down-regulation of hundreds of other genes via both AHR-dependent and AHR-independent mechanisms3–5,10–12.
Although not generally appreciated13,14, cigarette smoke (and chemicals therein) as well as other inhaled pollutants are taken up not only by the lung, but by far the majority of particles is swallowed; hence, our need to better understand the pharmacokinetics and pharmacodynamics of oral PAHs. To examine the role of CYP1A1 in the intact animal receiving daily oral BaP, this lab previously compared Cyp1a1(−/−) knockout mice with Cyp1(+/+) wild-type mice8,9. Based on many in vitro and cell culture studies showing that CYP1A1 metabolically activates BaP to undesirable oxygenated intermediates capable of causing toxicity and cancer, we had expected mice without any basal or inducible CYP1A1 to be more protected than Cyp1(+/+) wild-type mice. To our surprise, however, Cyp1a1(−/−) knockout mice died between 28 and 32 days of oral BaP (125 mg/kg/day), whereas wild-type and Cyp1b1(−/−) knockout mice remained healthy. By day 18 on this diet, Cyp1a1(−/−) mice exhibited overt immunosuppression and much higher levels of BaP-DNA adduct formation in liver, small intestine, spleen and bone marrow. Moreover, the total body burden of BaP was ~25 times greater in Cyp1a1(−/−) than in Cyp1(+/+) mice. This led us to conclude that, in the intact animal, oral BaP-induced CYP1A1 in the intestine appears to be more important in detoxication than metabolic activation8.
Cyp1a1/1b1(−/−) double-knockout mice, receiving the same dose of oral BaP, were about 80% “rescued” from immunosuppression; in other words, they responded more like wild-type mice9. The total body burden of BaP was about three times greater in Cyp1a1/1b1(−/−) than in Cyp1a1(−/−) mice, i.e. ~75 times greater in Cyp1a1/1b1(−/−) than in wild-type mice––demonstrating that the total body BaP burden can be dissociated from the clinical outcome. Hence, it was concluded that immunosuppression is prevented in Cyp1a1/1b1(−/−) mice, most likely because CYP1B1-mediated BaP metabolism is required in the immune cell before BaP-induced immunosuppression can occur9.
What about lower doses of oral BaP? Long ago, this lab had demonstrated that decreased levels of oral BaP (~12 and ~6 mg/kg/day) changed the clinical outcome from immunosuppression to immune cell malignancies15. Even in mice receiving oral BaP at 12.5 or 1.25 mg/kg/day for 18 days8, we found that BaP-DNA adducts are readily detectable and highly elevated in duodenum, liver, spleen and bone marrow of Cyp1a1(−/−)––compared with that of Cyp1(+/+) mice. Therefore, our hypothesis was that a lower oral dose of BaP might cause cancer rather than toxicity in Cyp1a1(−/−), but not Cyp1a1/1b1(−/−), mice. We examined the above-mentioned genotypes during a 16-week course of oral BaP at 12.5 mg/kg/day. Intriguingly, we discovered two specific types of cancer in different target organs: adenocarcinoma in the proximal small intestine (PSI), and squamous cell carcinoma in the preputial gland duct (PGD) epithelium––dependent on which Cyp1 genes were missing.
MATERIALS AND METHODS
Chemicals
BaP was purchased from Sigma (St. Louis, MO). All other chemicals and reagents were bought from either Aldrich Chemical Company (Milwaukee, WI) or Sigma Chemical Company as the highest available grades.
Mice
Generation of the Cyp1a1(−/−)16 and Cyp1b1(−/−)17 single-knockout and the Cyp1a1/1b1(−/−) double-knockout9 mouse lines have been described. The three genotypes have been backcrossed onto the C57BL/6J background for eight generations; this ensures that the knockout genotype resides in a genetic background that is >99.8% C57BL/6J18. Age-matched C57BL/6J Cyp1(+/+) wild-type mice, purchased from The Jackson Laboratory (Bar Harbor, ME), can therefore be used as comparative controls. All experiments with these four genotypes were carried out using males only and began at 6 ± 1 weeks of age. All animal experiments were conducted in accordance with the National Institutes of Health standards for the care and use of experimental animals and the University Cincinnati Medical Center Institutional Animal Care and Use Committee.
Treatment
All experiments in the present study used oral BaP at 12.5 mg/kg/day. Rodent chow (Harlan Teklad; Madison, WI) was soaked at least 24 h in BaP-containing corn oil (1.0 mg/ml) before presenting to the mice. By knowing the weight of the food ingested daily by a 20-g mouse and by using [3H]BaP in several earlier experiments19, the daily oral BaP dose had been estimated to be ~12.5 mg/kg/day. To start Day 1 of the experiment, the mice (after having been fasted overnight) were presented with the BaP-laced food; control mice received food soaked in corn oil alone. The mice eagerly ate corn oil-soaked chow immediately. Except for our initial survival studies carried out over 26 weeks, all other oral BaP experiments in this study were run for 4, 8, 12 or 16 weeks. Total body weight and liver weight were measured after 12 weeks on oral BaP.
Biohazard Precaution
BaP is highly toxic and regarded as a human carcinogen. All personnel were instructed in safe-handling procedures. Lab coats, gloves and masks were worn at all times, and contaminated materials were collected separately for disposal by the Hazardous Waste Unit of the University Cincinnati Medical Center or by independent contractors. BaP-treated mice were housed separately, and their carcasses were treated as contaminated biologic materials.
Mouse Tissue Histology
We collected mouse tissues––including proximal small intestine (PSI), liver, skin surrounding and including the preputial gland duct (PGD), brain, thymus, thyroid, lung, esophagus, forestomach, glandular stomach, ileum, large intestine, spleen, adrenal gland, kidney, and bone marrow. “PSI” refers to the first 4 cm of intestine beyond the pyloric junction, i.e. duodenum and proximal jejunum. In addition to gross inspection, paraformaldehyde-fixed tissues were dehydrated, embedded in paraffin, sectioned, and stained with hematoxylin and eosin; the sections were photographed at several magnifications.
For immunohistochemistry, antigen retrieval was achieved, when needed, by treating slide-mounted tissue sections with heated 0.01 M citrate solution, pH 6.0 (Poly Scientific; Bay Shore, NY). After quenching the endogenous peroxidase and incubating with blocking serum, sections were incubated with diluted anti-pan keratin (Ventana Medical Systems; Tucson, AZ) or anti-cleaved caspase-3 (Cell Signaling Technology; Danvers, MA) primary antibody for 18 h at 4°C. Sections were then washed with PBS and incubated with biotinylated secondary antibodies (Jackson Immunoresearch Labs; West Grove, PA) directed against the primary antibody type. After washing with PBS, slides were developed using the ABC/DAB method (Vector Labs; Burlingame, CA), counterstained with methyl green stain, and photographed using an RT Slider digital camera (Diagnostic Instruments Inc.; Sterling Heights, MI).
Total RNA Preparation
Total RNA was isolated from PSI and liver (N=3 mice per group) using the RNAgents Total RNA Isolation System™ (Promega; WI, USA).
Reverse Transcription
Mouse tissues were harvested and frozen in liquid nitrogen, and stored at −80°C until use. Total RNA (2 µg) was added to a reaction containing 3.8 µM oligo(dT)20 and 0.77 mM dNTP––to a final volume of 13 µl. Reactions were incubated 65°C for 5 min, then 4°C for 2 min. To the reaction mixture we added 7 µl of solution containing 14 mM dithiothreitol, 40 units RNaseOUT Recombinant RNase inhibitor™ (Invitrogen; CA, U.S.A), and 200 units SuperScript III™ (Invitrogen). Reactions were incubated at 50°C for 50 min, followed by 75°C for 10 min (to inactivate the reverse transcriptase). Distilled water (80 µl) was added to the isolated cDNA; these samples were then stored at –80°C until use.
Copy Number of mRNA
The mRNAs from each of the three Cyp1 genes plus the prostaglandin G synthase-2 gene (Ptgs2) were quantified by fitting qRT-PCR data to curves generated from cloned RNAs (cRNAs) for each cDNA. Briefly, templates for cRNA synthesis were produced by PCR on cDNA constructs from each CYP1 cDNA plus PTGS2 cDNA that had been cloned into pcDNA3.1(+) (Invitrogen). PCR was performed, using a forward primer including T7 promoter sequence and a reverse primer having an oligo(dT) that followed the stop codon. The amplified products were purified by electrophoresis. In vitro transcription was performed using T7 RiboMAX™ Express Large-Scale RNA Production System (Promega). After DNaseI treatment, the cRNAs were spectrophotometrically quantified. The cRNAs were used to generate a standard curve in quantitative real-time (qRT) PCR reactions from which mRNA copy number from qRT-RNA measurements (vide infra) could be extrapolated.
Quantitative Real-Time PCR (qRT-PCR)
The primers used are available upon request. The qRT-PCR was performed in the ABI PRISM 7000 Sequence Detection System™ (Applied Biosystems; Foster City, CA), using Power SYBR Green PCR Master Mix (Applied Biosystems). Individual mRNA abundance was determined, using the standard-curve method (from 101 to 108 copies/µl), as previously described by K.J. Livak (PE-ABI; Sequence Detector User; Bulletin #2 (Ref. 20). Each sample was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA concentration curves.
Microarray Hybridization
At each of four time-points (0, 4, 8 and 12 weeks of oral BaP), Cyp1a1(−/−) knockout mice (N=6) provided the RNA from the PSI; RNA from three mice was combined to make two groups. Gross and microscopic examination confirmed normal histology in all mice at zero-time and 4 weeks, varying degrees of dysplasia21 seen at 8 weeks, and (both intraepithelial and invasive) neoplasia at 12 weeks. Any mice that did not show the expected histology at any of these time-points (i.e. outliers) were excluded. Microarray experiments were carried out essentially as described elsewhere and referenced therein22. The mouse 70-mer MEEBO oligonucleotide library version 1.05 (25,130 unique gene symbols on the array; Invitrogen; Carlsbad, CA) was suspended in 3X SSC at 30 µM and printed at 22°C, with 65% relative humidity, on aminosilane-coated slides (Cel Associates, Inc.; Pearland, TX), using a high-speed robotic Omnigrid machine (GeneMachines; San Carlos, CA) with Stealth SMP3 pins (Telechem; Sunnyvale, CA). The complete gene list can be viewed at http://www.invitrogen.com. Spot volumes were 0.5 nl, and spot diameters 75–85 µm. The oligonucleotides were cross-linked to the slide substrate by exposure to 600 mJ of ultraviolet light.
Fluorescence-labeled cDNAs were synthesized from total RNA, using an indirect aminoallyl-labeling method via an oligo(dT)-primed reverse-transcriptase reaction. The cDNA was decorated with mono-functional reactive cyanine-3 and cyanine-5 dyes (Cy3 and Cy5; Amersham; Piscataway, NJ). The details and a complete description of the slide preparation can be found at http://microarray.uc.edu.
Imaging and data generation were carried out using a GenePix 4000A and GenePix 4000B (Axon Instruments; Union City, CA) and associated software from Axon Instruments, Inc. (Foster City, CA). The microarray slides were scanned with dual lasers having the wavelength frequencies to excite Cy3 and Cy5 fluorescence emittance. Images were captured in *.jpg and *.tif files, and DNA spots captured by the adaptive circle segmentation method. Information extraction for a given spot is based on the median value for the signal pixels and the median value for the background pixels, to produce a gene-set data file for all the DNA spots.
