Abstract
The conjugation of biomolecules such as proteins and peptides to semiconductor quantum dots (QD) is a critical step in the development of QD-based imaging probes and nanocarriers. Such protein-QD assemblies can have a wide range of biological applications including in vitro protein assays and live-cell fluorescence imaging. One conjugation scheme that has a number of advantages is the self-assembly of biomolecules on a QD surface via polyhistidine coordination. This approach has been demonstrated using QDs that have different coating types, resulting in different interactions between the biomolecule and QD surface. Here we report the use of a fluorescence resonance energy transfer (FRET) assay to evaluate the self-assembly of fluorescent proteins on the surface of QDs with eight distinct coatings, including two used in commercial preparations. The results of this systematic comparison can provide a basis for rational design of self-assembled biomolecule-QD complexes for biomedical applications.
INTRODUCTION
Since quantum dots (QDs) were first rendered water soluble, thereby making them available for biological applications (1, 2), extensive studies have been performed using QD-based approaches for imaging, biosensing and cellular delivery (3–6). However, assembly of biomolecule-QD constructs has proven to be a challenge even as orthogonal bioconjugation techniques become available (7). An optimal bioconjugation strategy would be quick and facile with high affinity, would not require subsequent purification, and would enable control over the conjugate valence, all while not disrupting the biological function of the biomolecule. Standard protocols include chemical coupling with covalent bonds (7), non-covalent interactions exploiting ligand-receptor affinities such as biotin-avidin binding (8), and more general affinity-driven self-assemblies using electrostatic interactions (9, 10) or metal chelation (11–16). Each of these techniques has advantages and disadvantages that must be assessed when choosing the bioconjugation method for a particular design of the biomolecule-QD construct.
Covalent coupling provides the most durable attachment of a biomolecule to a QD with carbodiimide (EDC) chemistry being the most commonly utilized. However, this often causes nanoparticle aggregation, may result in a relatively low number of biomolecules per nanoparticle compared to the reaction ratios, and often requires purification steps that are also a source of loss, depressing the overall yield of the functional construct (7). In addition, the heterogeneous conjugation may cause proteins to attach in an unproductive manner or to lose activity. Streptavidin-biotin binding is a common and useful coupling mechanism that utilizes exceptionally simple protocols, provided the constituent components are already functionalized with the required streptavidin and biotin moieties. With femtomolar binding affinities, the streptavidin-biotin interaction is often considered to be effectively as stable as a covalent bond (17). In some applications, however, the advantages of the streptavidin-biotin interaction may be overshadowed by the tetrameric nature of streptavidin, which can result in multiple binding events and induce aggregation, two undesired and detrimental outcomes (18). The addition of multiple streptavidin molecules (with a molecular weight of ca. 53 kD) to a single QD adds considerably to the overall size of the nanoparticle construct. This extra protein bulk could be detrimental in applications where the increase in size may impair function. Optical biosensors based on fluorescence resonance energy transfer (FRET), for example, require short distances between their fluorescent donors and acceptors. As the efficiency of energy transfer is inversely related to the donor-acceptor distance to the sixth power (19), the additional separation by a few nanometers due to the size of streptavidin can dramatically reduce FRET efficiency.
An alternative conjugation strategy could exploit the affinities between specific biomolecule domains and various substrates. These range in strength and specificity from electrostatic interactions to metal chelation to interactions culled from phage display. Although the binding affinities of these reactions cannot rival that of avidin-biotin, advantages arise because the minimal affinity tag can often be naturally incorporated into the biomolecule of interest and bind to the QD without necessitating additional functionalization of the nanoparticle. Self-assembly based coupling requires only small tags, is facile, is typically not intrusive on the other components of the nanoparticle system, is orthogonal to most biological structures and activity, provides reasonable control over the relative component stoichiometries, and utilizes well-established protocols. This is highly desirable since it fulfills many of the characteristics of ideal nanoparticle bioconjugation chemistry.