Microarray Data Normalization and Analysis
We sought to identify differentially-expressed PSI genes from oral BaP-treated Cyp1a1(−/−) mice––comparing 4 weeks with zero-time, 8 weeks with zero-time, 12 weeks with zero-time, 8 weeks with 4 weeks, 12 weeks with 4 weeks, and 12 weeks with 8 weeks. Two biological-replicate arrays were carried out. Analysis was performed using R statistical software and the limma Bioconductor package23. Data normalization was conducted in two steps for each microarray separately22. First, background-adjusted intensities were log-transformed and the differences (M) and averages (A) of log-transformed values were calculated as M = log2(X1) – log2(X2) and A = [log2(X1) + log2(X2)]/2, where X1 and X2 denote the Cy5 and Cy3 intensities, respectively. Second, normalization was performed by fitting the array-specific local regression model of M as a function of A. Normalized log-intensities for the two channels were then calculated by adding half the normalized ratio to A for the Cy5 channel and subtracting half the normalized ratio from A for the Cy3 channel. Statistical analysis was performed by first fitting the following analysis-of-variance model for each gene separately: Yijk = m + Ai + Sj + Ck+ eijk, where Yijk corresponds to the normalized log-intensity on the ith array, with the jth treatment, and labeled with the kth dye (k = 1 for Cy5, and 2 for Cy3); m denotes the overall mean log-intensity, Ai is the effect of the ith array, Sj is the effect of the jth treatment, Ck is the gene-specific effect of the kth dye, and eijk is the error term for the ith array with the jth treatment, and labeled with the kth dye. Estimated fold-changes were calculated from the ANOVA models, and resulting t-test statistics from each comparison were modified using an intensity-based empirical Bayesian method (IBMT)22. This method, an extension of an earlier technique23, obtains more precise estimates of variance by pooling information across genes and accounting for the dependency of variance on probe-intensity levels. Identification of significant genes was accomplished from two avenues. First, the false discovery rate (FDR) was calculated24; genes with an FDR value of ≤0.10 are considered as significantly differentially expressed. Next, discovery of gene categories enriched with differentially-expressed genes was performed using LRpath25. The biological process and molecular-function branches of the Gene Ontology (GO) database26 were tested for enrichment, and genes belonging to those GO terms having a calculated FDR ≤0.05 were considered for further analysis.
Other Statistical Methods
Statistical significance between groups was determined by chi-square analysis, analysis of variance among groups, or Student’s t-test. All assays were performed in duplicate or triplicate, and repeated at least twice. Statistical analyses were also performed with the use of SAS® statistical software (SAS Institute Inc.; Cary, NC) and Sigma Plot (Systat Software, Inc.; Point Richmond, Ca). All statistical hypothesis testing was performed at the α = 0.05 significance level, and all statistical tests were two-sided.
Results
Survival, Body Weight, and Liver Weight
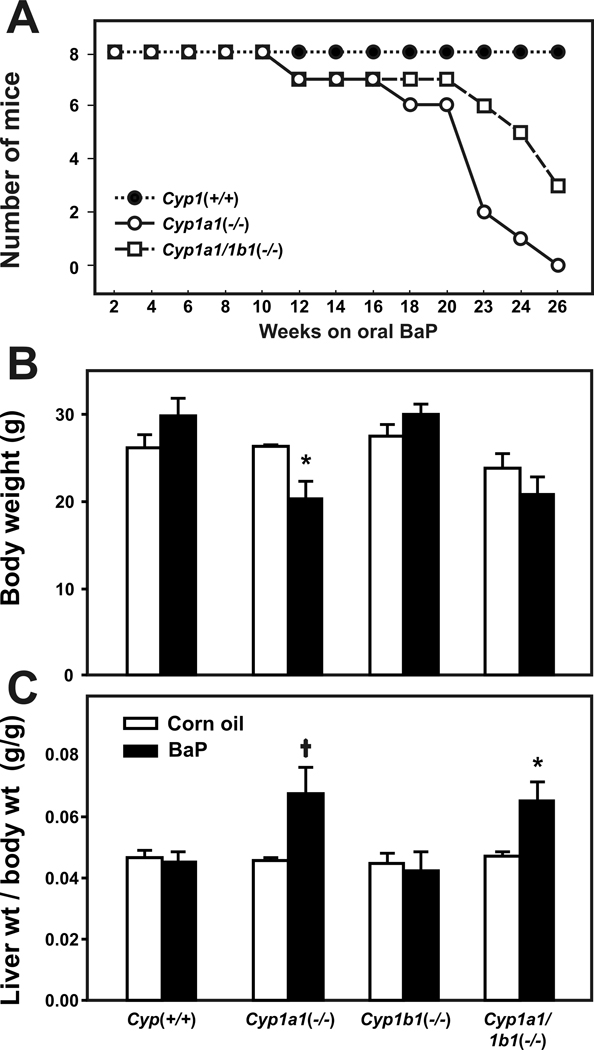
On a BaP diet of 12.5 mg/kg/day (Fig. 1A), Cyp1a1(−/−) knockout mice survived between 18 and 26 weeks. Cyp1a1/1b1(−/−) mice survived longer than Cyp1a1(−/−) mice, with some Cyp1a1/1b1(−/−) deaths beginning to occur especially beyond ~20 weeks of oral BaP. Virtually all of the deaths were associated with cancer and/or wasting. After 26 weeks of oral BaP, there was 100% survival of the Cyp1(+/+) wild-type mice (Fig. 1A) and Cyp1b1(−/−) mice (not shown).
Figure 1.
Survival rates, body weights, and liver-weight-per-total-body-weight ratio. Survival rates (A) during exposure to oral BaP at 12.5 mg/kg/day (N = 8 per genotype). Cyp1(+/+) wild-type (closed circles) are compared with Cyp1a1(−/−) knockout (open circles) and Cyp1a1/1b1(−/−) double-knockout (open squares) mice. At every oral BaP dose tested, Cyp1a1(−/−) mice always show significantly (P<0.001) more toxicity than Cyp1a1/1b1(−/−) mice9. This is because CYP1A1 is the major inducible CYP in the GI tract and, with oral BaP in Cyp1(+/+) wild-type mice, CYP1A1 becomes so highly induced that there is major detoxication and elimination of BaP metabolites in the feces. Without CYP1A1 in the GI tract in Cyp1a1(−/−) mice, at least 25-fold higher amounts of oral BaP reach distal tissues, including the spleen-thymus-bone marrow where BaP causes CYP1B1-dependent immunosuppression. Without CYP1A1 and CYP1B1 in the GI tract of Cyp1a1/1b1(−/−) mice, at least 75-fold higher amounts of oral BaP reach distal tissues; however, BaP is ineffective at causing immunosuppression because there is no CYP1B1 in the spleen-thymus-bone marrow to metabolize BaP to reactive oxygenated intermediates9. Similar to wild-type mice, no Cyp1b1(−/−) deaths were seen during the 26-week experiment (not shown). Body weights (B) and liver-weight-per-total-body-weight ratios (C) in the four genotypes, following corn oil alone (open bars) versus oral BaP at 12.5 mg/kg/day (closed bars) for 12 weeks. Cyp1a1(−/−) and Cyp1b1(−/−) single-knockout and Cyp1a1/1b1(−/−) double-knockout were compared with Cyp1(+/+) wild-type. *P<0.05. †P =.025.
Following 12 weeks of oral BaP at 12.5 mg/kg/day (Fig. 1B), Cyp1a1(−/−) mice showed significant (P <0.05) decreases in body weight, whereas no wasting was seen in the other three genotypes. Both Cyp1a1(−/−) (P =.025) and Cyp1a1/1b1(−/−) (P <0.05) mice exhibited substantial liver enlargement (Fig. 1C), as measured by the liver-weight-to-total-body-weight ratio; liver hypertrophy is believed to reflect chronic activation of the AHR––which of course occurs with a very high body burden of BaP in these two genotypes9. Cyp1(+/+) and Cyp1b1(−/−) mice did not show changes in either body weight or liver enlargement following 12 weeks of oral BaP at 12.5 mg/kg/day.
Adenocarcinoma of Proximal Small Intestine
Cyp1a1(−/−) mice showed a highly significant (P <0.001) increased risk for adenocarcinoma of the PSI (Table 1), detected by 12 weeks of oral BaP 12.5 mg/kg/day. Cyp1a1/1b1(−/−) double-knockout mice did not exhibit any PSI cancer on the BaP diet, whereas they had a highly significant (P <0.001) enhanced risk of squamous cell carcinoma of the PGD. No malignancies or other remarkable abnormalities were observed in any of the other 14 above-mentioned tissues examined.
Table 1.
Effects of oral BaP at 12.5 mg/kg/day for 12 weeks*
| Genotype | Clinical outcome | Number of mice in group | Number of mice with cancer |
|---|---|---|---|
| Cyp1(+/+) | Healthy | 16 | 0 |
| Cyp1a1(−/−) | Adenocarcinoma of PSI | 16 | 13 (P <0.001) |
| Cyp1b1(−/−) | Healthy | 16 | 0 |
| Cyp1a1/1b1(−/−) | Squamous cell carcinoma of PGD | 16 | 11 (P <0.001)† |
PSI, proximal small intestine; PGD, preputial gland duct. In these studies we did not include females, in which the clitoral gland is the equivalent of the preputial gland. Statistical values were calculated by chi-square analysis and Student’s two-tailed t-test.
One mouse of this genotype showed a micro adenoma of the PSI that was found to express high IgG and IgM levels.
Histology of the PSI Adenocarcinoma
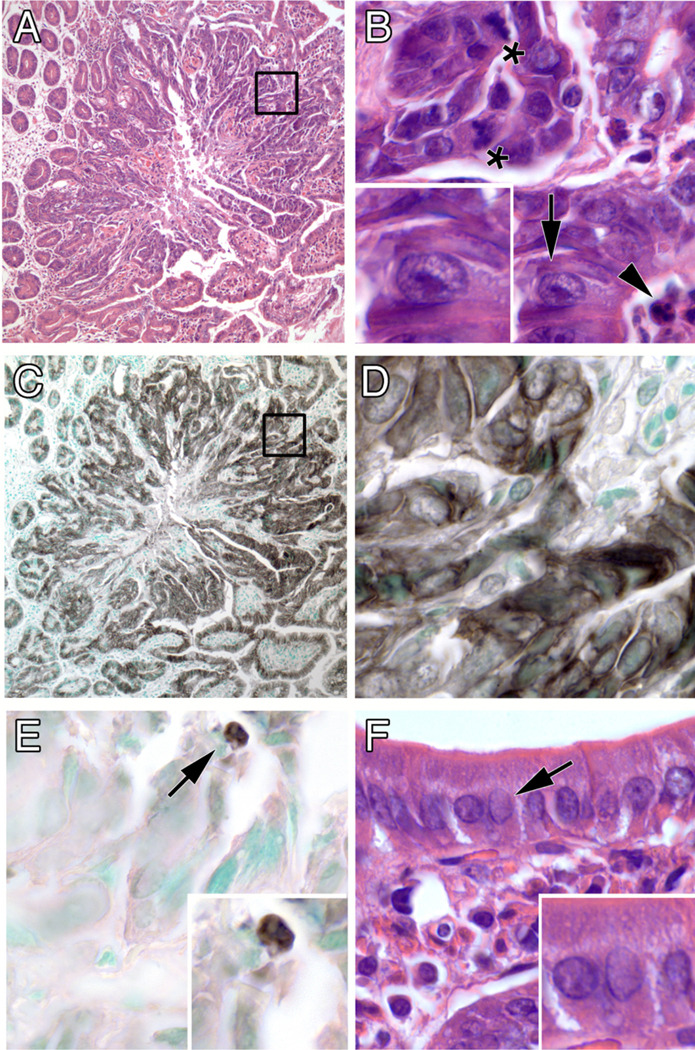
Histological sections of normal PSI (Fig. 2) were compared with areas involved with a lesional progression from dysplasia to invasive and metastatic carcinoma. Nests of abnormal and neoplastic cells were seen close to the lumen but in the lower half of the villus––the same region from which embryonic stem cells originate. The lower half of the villus is also where most of the CYP1 proteins (when present) reside in the PSI27. Moreover, if one compares the GI tract from tongue to rectum, the highest CYP1 mRNA levels, protein levels, and activity are localized in the PSI (27). Neoplastic cells can also be seen invading the stroma and lamina propria (Fig. 2, panels C through F). Reactive stroma (desmoplastic response), which is a histological marker for several types of invasive carcinomas including intestinal adenocarcinoma28, can be seen around the invasive cell nests. Nests of malignant cells were also identified in nearby lymph nodes (Fig. 2, panel F inset), which are the most common and earliest sites of metastasis from malignancies arising in epithelia29 and thus a key factor in the prognosis and treatment of patients with intestinal adenocarcinoma30.
Figure 2.
Progressive atypical histology of the PSI in Cyp1a1(−/−) knockout mice (following 12 weeks of oral BaP, 12.5 mg/kg/day), showing normal intestinal villi together with nests of neoplastic cells near the basement membrane. A, area of normal proximal small intestine. (B, C, & D) areas involved by a progression from epithelial dysplasia to carcinoma in situ (white arrows). (E & F) unequivocal areas of invasive carcinoma into the stroma and the muscle layer (white arrows). Inset: a small region of metastasis (white arrow) found in a nearby lymph node. Few (if any) lymphocytes or other immune cells implicating possible inflammation are present. Bar, upper right = 100 microns for all images.
Neoplastic cellular features of these tumors (Fig. 3) include: architectural disarray and a high nuclear-to-cytoplasmic volume ratio (compare Fig. 3, B & F), nuclear changes such as high mitotic activity, nuclear atypia with prominent enlarged nucleoli (Fig. 3, B), and occasional apoptotic bodies (Fig. 3, B & E). The immunohistochemical identity of this tumor was confirmed as an adenocarcinoma by its reactivity with a pan-keratin antibody (Fig. 3, C & D).
Figure 3.
Features of a typical PSI tumor. (A) low magnification showing expanding boundary of tumor (inset enlarged in B). (B) high magnification of disarrayed tumor cells showing mitotic figures (asterisks), enlarged nuclei with prominent nucleoli (arrow; enlarged in inset), and an apoptotic body (arrowhead). (C & D) immunohistochemical staining for a keratin epithelial marker, low and high magnification, respectively. (E) immunohistochemical staining for the apoptosis marker cleaved caspase-3. (F) normal-appearing intestinal epithelium away from the tumor, showing orderly cell arrangement and small nuclei without enlarged nucleoli (compare insets of panelsF & B). Original optical magnifications 100x and 1000x.
Gene Expression Comparing Oral BaP-Treated (4 weeks) with Untreated Mice
Microarray analysis was carried out at the treatment time-points of 0, 4, 8, and 12 weeks––during which PSI histology proceeds from normal to neoplasia. Comparing any oral BaP treatment-time with zero-time untreated mice (Table S1, first three rows), we found ~10-fold to ~60-fold more genes significantly (FDR ≤0.10) up- and down-regulated than we did when comparing one oral BaP-treated group with another oral BaP-treated group. This observation is perhaps not surprising, because oral BaP administration for any number of weeks (compared to no BaP) would greatly perturb homeostasis in the PSI; in contrast, comparing 8 weeks with 4 weeks of oral BaP, etc., would reflect a much smaller number of genes, but perhaps the genes up- and down-regulated during the 4- vs 8- vs 12-week times might be more likely to be associated with the developing adenocarcinoma.
Table S2 & Table S3 list the top 50 most significantly up- and down-regulated genes, respectively, when comparing 4 weeks of oral BaP with untreated mice; the first portions of Table 2 & Table 3 list a subset of these genes. The highest induction (206-fold) occurred with the Non-agouti gene product; besides its role in coat color, the a locus is associated with effects on the adrenal cortex31 and with the Itch gene (encoding an E3 ubiquitin ligase) involved in an immunological disease that includes lung and stomach inflammation and hyperplasia of lymphoid and hematopoietic cells32; interestingly, the Itch gene is also highly significantly (FDR = 0.00125; P-value = 0.00001) up-regulated in the PSI, when one compares oral BaP for 4 weeks with untreated mice (Table 2). The 40-fold increase in the β-2 microglobulin gene is worth noting, because a rise in this gene is associated with the acute-phase response. Pertinent to the present study is the 25-fold increase in CYP1B1 mRNA. Table S4 ranks the most significant gene ontology (GO) classes affected after 4 weeks of oral BaP, compared with untreated mice. Genes––having “various receptor activities, perception of chemical stimuli, and mitochondrial and membrane-bound organelle, metabolic, and ribonucleoprotein functions”––are among those most highly perturbed at this 4-week time-point, compared with that at zero-time. All data comparing 8 weeks of BaP with zero-time and 12 weeks of BaP with zero-time are not shown or discussed here because of space limitations, but are available to anyone who wishes to scrutinize them.
Table 2.
Microarray analysis of PSI: selection of a subset of genes (other than Ig genes) most significantly up-regulateda
| Symbol | Gene biochemical name | Fold increase | P-value | FDRb | |
|---|---|---|---|---|---|
| Comparison of 4 weeks of oral BaP treatment (12.5 mg/kg/day) with zero time-point | |||||
| Olfr710 | Olfactory receptor 710 | +37.2 | 1.2 × 10−9 | 2.8 × 10−5 | |
| Drr1 | Developmentally-regulated repeat element-containing transcript-1 | +17.0 | 2.2 × 10−9 | 5.8 × 10−5 | |
| Etfa | Electron-transferring flavoprotein, α-polypeptide | +17.2 | 2.9 × 10−9 | 6.3 × 10−5 | |
| Ece1 | Endothelin-converting enzyme-1 | +24.1 | 1.2 × 10−8 | 0.00013 | |
| Polr2a | Polymerase (RNA) II (DNA-directed) polypeptide A | +55.2 | 4.1 × 10−8 | 0.00018 | |
| a | Non-agouti | +206 | 4.9 × 10−8 | 0.00018 | |
| Csl | Citrate synthase-like | +20.1 | 8.8 × 10−8 | 0.00020 | |
| B2m | β-2 microglobulin | +40.2 | 1.6 × 10−7 | 0.00024 | |
| Itch | Itchy, E3 ubiquitin protein ligase | +4.47 | 0.00001 | 0.00125 | |
| Cyp1b1 | Cytochrome P450, family 1, subfamily b, polypeptide 1 | +25 | 1.1 × 10−5 | 0.00178 | |
| Comparison of 8 weeks with 4 weeks of oral BaP treatment (12.5 mg/kg/day) | |||||
| Xist | Inactive Chr X-specific transcripts | +70.0 | 6.3 × 10−7 | 0.01069 | |
| Serpina1a | Serine (or cysteine) peptidase inhibitor, clade A, member 1a | +14.8 | 4.0 × 10−6 | 0.01829 | |
| Nr0b2 | Nuclear receptor, family 0, member-2 (tumor suppressor function) | +8.74 | 4.7 × 10−5 | 0.04364 | |
| Serpina1b | Serine (or cysteine) peptidase inhibitor, clade A, member 1b | 6.16 | 0.00012 | 0.05701 | |
| Tff1 | Trefoil factor-1 | +9.96 | 9.7 × 10−5 | 0.05763 | |
| Serpina1d | Serine (or cysteine) peptidase inhibitor, clade A, member 1d | +5.80 | 0.00011 | 0.05871 | |
| Fabp6 | Fatty acid binding protein 6, ileal (gastrotropin) | +188 | 0.00036 | 0.09327 | |
| Comparison of 12 weeks with 4 weeks of oral BaP treatment (12.5 mg/kg/day) | |||||
| Nr0b2 | Nuclear receptor, family 0, member-2 (tumor suppressor function) | +8.41 | 6.9 × 10−6 | 0.02113 | |
| Slpi | secretory leukocyte peptidase inhibitor | +4.43 | 2.7 × 10−5 | 0.03701 | |
| Nupr1 | Nuclear protein-1 | +4.43 | 3.2 × 10−5 | 0.04021 | |
| Hmgcs2 | 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase-2 | +8.04 | 3.9 × 10−5 | 0.04085 | |
| Slc40a1 | Solute carrier (basolateral iron transporter), family 40, member 1 | +9.02 | 0.00010 | 0.05778 | |
| Fgf13 | Fibroblast growth factor-13 | +3.40 | 0.00013 | 0.06287 | |
| Acot1 | Acyl-Coenzyme A thioesterase-1 | +5.90 | 0.00013 | 0.06318 | |
| Cabc1 | Chaperone, ABC1-like activity of BCL complex (S. pombe) | +2.77 | 0.00014 | 0.06580 | |
| Itpr1 | Inositol 1,4,5-triphosphate receptor-1 | +2.98 | 0.00014 | 0.06580 | |
| Acot2 | Acyl-Coenzyme A thioesterase-2 | +3.22 | 0.00014 | 0.06596 | |
| Bok | BCL2-related ovarian killer protein | +2.93 | 0.00015 | 0.06596 | |
| Dab1 | Disabled homolog-1 (Drosophila) | +3.13 | 0.00017 | 0.06694 | |
| Rnf186 | Ring-finger protein-186 | +2.67 | 0.00022 | 0.07438 | |
| Klk1b3 | Kallikrein 1-related peptidase b3 | +2.98 | 0.00023 | 0.07585 | |
| Ranbp3l | RAN-binding protein-3-like | +5.01 | 0.00024 | 0.07863 | |
| Meis2 | Meis homeobox 2, Chr 2 | +3.01 | 0.00026 | 0.08017 | |
| Spon2 | Spondin-2, extracellular matrix protein | +2.54 | 0.00027 | 0.08086 | |
| Cyp4f16 | Cytochrome P450, family 4, subfamily F, 16 (fatty acid, eicosanoid metabolism) | +2.57 | 0.00029 | 0.08526 | |
| Comparison of 12 weeks with 8 weeks of oral BaP treatment (12.5 mg/kg/day) | |||||
| Eif2s3y | Eukaryotic translation initiation factor 2, subunit 3, Chr Y | +34.2 | 3.0 × 10−7 | 0.01069 | |
| Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide-3, Chr Y | +17.0 | 6.5 × 10−7 | 0.01069 | |
| Jarid1d | Jumonji, AT-rich interactive domain 1D (Rbp2-like), Chr Y | +8.39 | 1.0 × 10−5 | 0.02270 | |
| Rab30 | RAB30, member of RAS oncogene family | +4.17 | 3.5 × 10−5 | 0.04041 | |
| Olfr978 | Olfactory receptor 978 | +4.81 | 4.6 × 10−5 | 0.04364 | |
| Kcnn3 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 | +4.21 | 9.2 × 10−5 | 0.05668 | |
| Igfbp3 | Insulin-like growth factor binding protein-3 | +3.57 | 0.00013 | 0.05808 | |
| V1rg11 | Vomeronasal-1 receptor G11 | +3.28 | 0.00011 | 0.05987 | |
| Cyp3a41 | Cytochrome P450, family 3, subfamily A, 41 (drug, eicosanoid metabolism) | +3.64 | 0.00013 | 0.06305 | |
| Ces1 | Carboxylesterase-1 | +3.28 | 0.00013 | 0.06318 | |
| Leap2 | Liver-expressed antimicrobial peptide-2 | +8.31 | 0.00013 | 0.06381 | |
| G0s2 | G0/G1 switch gene 2 | +4.70 | 0.00014 | 0.06580 | |
| Adh4 | Alcohol dehydrogenase-4 (class II), pi polypeptide | +15.4 | 0.00020 | 0.07131 | |
| Cyp2c29 | Cytochrome P450, family 2, subfamily C, 29 (drug, eicosanoid metabolism) | +4.10 | 0.00020 | 0.07131 | |
This is a partial list of selected genes; the entire list of up-regulated genes for all three comparisons can be found in Tables S2, S6, S9 & S12 of the Supplementary Data.