A specific example of one such small affinity tag is the polyhistidine sequence (his-tag), which has an affinity for divalent cations. It was discovered that his-tag chelation of metal ions, such as Ni2+, Zn2+, or Cu2+, could be used to purify recombinant proteins using immobilized metal affinity chromatography (IMAC) (20). More recently, his-tags have been used to bind a range of biomolecules to CdSe-ZnS core-shell QDs which inherently display zinc ions on their surface (11–16). Although this coupling technique has been used with success in several QD-based sensing applications, the characteristics of the QD-polyhisitidine interaction appear to vary depending on the particular organic coating used to confer water solubility of the QDs (21).
Conjugation of biomolecules (including proteins) to inorganic nanoparticles plays an essential role in the development and application of nanomedicine, thus a more thorough understanding of the factors that affect his-tag mediated bioconjugation and self-assembly may enable the optimal design of biomolecule-nanoparticle hybrids for a broad range of applications. For example, in molecular imaging applications of QDs, it is critical to establish a robust assay to conjugate purified proteins to the surface of QDs with high affinity, and his-tag mediated bioconjugation is very attractive in that his-tag is typically present on the protein for purification. However, the applicability of this conjugation strategy depends on the specific coating on QDs, and the comparison of his-tag binding to different QDs will provide important insight. Further, the development of quantum dot-fluorescent protein biosensors will be greatly facilitated by a better understanding of the effects of surface ligand on metal-affinity coordination to quantum dots. In addition to expanding our knowledge of his-tag based self-assembly strategy, our demonstration of how fluorescent proteins can be used in FRET assays to study biomolecule-QD association may have a significant implication to the design of other nanoparticle bioconjugates.
In this study, we systematically evaluated the his-tag-mediated self-assembly of biomolecules onto the surface of CdSe/ZnS QDs using a FRET-based assay (Figure 1). The QDs studied here have several different coatings, including phospholipid-PEG and amphiphilic block co-polymers (both rely on hydrophobic interactions to passivate the QD surface), and DHLA-based moieties, which bind the ZnS surface with a thiolate during ligand-exchange. The advantages and disadvantages of using each QD coating scheme in nanoprobe self-assembly are discussed.
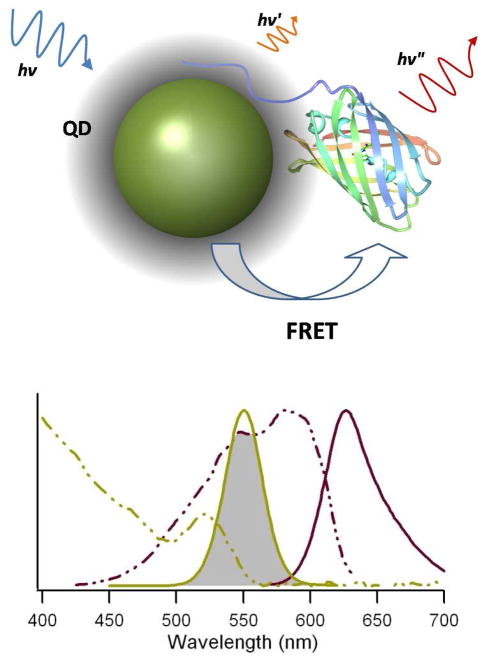
Figure 1.
FRET-based assessment of QD-FP self-assembly. Top—Schematic of the FRET assay used to investigate the his-tag self-assembly on quantum dots. An N-terminal polyhistidine sequence is used to assemble mCherry (PDB 2H5Q) on the surface of the quantum dot. If assembly is successful, the proximity of the QD and FP will facilitate energy transfer and a measurable FRET signal will be detected. Bottom—Absorbance (dashed lines) and emission (solid lines) spectra of a quantum dot (540 nm T2-MP carboxyl-functionalized EviTag (Qd6) shown in green) and mCherry (in red). The shaded area depicts the spectral overlap of the donor emission and acceptor absorbance.