False discovery rate, or adjusted P-value. One out of ten adjusted P-values ≤0.10 would be expected to be a false positive. In this and the following table, the P-values are dependent on both the measurements of fold-change, as well as how consistent they are (variance).
Table 3.
Microarray analysis of PSI: selection of a subset of genes (other than Ig genes) most significantly down-regulateda
| Symbol | Gene biochemical name | Fold decrease | P-value | FDRb | |
|---|---|---|---|---|---|
| Comparison of 4 weeks of oral BaP treatment (12.5 mg/kg/day) with zero time-point | |||||
| Esm1 | Endothelial cell-specific molecule-1 | −6.97 | 2.2 × 10−7 | 0.00028 | |
| Igfbp3 | insulin-like growth factor-binding protein-3 | −10.5 | 3.0 × 10−7 | 0.00030 | |
| Tnfrsf13b | Tumor necrosis factor receptor superfamily, member 13b | −23.6 | 3.4 × 10−7 | 0.00032 | |
| Zfyve19 | Zinc finger, FYVE domain-containing-19 | −9.96 | 8.5 × 10−7 | 0.00051 | |
| Dullard | Dullard homolog (Xenopus laevis) | −6.25 | 1.3 × 10−6 | 0.00060 | |
| Safb | Scaffold attachment factor B | −9.40 | 1.3 × 10−6 | 0.00061 | |
| Pink1 | PTEN-induced putative kinase-1 | −4.88 | 2.3 × 10−6 | 0.00081 | |
| Edf1 | Endothelial differentiation-related factor-1 | −6.09 | 2.6 × 10−6 | 0.00085 | |
| Comparison of 8 weeks with 4 weeks of oral BaP treatment (12.5 mg/kg/day) | |||||
| Eif2s3y | Eukaryotic translation initiation factor-2, subunit 3, Chr Y | −30.8 | 2.8 × 10−6 | 0.01556 | |
| Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Chr Y | −16.5 | 5.3 × 10−6 | 0.01959 | |
| Olfr978 | Olfactory receptor 978 | −6.34 | 0.00013 | 0.06423 | |
| Bcmo1 | Beta-carotene 15,15’-monooxygenase | −5.48 | 0.00014 | 0.06580 | |
| Jarid1d | Jumonji, AT-rich interactive domain 1D (Rbp2-like), Chr Y | −6.28 | 0.00017 | 0.06694 | |
| Rab30 | RAB30, member of RAS oncogene family | −4.63 | 0.00021 | 0.07206 | |
| Osbpl1a | Oxysterol binding protein-like 1A | −4.29 | 0.00030 | 0.08555 | |
| Comparison of 12 weeks with 4 weeks of oral BaP treatment (12.5 mg/kg/day) | |||||
| Bcmo1 | Beta-carotene 15,15’-monooxygenase | −17.4 | 9.4 × 10−7 | 0.01069 | |
| Ccbl1 | Cysteine conjugate-beta lyase 1 | −5.50 | 7.6 × 10−6 | 0.02150 | |
| Slc13a2 | Solute-carrier (Na+/(SO4)2–/carboxylate cotransporter), family 13, member 2 | −5.74 | 8.7 × 10−6 | 0.02221 | |
| Acta1 | Actin, alpha 1, skeletal muscle | −10.9 | 1.9 × 10−5 | 0.03236 | |
| Hist1h4d | Histone cluster 1, H4d | −4.49 | 2.3 × 10−5 | 0.03745 | |
| Cyp27a1 | Cytochrome P450, family 27, subfamily A, member 1 (bile acid synthesis) | −3.91 | 3.6 × 10−5 | 0.04041 | |
| Susd2 | Sushi domain-containing-2 | −3.57 | 5.4 × 10−5 | 0.04449 | |
| Hist2h2be | Histone cluster 2, H2be | −3.32 | 5.9 × 10−5 | 0.04530 | |
| Ahrr | Aryl hydrocarbon receptor repressor | –6.25 | 6.1 × 10−5 | 0.04586 | |
| Gm1186 | Unknown gene associated with advanced gastrointestinal stromal tumor | −4.37 | 6.4 × 10−5 | 0.04665 | |
| Hist1h4b | Histone cluster, 1, H4b | −3.06 | 9.9 × 10−5 | 0.05778 | |
| Oas1e | 2’-5’-Oligoadenylate synthetase 1E | −3.73 | 0.00010 | 0.05778 | |
| Kcnj3 | Potassium inwardly-rectifying channel, subfamily J, member 3 | −3.70 | 0.00010 | 0.05778 | |
| Nr1d2 | Nuclear receptor, family 1, subfamily D, member 2 (heme ligand-regulated component of mammalian clock) | −3.26 | 0.00011 | 0.06044 | |
| Hist1h2bb | Histone cluster, 1, H2bb | −3.26 | 0.00013 | 0.06287 | |
| Hist1h4k | Histone cluster, 1, H4k | −2.83 | 0.00014 | 0.06596 | |
| Hist1h4h | Histone cluster, 1, H4h | −3.09 | 0.00015 | 0.06596 | |
| Edc3 | Enhancer of mRNA decapping-3 homolog (S. cerevisiae) | –3.75 | 0.00015 | 0.06596 | |
| Hist1h4f | Histone cluster, 1, H4f | −2.77 | 0.00021 | 0.07206 | |
| Hist1h4i | Histone cluster, 1, H4i | −2.76 | 0.00022 | 0.07348 | |
| Hist1h3d | Histone cluster, 1, H3d | −2.99 | 0.00023 | 0.07534 | |
| Hist1h4c | Histone cluster, 1, H4c | −3.04 | 0.00025 | 0.07918 | |
| H2-ab1 | Histocompatibility 2, class II antigen A, beta-1 | −2.84 | 0.00025 | 0.07936 | |
| H2-DMb1 | Histocompatibility 2, class II, locus Mb1 | −2.43 | 0.00036 | 0.09327 | |
| Ccng1 | Cyclin G1 | −2.61 | 0.00036 | 0.09327 | |
| Comparison of 12 weeks with 8 weeks of oral BaP treatment (12.5 mg/kg/day) | |||||
| Xist | Inactive Chr X specific transcripts | −67.7 | 8.3 × 10−8 | 0.00685 | |
| Serpina1a | Serine (or cysteine) peptidase inhibitor, clade A, member 1a | −9.73 | 1.4 × 10−6 | 0.01069 | |
| Serpina1b | Serine (or cysteine) peptidase inhibitor, clade A, member 1b | −7.32 | 9.2 × 10−6 | 0.02204 | |
| Serpina1d | Serine (or cysteine) peptidase inhibitor, clade A, member 1d | −6.16 | 1.2 × 10−5 | 0.02311 | |
| Gfap | Glial fibrillary acidic protein | −4.79 | 2.4 × 10−5 | 0.03475 | |
| Defcr20 | Defensin-related cryptdin 20 | −14.1 | 3.5 × 10−5 | 0.04041 | |
| Acta1 | Actin, alpha-1, skeletal muscle | −8.37 | 3.8 × 10−5 | 0.04041 | |
| Fabp6 | Fatty acid-binding protein-6, ileal (gastrotropin) | −176 | 5.6 × 10−5 | 0.04515 | |
| Tns4 | Tensin-4 | −3.35 | 5.9 × 10−5 | 0.04530 | |
| Oprd1 | Opioid receptor, delta-1 | −3.39 | 6.2 × 10−5 | 0.04654 | |
| Ahrr | Aryl hydrocarbon receptor repressor | –6.07 | 6.7 × 10−5 | 0.04675 | |
| Cyb5r2 | Cytochrome b5 reductase-2 | −4.41 | 6.8 × 10−5 | 0.04675 | |
| Hist1h4k | Histone cluster, 1, H4k | −3.03 | 0.00010 | 0.05778 | |
| Tff1 | Trefoil factor-1 | −4.72 | 0.00013 | 0.06287 | |
| Slc13a2 | Solute-carrier, family 13, subfamily A, member 2 | −2.95 | 0.00014 | 0.06502 | |
| Mxd1 | MAX dimerization protein-1 | −2.93 | 0.00015 | 0.06596 | |
| Cyp2e1 | Cytochrome P450, family 2, subfamily E, 1 (alcohol, eicosanoid metabolism) | −2.98 | 0.00017 | 0.06717 | |
| Bcmo1 | Beta-carotene 15,15’-monooxygenase | −3.18 | 0.00018 | 0.06768 | |
| Hist1h4b | Histone cluster, 1, H4b | −2.75 | 0.00018 | 0.06787 | |
| Hist1h4m | Histone cluster, 1, H4m | −2.56 | 0.00031 | 0.08674 | |
This is a partial list of selected genes; the entire list of down-regulated genes for all three comparisons can be found in Tables S3, S7, S10 & S13 of the Supplementary Data.
False discovery rate, or adjusted P-value. One out of ten adjusted P-values ≤0.10 would be expected to be a false positive.
Oral BaP treatment would be expected to induce CYP1B1 and many other AHR-regulated BaP-metabolizing enzymes. Table S5 lists BaP metabolism-related genes in the PSI that were found to be significantly up- or down-regulated, comparing oral BaP treatment for 4 weeks with untreated mice. Including Cyp1b1, there was a total of eleven P450 genes up- and two down-regulated. There were also nine UDP glucuronosyltransferase genes up-regulated, three glutathione S-transferase genes up-and four down-regulated, four aldehyde dehydrogenase genes up- and one down-regulated, three aldoketoreductase genes up-regulated, the AHR and the AHR repressor genes both up-regulated, and the Nqo2 gene down-regulated. Neither Cyp1a2 nor Nqo1 was on these lists.
Gene Expression during Development of PSI Adenocarcinomas
Comparing 8 weeks of oral BaP (dysplasia seen) with 4 weeks of oral BaP (histology normal) in the Cyp1a1(−/−) mouse (Table S6 & Table S7), we found (FDR ≤0.10; P <0.00031) 14 genes up-regulated, six of which (43%) were immunoglobulin (Ig) genes; increases ranged between 4.2- and 188-fold. We found 16 genes down-regulated, nine of which (56%) were Ig genes; decreases ranged between 3.7- and 31-fold. GO classes of genes that were most perturbed (Table S8) included 97 classes with FDR ≤0.050 and 27 classes with FDR ≤0.0001; the most significant categories (P ≤6.1 × 10−7) included “extracellular region, defense response to bacteria, responses to stimulus and wounding, oxidoreductase activity, and acute-inflammatory and acute-phase response”.
Comparing 12 weeks of oral BaP (neoplasia) with 4 weeks of oral BaP (normal histology) in the Cyp1a1(−/−) mouse (Table S9 & Table S10), we found (FDR ≤0.10; P ≤0.00039) 90 genes up-regulated, 66 of which (73%) were immunoglobulin Ig genes; two-thirds of these Ig genes were kappa chain (Igk) genes while most of the remaining were heavy chain (Igh) genes; increases ranged between 2.4- and 21-fold. We found 44 genes down-regulated, with striking decreases in only one Igk and three histocompatibility-2 (H2) genes but eleven histone cluster genes; decreases ranged between 2.4- and 17-fold. GO classes of genes that were most perturbed (Table S11) included 102 classes with FDR ≤0.050 and 61 classes with FDR ≤0.0001; the most highly significant categories changed markedly between the 8-week-vs-4-week comparison and the 12-week-vs-4-week comparison. The most significant categories (P ≤9.9 × 10−11) now included “nucleosome assembly and DNA-packaging, chromatin assembly or disassembly, protein-DNA complex assembly and chromosomes, and chromosomal organization and biogenesis”.
Comparing 12 weeks of oral BaP (neoplasia) with 8 weeks of oral BaP (dysplasia seen) in the Cyp1a1(−/−) mouse (Table S12 & Table S13), we found (FDR of ≤0.10; P ≤0.00038) 114 genes up-regulated, 79 of which (69%) were Ig genes; ~70% of these Ig genes were Igk genes and 27% were Igh genes; increases ranged between 2.5- and 34-fold. We found 40 genes down-regulated, with striking decreases in two Ig–related genes and four histone cluster genes; decreases ranged between 2.6- and 68-fold. GO classes of genes that were most perturbed (Table S14) included 187 classes with FDR ≤0.050 and 68 classes with FDR ≤0.0001. The most highly significant categories (P ≤1.4 × 10−13) between the 12-week-vs-8-week comparison were quite different from either the 8-week-vs-4-week comparison or the 12-week-vs-4-week comparison. GO categories for the 12-week-to-8-week comparison (Table S14) included “oxidoreductase and monooxygenase catalytic activities, iron ion-binding, six mitochondrial categories, nucleosome assembly, and organic acid and carboxylic acid metabolism”.
Selected subsets of up- and down-regulated genes other than all the immunoglobulin genes for the three oral BaP-treated time-point comparisons are listed in Table 2 & Table 3. Twenty-eight of the highly significant up- and down-regulated genes detected by microarray (at 8 weeks vs 4 weeks, 12 weeks vs 4 weeks, and 12 weeks vs 8 weeks oral BaP) were further quantified using qRT-PCR (Table 4). Out of 40 time-points, 31 were significant (P <0.05) and consistent with the microarray data. Please see the “Discussion” section for speculation about possible genetic networks associated with this form of oral BaP-mediated tumorigenesis.
Table 4.
Results of selected genes up- and down-regulated, as confirmed by qRT-PCRa
| Amount of mRNA at first time-point minus amount of mRNA at second time-point | |||
|---|---|---|---|
| Genes | 8 wks vs 4 wks (P value) |
12 wks vs 4 wks (P value) |
12 wks vs 8 wks (P value) |
| Xist | 6.8 (0.03) | 0.87 (0.77) | |
| Serpina1a | 1.2 (0.8) | 14 (0.006) | |
| Serpina1b | 1.1 (0.04) | 5.7 (0.09) | |
| Serpina1d | 0.71 (0.6) | 3.2 (0.01) | |
| Tff1 | 630.35 (0.001) | 0.01 (0.004) | |
| Eif2s3y | 0.63 (0.42) | 4.0 (0.03) | |
| Ddx3y | 0.23 (0.008) | 4.6 (0.004) | |
| Jarid1d | 1.1 (0.097) | 1.2 (0.039) | |
| Rab30 | 0.35 (0.009) | 10 (0.003) | |
| Nr0b2 | 41 (0.0001) | 5.5 (0.0001) | |
| Bcmo1 | 0.48 (0.5) | 0.77 (0.7) | 0.37 (0.3) |
| Nupr1 | 3.6 (0.02) | ||
| Slpi | 2.9 (0.07) | ||
| Klk1b3 | 7.2 (0.006) | ||
| Hist1h2bb | 0.05 (0.0005) | ||
| Hist1h4d | 0.08 (0.0002) | ||
| Hist2h2be | 0.04 (0.002) | ||
| Hist1h4b | 0.18 (0.001) | ||
| Hist1h4k | 0.06 (0.0006) | ||
| Hist1h4h | 0.07 (0.0003) | ||
| H2-ab1 | 0.03 (0.002) | ||
| H2-dmb2 | 0.05 (0.009) | ||
| H2-dmb1 | 0.05 (0.001) | ||
| Hist1h4f | 0.06 (0.0007) | ||
| Hist1h4i | 0.10 (0.006) | ||
| Hist1h4c | 0.06 (0.0004) | ||
| Hist1h3d | 0.08 (0.0001) | ||
| Hist1h3b | 0.08 (0.001) | ||
Primers selected for each of these genes are available upon request. P-values > 0.05 are shown in blue.
CYP1 and PTGS2 mRNA Levels in PSI and Liver
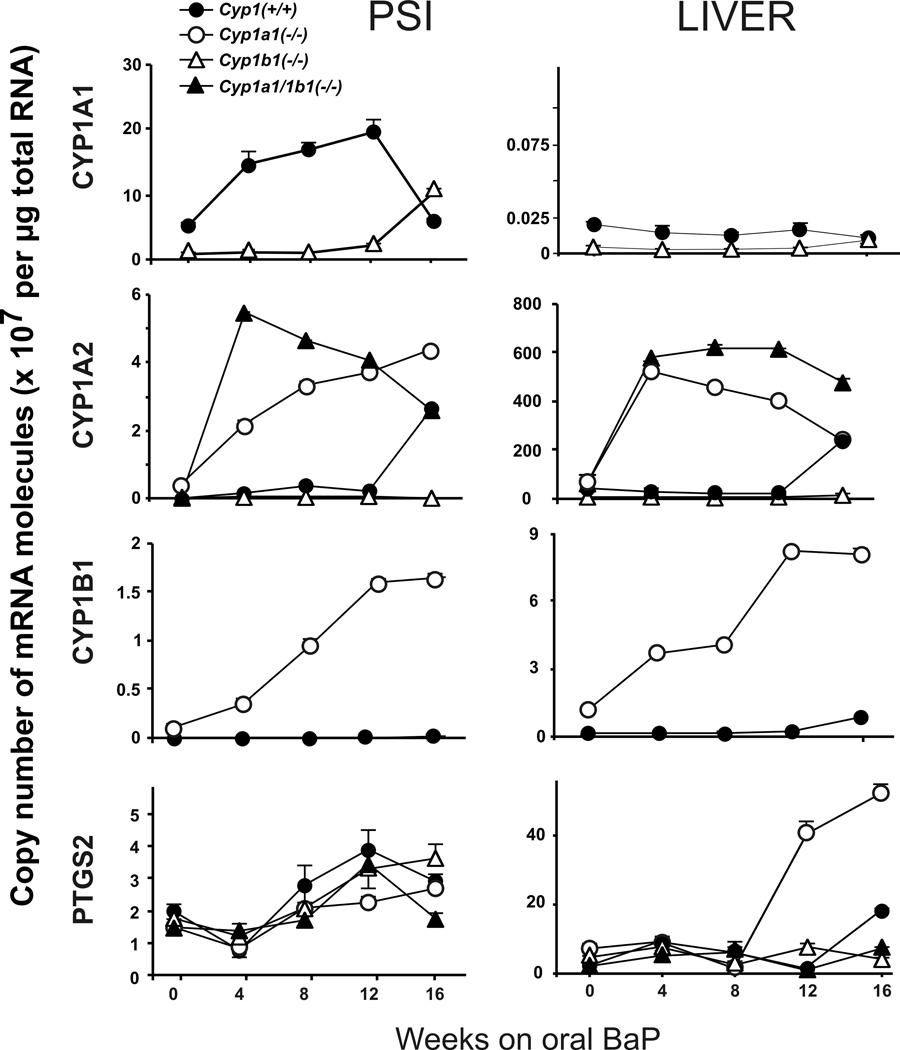
CYP1B1 mRNA levels in the stomach and PSI of Cyp1a1(−/−) knockout mice receiving oral BaP exhibit an apparent compensatory up-regulation that is 15- to 25-fold higher than that in Cyp1(+/+) wild-type mice27. Because cyclooxygenases-2 (PTGS2) is also known to be inducible by BaP and to metabolically activate BaP to its highly mutagenic and carcinogenic 7,8-diol-trans-9,10-epoxide33, we chose to examine the expression levels of this mRNA as well. The four mRNA levels in PSI vs liver from the four genotypes were compared (Fig. 4), over the course of 16 weeks of oral BaP. The increases and decreases of these mRNAs have previously been shown to very highly correlate with increased or decreased levels of the corresponding proteins27.
Figure 4.
Time-course of CYP1A1, CYP1A2, CYP1B1, and PTGS2 mRNA levels in the PSI, liver and PGD from Cyp1(+/+), Cyp1a1(−/−) and Cyp1a1/1b1(−/−) mice over the course of 16 weeks of oral BaP at 12.5 mg/kg/day. Brackets denote S.E.M.; when bracket cannot be seen, the S.E.M. is within the size of the symbol.
Confirming the previous study27, CYP1A1 mRNA became strikingly elevated in the PSI of oral BaP-treated Cyp1(+/+) mice (Fig. 4, top row); at zero-time, CYP1A1 mRNA levels were detectable above baseline and presumably reflect inducers in the lab chow. It is noteworthy that CYP1A1 mRNA levels in liver are virtually undetectable in wild-type and Cyp1b1(−/−) mice, at these time-points. In a kinetics-of-CYP1A1-induction study (Shi and Nebert, in preparation), it has been found that intestinal CYP1A1 induction is so robust in oral BaP-treated Cyp1(+/+) mice that BaP is detoxified and cleared rapidly––thereby resulting in negligible amounts of BaP inducer reaching the liver, thus resulting in CYP1A1 de-induction, following an initial hepatic CYP1A1 induction. In Cyp1b1(−/−) mice, CYP1A1 mRNA was very low but appears to increase in the PSI between 12 and 16 weeks on oral BaP. No CYP1A1 mRNA was detectable in either Cyp1a1(−/−) or Cyp1a1/1b1(−/−) mice at any time-point.
Consistent with the previous study27, oral BaP-induced CYP1A2 mRNA shows a compensatory up-regulation in PSI and liver of both Cyp1a1(−/−) and Cyp1a1/1b1(−/−), but not Cyp1b1(−/−) mice (Fig. 4, 2nd row). An explanation for this phenomenon remains unknown. In both PSI and liver, CYP1A2 mRNA peculiarly became significantly elevated at the 16-week time-point of oral BaP treatment in Cyp1(+/+) mice.