EXPERIMENTAL PROCEDURES
Materials
In-house synthesis and coating of QDs
CdSe/ZnS QDs were synthesized and surface modified with DHLA (QD1), DHLA-PEG600-OH (DHLA-PEG; QD2), or DHLA-PEG750-OCH3 (DHLA-mPEG; QD3) as previously described (22–24). The inorganic core/shell QDs were synthesized in a step-wise reaction using organometallic precursors (e.g., trioctylphosphine selenium (TOP:Se), cadmium acetylacetonate, diethylzinc, and hexamethyldisilathiane) at high temperatures (25–28). Since these nanoparticles are grown and coated with a mixture of trioctylphosphine, trioctylphosphine oxide, and hexadecylamine, they are water insoluble. In order to render them water soluble, the native ligands (TOP/TOPO/HDA) were replaced with the DHLA-based ligands (29).
Commercially-available QDs
Commercial CdSe/ZnS quantum dots (QDs) were acquired from two sources. Qdot® 545 nm ITK carboxyl (QD4) and amino-PEG (QD5) quantum dots were bought from Invitrogen (now Life Technologies, Carlsbad, CA). Three varieties of the T2-MP EviTags™ were purchased from Evident Technologies (Troy, NY). The carboxyl-functionalized (QD6), amine-functionalized (QD7), and non-functionalized EviTags (QD8) vary only in their lipid-PEG terminal group (Note: EviTags are now sold as eFluor™ Nanocrystals by eBioscience in San Diego, CA).,
Rhodamine 6G was acquired from Invitrogen. Luria Bertani (LB) agar, LB broth, ampicillin (Amp), chloramphenicol (Cam), and isopropyl-β-D-thiogalactoside (IPTG) were purchased from US Biological (Swampscott, MA). Borax, Tris-EDTA (TE) buffer, and NiCl2 were procured from Sigma Aldrich (St. Louis, MO).
Quantum Dot Characterization
The hydrodynamic diameter and zeta potential of all eight of the QD varieties were measured, as was their quantum yield (QY). Dynamic light scattering (DLS) was used to measure the hydrodynamic diameter and zeta potential using the Nicomp 280 from Particle Sizing Systems (PSS; Goleta, CA). Each QD stock solution was diluted with distilled water and filtered through a 0.22 μm syringe filter before the hydrodynamic diameter was measured. The same sample was then further diluted with distilled water for measurement of the Zeta potential at pH 7. Subsequently, a KOH solution in water was added to the sample to raise the pH to 9.5 before the Zeta potential was measured again.
The QD quantum yields were measured against rhodamine 6G in water. Absorption and emission spectra of the QDs were measured using an Ultrospec 2100 pro UV-Vis Spectrophotometer (GE Healthcare, Piscataway, NJ) and a Tecan Safire2 multiplate reader (Männedorf, Switzerland), respectively.
Mutagenesis, Expression, and Purification of the Fluorescent Protein mCherry
Engineering and production of the fluorescent protein (FP) mCherry proceeded as previously described (11). A pRSET-B plasmid containing the insert for the fluorescent protein mCherry was kindly provided by Roger Tsien’s laboratory at the University of California, San Diego. This plasmid was modified using PCR mutagenesis to produce the control mCherry, which lacks any terminal polyhistidine sequence, and His6-mCherry, which contains a His6-tag linked to the fluorescent protein with three glycines. Both mutagenesis reactions, which were performed using Phusion High-Fidelity Master Mix (New England Biolabs, Ipswich, MA) in PCR reactions with primers custom synthesized by Integrated DNA Technologies, Inc. (IDT; Coralville, IA), also removed any unnecessary amino acids from the original pRSET-B plasmid, including an existing 6xHis, an Xpress Epitope, an enterokinase site, and extra amino acids arising from the multiple cloning site. The His6-mCherry plasmid was also mutated to create a protein with three site-mutations, S148C I165N Q167M, resulting in a non-fluorescent, non-chromogenic GFP-like protein termed His6-mCherry-NF.