With regard to PTGS2 mRNA levels, there were no remarkable differences in the PSI from any of the four genotypes studied (Fig. 4, bottom row). Curiously, in the liver, very large increases in PTGS2 mRNA occurred in Cyp1a1(−/−) mice between 8 weeks and 16 weeks on oral BaP.
Consistent with the previous report27, CYP1B1 mRNA levels (normally very low in PSI and liver) were strikingly up-regulated in the absence of the Cyp1a1 gene, compared with no changes in CYP1B1 mRNA levels in wild-type mice (Fig. 4, 3rd row); this was seen both in PSI––the location where the oral BaP-induced adenocarcinoma forms––and in liver. It is possible that CYP1B1 overexpression in Cyp1a1(−/−) knockout mice might be involved in causation of the PSI tumorigenesis. However, CYP1B1 mRNA was even more elevated in liver than in PSI of Cyp1a1(−/−) mice, but no hepatic tumors were found during this oral BaP regimen.
Characteristics of Squamous Cell Carcinoma of PGD
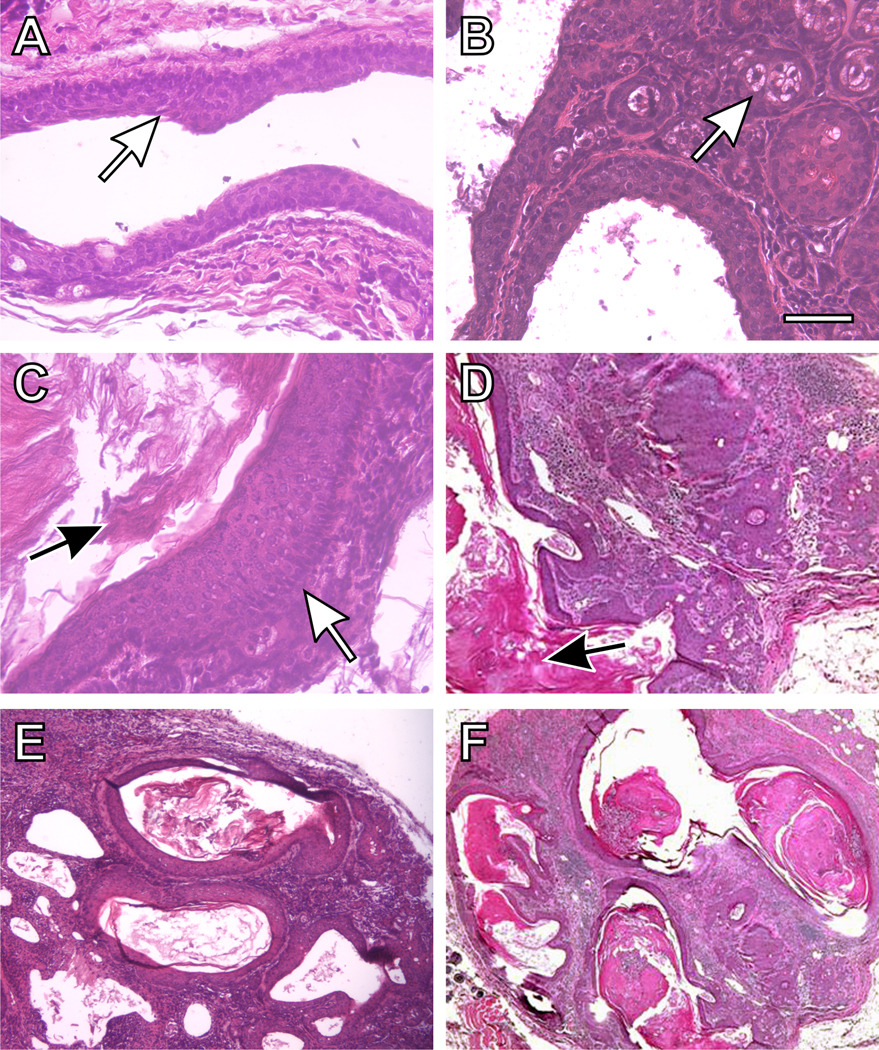
The normal duct is illustrated (Fig. 5, panels A & B) from the preputial gland to the skin surface: there is stratified squamous epithelium with low-cuboidal adjacent secretory acinar cells. Upon irritation or inflammation in Cyp1a1/1b1(−/−) mice receiving oral BaP for 12 weeks, the cuboidal epithelial cells appear to undergo alteration to form stratified squamous epithelia––which produce excessive amounts of keratin (Fig. 6, panels C through F). There are multifocal squamous epithelial cell islands with severe hyperkeratosis, and multiple small clusters of squamous epithelial cells breaking off in the larger keratin-laden cysts. Subcutaneous suppurative inflammation can also be observed, which appears to be associated with keratin disposition. The oral BaP-induced squamous cell carcinomas were sometimes accompanied by secondary abscesses.
Figure 5.
Histology of a typical preputial gland duct of Cyp1a1/1b1(−/−) double-knockout mice (following 12 weeks of oral BaP, 12.5 mg/kg/day), showing squamous cell carcinoma of the duct with invasion into the surrounding tissues. (A) normal preputial duct morphology with a stratified squamous cell epithelial lining of the larger ducts (white arrow). (B) cuboidal holocrine secretory acinar cells (white arrow). (C) a site of thickening of the squamous epithelium (white arrow), and increased keratin production can be seen (C & D black arrows). {D, E & F) excessive accumulation of keratin within ducts and resulting dilatation, and an epithelial morphology characteristic of squamous cell carcinoma. There is thickening of the lining of the tubular glands, with an abscess and excessive inflammation. The overall architecture is disorganized, and the amount of stroma between glands is increased. Bar = 100 microns for all images.
Discussion
This study shows how “complicated” environmentally-caused cancer can be. While receiving the same dose (12.5 mg/kg/day) of the same procarcinogen BaP by the same route-of-administration (and oral BaP is the most common route of entry into humans, whether smoking cigarettes or eating charcoal-grilled meat): the wild-type mouse shows no malignancies, removal of one gene (Cyp1a1) leads to an unusual form of adenocarcinoma in the PSI (Fig. 2 & Fig. 3); removal of two genes (Cyp1a1 plus Cyp1b1) protects against adenocarcinoma of the PSI, but the mice now develop squamous cell carcinoma of the PGD (Fig. 5). When the Cyp1b1 gene alone is absent, the clinical outcome is no different from that of the wild-type mouse.
There is no CYP1B1 mRNA or functional enzyme in the Cyp1a1/1b1(−/−) double-knockout (Fig. 4, 3rd row). Thus, it is possible that the compensatory up-regulation of CYP1B1 levels in the PSI of Cyp1a1(−/−) mice is associated with the initiation and/or progression of PSI adenocarcinoma––perhaps via BaP metabolism by the CYP1B1 enzyme; this hypothesis is being explored further in our lab. This hypothesis is further supported by absence of CYP1B1 in the Cyp1a1/1b1(−/−) mouse resulting in no PSI tumors. However, why there are no liver tumors in oral BaP-treated Cyp1a1(−/−) mice, despite a compensatory up-regulation of hepatic CYP1B1 (Fig. 4, 3rd row), will require further study.
PAHs are known to cause skin papillomas and epithelial squamous cell carcinomas, in numerous locations where the procarcinogen is applied topically2. PAHs can also produce subcutaneous fibrosarcomas, following a subcutaneously-administered PAH1,34,35. PAHs are well known to cause lung cancer, when the procarcinogen is presented by inhalation or intratracheal administration1,36,37. Moreover, PAHs are known to cause urinary bladder papillomas and epithelial cancers, when the chemical is in direct contact with the urothelial cells38. Epidemiological studies have shown that human cigarette smokers exhibit an increased risk of lung, head-and-neck, pancreas and urinary tract neoplasms39. PAHs given orally to mice can cause tumors of the oral mucosa, tongue, esophagus and forestomach40,41. Most of these examples involve the parent PAH in direct contact with the tissue or cell type in which the malignancy develops. One tissue that has always appeared remarkably recalcitrant to PAH-induced cancer, however, is the small intestine2,42. Development of PSI adenoma and adenocarcinoma in oral BaP-treated Cyp1a1(−/−) mice, as reported here, therefore appears to be unique.
There are numerous pathways involving genes found to be most prominently up- and down-regulated in the Cyp1a1(−/−) PSI, as tumorigenesis progresses between 4 weeks and 12 weeks of oral BaP (Table 2 & Table 3; Table S6–Table S14). Comparing 8 weeks with 4 weeks of treatment, Xist expression is 70-fold increased, three Serpin (clade A) genes are 6- to 15-fold increased, and Tff1 expression is 10-fold increased. Comparing 12 weeks with 8 weeks of oral BaP, Xist is 68-fold decreased, the three Serpina1 genes are 6- to 10-fold decreased, and Tff1 is 4.7-fold decreased. During these same times, Eif2s3y, Ddx3y, Jarid1d and Rab30 are reciprocally 5- to 30-fold decreased (comparing 8 weeks with 4 weeks), and then reciprocally 4.2- to 34-fold increased (comparing 12 weeks with 8 weeks). These data (summarized in Table 4) are consistent with extremely high levels of Xist, Serpina1 and Tff1 expression, compared with extremely low levels of Eif2s3y, Ddx3y, Jarid1d and Rab30 expression––occurring around the 8-week time-point.
The Xist gene is located on the X chromosome, whereas the Eif2s3y, Ddx3y and Jarid1d genes are located on the Y chromosome. Large noncoding RNAs such as XIST are required for epigenetic silencing of multiple genes not only in cis but also possibly in trans43. Perhaps BaP reactive metabolites (formed by elevated CYP1B1 in the PSI) activate the Xist gene by an epigenetic mechanism, which then silences these three genes on the Y chromosome. XIST works as a functional RNA that recruits repressive chromatin factors, by altering histone-tail modifications and DNA methylation at the Xist promoter44; the presence of unmethylated Xist promoter is frequently associated with tumorigenesis, especially stem-cell-derived tumors45, which the PSI adenocarcinoma might be. Jarid1d1 is known to be X-inactivated and encodes a histocompatibility Y-antigen46; perhaps related to this finding, six histone cluster genes and three H2 genes are also significantly down-regulated when comparing 12 weeks with 4 weeks of BaP treatment (Table 4; Table S10).
Interestingly, the striking up- and down-regulation of the Serpina1a, Serpina1b and Serpina1d genes (located on chromosome 12) go hand-in-hand with the Xist and Tff1 genes (Table S10). Serpins (serum proteases) are components of the immune system––playing a role in migration, phagocytosis and elimination of cancerous cells; some serpins help combat tissue damage and cell death47. Trefoil factor-1 (TFF1) is a tumor suppressor product involved in the GI tract, protects gut mucosa from environmental insults, maintains the mucosal barrier, and participates in folding secreted proteins inside the endoplasmic reticulum48,49. Trefoil factors promote cell survival, growth, and motility and are regulated by various cancer-causing stimuli50. Topically-applied PAHs are well known to cause irritation, inflammation, and the acute-phase response. Rapid repair of GI tract mucous epithelia is essential for preventing inflammation, which can be a critical component in cancer progression51. Of note, Tff1 expression is highest at the 8-week time-point of oral BaP (Table 2 & Table 3)
The Rab30 gene52,53 (member of the RAS family) is 4.6-fold down-regulated between the 8-week and 4-week time-points and 4.2-fold up-regulated between the 12-week and 8-week time-points (Table S7 & Table S12). Conversely, expression of the Nr0b2 tumor suppressor gene (member of the RAS family and downstream target of the farnesoid X receptor)54 is 8.8-fold up-regulated between the 8-week vs 4-week time-points and 8.4-fold up-regulated between the 12-week vs 4-week time-points (Table S6 & Table S9). These data are consistent with the Rab30 oncogene expression being activated at 4 and 12 weeks, and the Nr0b2 tumor suppressor gene expression is low at 4 weeks and increased at both 8 and 12 weeks, of oral BaP.
Comparing 8 weeks with 4 weeks of oral BaP, one sees many genes up-regulated that are associated with bacterial infection, wounding, and inflammatory and acute-phase responses. A model has been proposed55 which defines six properties that a tumor acquires––including uncontrolled growth, immortality, and the capacity to invade other tissues. It has been suggested that this model should be revised to include cancer-related inflammation56,57.
It is curious that one often finds olfactory receptor genes both highly up- and down-regulated in the PSI (Table S2, Table S7, Table S10 & Table S12). These data support the findings of others and suggest that these receptors might participate in critically important life functions (e.g. perception of foreign chemicals) beyond their role of olfaction in nasal olfactory epithelial cells58.
Three genes worthy of mention are highly increased, only when comparing 12 weeks with 4 weeks of oral BaP (Table 2 & Table 4; Table S9). Nupr1 expression is 4.4-fold elevated. NUPR1, first identified during acute pancreatitis in the rat and then as a gene product up-regulated in human metastatic breast cancer, has begun to be appreciated as playing a role in the progression of several malignancies including those of breast, thyroid, brain and pancreas59. Slpi expression is 4.4-fold augmented. Secretory leukocyte peptidase inhibitor (SLPI), produced by epithelial cells of the GI tract, has antimicrobial and anti-protease functions; Slpi expression is up-regulated during wound-healing and also often up-regulated during tumorigenesis60. Klk1b3 expression is increased 3-fold; KLK1B3 is a member of the kallikreins, a subgroup of extracellular serine proteases that are often up-regulated by serpins during various types of stress, inflammation and tumorigenesis61.
Perhaps the most unexpected finding in the microarray expression data (Table S6–Table S12) includes the up-regulation of immune genes during development of the PSI neoplasm: 73% of 90 genes most up-regulated were Ig genes with 67% Igk and 29% Igh genes, when comparing 12 weeks with 4 weeks of oral BaP; and 69% of 114 genes most up-regulated were Ig genes with 70% Igk and 27% Igh, when comparing 12 weeks with 8 weeks of oral BaP. Also, of 44 genes most down-regulated when comparing 12 weeks with 4 weeks of oral BaP, there were three H2 genes and 11 histone cluster genes. This pattern would suggest infiltration by B-lymphocytes, or perhaps by plasmacytoid dendritic cells62. However, the Fig. 3 data show no suggestion of immune-derived cells and confirm that this PSI tumor is indeed an adenocarcinoma. These findings are consistent with the growing evidence that Igk and Igh gene expression can be paradoxically expressed in a variety of malignancies of epithelial origin63.
It has become increasingly appreciated that the mucosal immune system of the GI tract has evolved as a distinct immune organ––functioning in parallel with its systemic counterpart64. There is also accumulating evidence that Paneth cells participate in intestinal inflammation and might be involved in Ig-mediated acquired immunity of the GI tract65; in fact, interleukin-17, a pro-inflammatory cytokine known to be produced principally by activated T-cells, has recently been shown to be rapidly released from the granules inside Paneth cells66. It is also perhaps relevant that oral high-dose BaP (125 mg/kg/day) causes immunosuppression within 2–3 weeks8,9, whereas with oral BaP at one-tenth of the high dose (in the present study), these PSI adenocarcinomas exhibit striking overexpression of numerous Igk and Igh genes.
Lastly, tumors of the preputial gland commonly occur in rodents, when tested with a large number of environmental chemicals67. PAHs such as BaP specifically cause hyperkeratosis of the PGD epithelium. We speculate that keratin-plugging of secretions from the preputial gland could then lead to pruritus, followed by the mouse scratching itself and, thus, secondary infections. BaP-induced hyperkeratinization, plus BaP being a well-known tumor promoter, plus secondary inflammation––might all combine to cause both initiation and promotion of squamous cell carcinoma in the PGD. In some ways, BaP-induced hyperkeratinization of the preputial gland duct might be similar to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced hyperkeratinization of human sebaceous glands, which results in the exemplary model of chloracne in dioxin-exposed populations68.
In the PGD epithelium, it would appear that CYP1B1 metabolism might be beneficial, i.e. causing BaP detoxication, because these tumors only appear when the Cyp1b1 gene has been ablated. When the body burden of BaP is high, as in the Cyp1a1(−/−) mouse, BaP metabolism by CYP1B1 in the PGD might function to detoxify BaP. When the body burden of BaP is even 3-fold higher, as in the Cyp1a1/1b1(−/−) mouse, both CYP1A1 and CYP1B1 are absent in the PGD; thus, neither enzyme can detoxify the PAH parent compound. It is postulated that either (i) there is compensatory up-regulation of a new gene in the PGD that encodes an enzyme responsible for metabolic activation of BaP, or (ii) chronic irritation and/or inflammation caused by the parent BaP molecule rather than BaP metabolites lead to this squamous cell carcinoma of the PGD. Plans are underway to examine closely cDNA microarray expression of genes (also qRT-PCR of selected mRNAs and Western immunoblots of selected proteins) in the PGD, as we have done for the PSI, during this same 12-week regimen of oral BaP at 12.5 mg/kg/day in the Cyp1a1/1b1(−/−) mouse versus the Cyp1a1(−/−) mouse. We also plan to extend these studies to mice receiving oral BaP at 1.25 mg/kg/day.
Supplementary Material
Table 5.
Summary of genes most strikingly up- and down-regulated, when comparing the 4-, 8- and 12-week oral BaP treatment time-points
| 8 wks/4 wks | 12 wks/4 wks | 12 wks/8 wks |
|---|---|---|
| Xist | Xist | |
| Serpina1a, a1b, a1d | Serpina1a, a1b, a1d | |
| Tff1 | Tff1 | |
| HIGHLY UP | HIGHLY DOWN | |
| Eif2s3y | Eif2s3y | |
| Ddx3y | Ddx3y | |
| Jarid1d | Jarid1d | |
| Rab30 | Rab30 | |
| HIGHLY DOWN | HIGHLY UP | |
| Nr0b2UP | Nr0b2UP | |
| Bcmo1DOWN | Bcmo1DOWN | Bcmo1DOWN |
| Nupr1UP | ||
| SlpiUP | ||
| Klk1b3UP | ||
| ElevenHistgenes DOWN | ||
| ThreeH2genes DOWN | ||
Acknowledgments
We thank Drs. Ying Chen, Timothy P. Dalton, Ron Jandacek, Chris L. Karp, Susan Kasper, and Patrick Tso for valuable discussions and careful readings of this manuscript. We thank Mary Rolfes and Amy Opoka for technical assistance with the immunohistochemistry. We appreciate the microarray assistance and statistical support of Danielle Halbleib, Saikumar Karyala, Mario Medvedovic, and Maureen A Sartor. This work was supported by NIH Grants R01 ES014403 and P30 ES06096.
Contributor Information
Zhanquan Shi, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056.
Nadine Dragin, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056.
Marian L. Miller, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056
Keith F. Stringer, Department of Pathology, Cincinnati Children’s Hospital Medical Center, Cincinnati OH 45229
Elisabet Johansson, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056.
Jing Chen, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056.
Shigeyuki Uno, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056.
Frank J. Gonzalez, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute National Institutes of Health, Bethesda, MD 20892
Carlos A. Rubio, Gastrointestinal and Liver Pathology Research Laboratory, Karolinska Institute, 171 76 Stockholm, Sweden
Daniel W. Nebert, Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, P.O. Box 670056, Cincinnati OH 45267-0056.
References
- 1.Pelkonen O, Nebert DW. Metabolism of polycyclic aromatic hydrocarbons: etiologic role in carcinogenesis. Pharmacol Rev. 1982;34:189–222. [PubMed] [Google Scholar]
- 2.Conney AH, Chang RL, Jerina DM, Wei SJ. Studies on the metabolism of benzo[a]pyrene and dose-dependent differences in the mutagenic profile of its ultimate carcinogenic metabolite. Drug Metab Rev. 1994;26:125–163. doi: 10.3109/03602539409029788. [DOI] [PubMed] [Google Scholar]
- 3.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 4.Nebert DW. The Ah locus: genetic differences in toxicity, cancer, mutation, and birth defects. Crit Rev Toxicol. 1989;20:153–174. doi: 10.3109/10408448909017908. [DOI] [PubMed] [Google Scholar]
- 5.Miller KP, Ramos KS. Impact of cellular metabolism on the biological effects of benzo[a]pyrene and related hydrocarbons. Drug Metab Rev. 2001;33:1–35. doi: 10.1081/dmr-100000138. [DOI] [PubMed] [Google Scholar]
- 6.Rubin H. Synergistic mechanisms in carcinogenesis by polycyclic aromatic hydrocarbons and by tobacco smoke: a bio-historical perspective with updates. Carcinogenesis. 2001;22:1903–1930. doi: 10.1093/carcin/22.12.1903. [DOI] [PubMed] [Google Scholar]
- 7.Yoshino I, Maehara Y. Impact of smoking status on the biological behavior of lung cancer. Surg Today. 2007;37:725–734. doi: 10.1007/s00595-007-3516-6. [DOI] [PubMed] [Google Scholar]
- 8.Uno S, Dalton TP, Derkenne S, Curran CP, Miller ML, Shertzer HG, Nebert DW. Oral exposure to benzo[a]pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol Pharmacol. 2004;65:1225–1237. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- 9.Uno S, Dalton TP, Dragin N, Curran CP, Derkenne S, Miller ML, Shertzer HG, Gonzalez FJ, Nebert DW. Oral benzo[a]pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Mol Pharmacol. 2006;69:1103–1114. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- 10.Puga A, Maier A, Medvedovic M. The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol. 2000;60:1129–1142. doi: 10.1016/s0006-2952(00)00403-2. [DOI] [PubMed] [Google Scholar]
- 11.Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- 12.Nebert DW, Karp CL. Endogenous functions of the aryl hydrocarbon receptor: intersection of cytochrome P450 (CYP)1-metabolized eicosanoids and AHR biology. J Biol Chem. 2008;283:36061–36065. doi: 10.1074/jbc.R800053200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Järup L. Hazards of heavy metal contamination. Br Med Bull. 2003;68:167–182. doi: 10.1093/bmb/ldg032. [DOI] [PubMed] [Google Scholar]
- 14.Rozman KK, Klaassen CD. Absorption, distribution, and excretion of toxicants. In: Klaassen CD, editor. Casarett & Doull's Toxicology: The Basic Science of Poisons. 7 ed. New York, NY: McGraw-Hill; 2007. pp. 107–132. [Google Scholar]
- 15.Nebert DW, Jensen NM. Benzo[a]pyrene-initiated leukemia in mice. Association with allelic differences at the [Ah] locus. Biochem Pharmacol. 1979;27:149–151. doi: 10.1016/0006-2952(79)90284-3. [DOI] [PubMed] [Google Scholar]
- 16.Dalton TP, Dieter MZ, Matlib RS, Childs NL, Shertzer HG, Genter MB, Nebert DW. Targeted knockout of Cyp1a1 gene does not alter hepatic constitutive expression of other genes in the mouse [Ah] battery. Biochem Biophys Res Commun. 2000;267:184–189. doi: 10.1006/bbrc.1999.1913. [DOI] [PubMed] [Google Scholar]
- 17.Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Cytochrome P450 CYP1B1 determines susceptibility to 7,12-dimethylbenz[a]anthracene-induced lymphomas. Proc Natl Acad Sci U S A. 1999;96:1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nebert DW, Dalton TP, Stuart GW, Carvan MJ., III "Gene-swap knock-in" cassette in mice to study allelic differences in human genes. Ann N Y Acad Sci. 2000;919:148–170. doi: 10.1111/j.1749-6632.2000.tb06876.x. [DOI] [PubMed] [Google Scholar]
- 19.Robinson JR, Felton JS, Levitt RC, Thorgeirsson SS, Nebert DW. Relationship between "aromatic hydrocarbon responsiveness" and the survival times in mice treated with various drugs and environmental compounds. Mol Pharmacol. 1975;11:850–865. [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative Ct method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Rubio CA. Gastric duodenal metaplasia in duodenal adenomas. J Clin Pathol. 2007;60:661–663. doi: 10.1136/jcp.2006.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sartor MA, Schwanekamp J, Halbleib D, Mohamed I, Karyala S, Medvedovic M, Tomlinson CR. Microarray results improve significantly as hybridization approaches equilibrium. Biotechniques. 2004;36:790–796. doi: 10.2144/04365ST02. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear models and empirical Bayesian methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 24.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate-controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 25.Sartor MA, Leikauf GD, Medvedovic M. LRPath: A logistic regression analysis for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uno S, Dragin N, Miller ML, Dalton TP, Gonzalez FJ, Nebert DW. Basal and inducible CYP1 mRNA quantitation and protein localization throughout the mouse gastrointestinal tract. Free Radic Biol Med. 2008;44:570–583. doi: 10.1016/j.freeradbiomed.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M, Spitler J, Rowley DR. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003;9:4792–4801. [PubMed] [Google Scholar]
- 29.Gendreau KM, Whalen GF. What can we learn from the phenomenon of preferential lymph node metastasis in carcinoma? J Surg Oncol. 1999;70:199–204. doi: 10.1002/(sici)1096-9098(199903)70:3<199::aid-jso11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Bilimoria KY, Bentrem DJ, Stewart AK, Talamonti MS, Winchester DP, Russell TR, Ko CY. Lymph node evaluation as a colon cancer quality measure: a national hospital report card. J Natl Cancer Inst. 2008;100:1310–1317. doi: 10.1093/jnci/djn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka S, Kuwahara S, Nishijima K, Ohno T, Matsuzawa A. Genetic association of mutation at agouti locus with adrenal X zone morphology in BALB/c mice. Exp Anim. 2006;55:343–347. doi: 10.1538/expanim.55.343. [DOI] [PubMed] [Google Scholar]
- 32.Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M, Scialpi F, Malatesta M, Zocchi L, Browne G, Ciechanover A, Bernassola F. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15:1103–1112. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- 33.Marnett LJ, Panthananickal A, Reed GA. Metabolic activation of 7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene during prostaglandin biosynthesis. Drug Metab Rev. 1982;13:235–247. doi: 10.3109/03602538209029998. [DOI] [PubMed] [Google Scholar]
- 34.Kouri RE, Rude TH, Joglekar R, Dansette PM, Jerina DM, Atlas SA, Owens IS, Nebert DW. 2,3,7,8-Tetrachlorodibenzo-p-dioxin as cocarcinogen causing 3-methylcholanthrene-initiated subcutaneous tumors in mice genetically "nonresponsive" at Ah locus. Cancer Res. 1978;38:2777–2783. [PubMed] [Google Scholar]
- 35.Lubet RA, Connolly GM, Nebert DW, Kouri RE. Dibenz[a,h]anthracene-induced subcutaneous tumors in mice Strain sensitivity and the role of carcinogen metabolism. Carcinogenesis. 1983;4:513–517. doi: 10.1093/carcin/4.5.513. [DOI] [PubMed] [Google Scholar]
- 36.Conney AH. Induction of microsomal enzymes by foreign chemicals, carcinogenesis by polycyclic aromatic hydrocarbons: G.H.A Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 37.Armstrong B, Hutchinson E, Unwin J, Fletcher T. Lung cancer risk after exposure to polycyclic aromatic hydrocarbons: a review and meta-analysis. Environ Health Perspect. 2004;112:970–978. doi: 10.1289/ehp.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langenbach R, Malick L, Nesnow S. Rat bladder cell-mediated mutagenesis of Chinese hamster V79 cells and metabolism of benzo[a]pyrene. J Natl Cancer Inst. 1981;66:913–917. [PubMed] [Google Scholar]
- 39.Carbone D. Smoking and cancer. Am J Med. 1992;93:13S–17S. doi: 10.1016/0002-9343(92)90621-h. [DOI] [PubMed] [Google Scholar]
- 40.Culp SJ, Gaylor DW, Sheldon WG, Goldstein LS, Beland FA. A comparison of the tumors induced by coal tar and benzo[a]pyrene in a 2-year bioassay. Carcinogenesis. 1998;19:117–124. doi: 10.1093/carcin/19.1.117. [DOI] [PubMed] [Google Scholar]
- 41.Kim TW, Chen Q, Shen X, Regezi JA, Ramos DM, Tanaka H, Jordan RC, Kramer RH. Oral mucosal carcinogenesis in SENCAR mice. Anticancer Res. 2002;22:2733–2740. [PubMed] [Google Scholar]
- 42.Buening MK, Wislocki PG, Levin W, Yagi H, Thakker DR, Akagi H, Koreeda M, Jerina DM, Conney AH. Tumorigenicity of the optical enantiomers of the diastereomeric benzo[a]pyrene 7,8-diol-9,10-epoxides in newborn mice: exceptional activity of (+)-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. Proc Natl Acad Sci USA. 1978;75:5358–5361. doi: 10.1073/pnas.75.11.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umlauf D, Fraser P, Nagano T. The role of long non-coding RNAs in chromatin structure and gene regulation: variations on a theme. Biol Chem. 2008;389:323–331. doi: 10.1515/BC.2008.047. [DOI] [PubMed] [Google Scholar]
- 44.Shibata S, Wutz A. Transcript versus transcription? Epigenetics. 2008;3:246–249. doi: 10.4161/epi.3.5.6990. [DOI] [PubMed] [Google Scholar]
- 45.Lind GE, Skotheim RI, Lothe RA. The epigenome of testicular germ cell tumors. APMIS. 2007;115:1147–1160. doi: 10.1111/j.1600-0463.2007.apm_660.xml.x. [DOI] [PubMed] [Google Scholar]
- 46.Kent-First MG, Maffitt M, Muallem A, Brisco P, Shultz J, Ekenberg S, Agulnik AI, Agulnik I, Shramm D, Bavister B, bdul-Mawgood A, VandeBerg J. Gene sequence and evolutionary conservation of human SMCY. Nat Genet. 1996;14:128–129. doi: 10.1038/ng1096-128. [DOI] [PubMed] [Google Scholar]
- 47.Mangan MS, Kaiserman D, Bird PI. The role of serpins in vertebrate immunity. Tissue Antigens. 2008;72:1–10. doi: 10.1111/j.1399-0039.2008.01059.x. [DOI] [PubMed] [Google Scholar]
- 48.Tomasetto C, Rio MC. Pleiotropic effects of Trefoil Factor 1 deficiency. Cell Mol Life Sci. 2005;62:2916–2920. doi: 10.1007/s00018-005-5479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki M, Ikeda H, Nakanuma Y. Expression profiles of MUC mucins and trefoil factor family (TFF) peptides in the intrahepatic biliary system: physiological distribution and pathological significance. Progr Histochem Cytochem. 2007;42:61–110. doi: 10.1016/j.proghi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Perry JK, Kannan N, Grandison PM, Mitchell MD, Lobie PE. Are trefoil factors oncogenic? Trends Endocrinol Metab. 2008;19:74–81. doi: 10.1016/j.tem.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann W. Trefoil factors TFF (trefoil factor family) peptide-triggered signals promoting mucosal restitution. Cell Mol Life Sci. 2005;62:2932–2938. doi: 10.1007/s00018-005-5481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinka R, Gillingham AK, Kondylis V, Munro S. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol. 2008;183:607–615. doi: 10.1083/jcb.200808018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 56.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 58.Olender T, Lancet D, Nebert DW. Update on the olfactory receptor (OLFR) gene superfamily. Hum Genomics. 2008;3:87–97. doi: 10.1186/1479-7364-3-1-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chowdhury UR, Samant RS, Fodstad O, Shevde LA. Emerging role of nuclear protein-1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 2009;28:225–232. doi: 10.1007/s10555-009-9183-x. [DOI] [PubMed] [Google Scholar]
- 60.Nukiwa T, Suzuki T, Fukuhara T, Kikuchi T. Secretory leukocyte peptidase inhibitor and lung cancer. Cancer Sci. 2008;99:849–855. doi: 10.1111/j.1349-7006.2008.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pampalakis G, Sotiropoulou G. Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim Biophys Acta. 2007;1776:22–31. doi: 10.1016/j.bbcan.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Kelsall B. Recent progress in understanding the phenotype and function of intestinal dendritic cells and macrophages. Mucosal Immunol. 2008;1:460–469. doi: 10.1038/mi.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu D, Zheng H, Liu H, Li M, Ren W, Liao W, Duan Z, Li L, Cao Y. Immunoglobulin expression and its biological significance in cancer cells. Cell Mol Immunol. 2008;5:319–324. doi: 10.1038/cmi.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wershil BK, Furuta GT. Gastrointestinal mucosal immunity. J Allergy Clin Immunol. 2008;121(2 Suppl):S380–S383. doi: 10.1016/j.jaci.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 65.Tang QJ, Wang LM, Tao KZ, Ge CR, Li J, Peng YL, Jiang CL, Geng MY. Expression of polymeric immunoglobulin receptor mRNA and protein in human paneth cells: Paneth cells participate in acquired immunity. Am J Gastroenterol. 2006;101:1625–1632. doi: 10.1111/j.1572-0241.2006.00605.x. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi N, Vanlaere I, de RR, Cauwels A, Joosten LA, Lubberts E, van den Berg WB, Libert C. IL-17 produced by Paneth cells drives TNF-induced shock. J Exp Med. 2008;205:1755–1761. doi: 10.1084/jem.20080588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitsumori K, Elwell MR. Proliferative lesions in the male reproductive system of F344 rats and B6C3F1 mice: incidence and classification. Environ Health Perspect. 1988;77:11–21. doi: 10.1289/ehp.887711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pesatori AC, Consonni D, Bachetti S, Zocchetti C, Bonzini M, Baccarelli A, Bertazzi PA. Short- and long-term morbidity and mortality in the population exposed to dioxin after the "Seveso accident". Industr Health. 2003;41:127–138. doi: 10.2486/indhealth.41.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.