Plasmids coding for mCherry, His6-mCherry, and His6-mCherry-NF were expressed in Rosetta 2 (DE3)s using IPTG induction. Protein purification was performed on an ÄKTAprime plus system (GE Healthcare). The his-tagged proteins were purified on a 1 mL HisTrap HF column, while the untagged protein was purified with a hydrophobic column (HiPrep 16/10 Butyl FF) followed by size-exclusion chromatography (HiPrep 16/60 Sephacryl S-300 HR). The purified proteins were concentrated and buffer-exchanged into PBS using centrifugal filtration devices (Centricon Plus-20; Millipore, Billerica, MA). The proteins were aliquoted into PCR tubes and snap frozen in liquid nitrogen as previously described (30). Protein purity was checked with SDS-PAGE, and protein concentrations were determined using a BCA Assay (Pierce, Rockford, IL).
Calculation of the Spectral Overlap Integrals and Förster Distance
Igor Pro was used as previously described to calculate the overlap integral and Förster distances for each of the possible FRET pairs (19, 31). The spectral overlap integral:
| (1) |
describes the degree of coincidence between the donor emission and the acceptor absorption, where FD-corr designates the normalized emission spectrum of the donor, εA is the molar extinction coefficient of the acceptor, and λ is the wavelength in nanometers. Once the overlap integral was calculated, the Förster distance, Ro, or the distance between the donor and acceptor at which the FRET efficiency is 50%, was determined using the equation:
| (2) |
where k2 is the dipole orientation factor, assumed to be 2/3, QD is the quantum yield of the donor, and n is the refractive index of the medium. Both of these equations were inputted into Igor Pro along with the normalized donor and acceptor spectra and the overlap integral, to calculate the Förster distance for each FRET pair.
FRET Assays
Assays assessing the binding of his-tagged mCherry to diverse QDs were conducted by measuring the FRET efficiency of the various pairs in black, flat-bottomed, non-binding 384-well plates (Corning). QDs and FPs were mixed directly in the well plates with a final QD concentration of 50 nM and 0 to 12 molar equivalents of protein per QD in either 10 mM tetraborate buffered saline, pH 9.5, or 10 mM tetraborate buffer, 1M NaCl, pH 9.5, as indicated. After allowing at least 15 minutes for self-assembly, the emission spectra of each FRET pair was measured in a Tecan Safire2 multiplate reader with an excitation wavelength of 400 nm, excitation bandwidth of 10 nm, emission bandwidth of 5 nm, integration time of 100 μs, and a stepsize of 2 nm. All of the assays were performed in triplicate, as were FP-only controls. For measurements involving the use of supplemented Ni2+, the spectra were measured first with just the QDs and FPs present, then 1 μL of 20 mM NiCl2 was added to the 80 μL reaction volume, resulting in a final Ni2+ concentration around 250 μM. The solutions were incubated at room temperature for at least fifteen minutes to allow binding to reach equilibrium and then the emission spectra were again measured. In this study, the donor-acceptor distance is assumed to be the distance between the center of the QD and the fluorescent protein chromophore. Since FRET signal is generated only when the QD and fluorescent proteins are within ~20 nm, unbound fluorescent proteins contribute little to the overall signal, thus were not removed from the sample prior to measuring the FRET signal.
The background emission resulting from detector gain and any direct excitation of mCherry was subtracted from each of the FRET spectra at that same mCherry concentration. Each background-subtracted spectrum was then deconvolved using PeakFit software (v4.12, Systat, San Jose, CA); the symmetrical quantum dot emission was fit to a Voigt Area peak, while an exponentially modified Gaussian (EMG) curve was used to account for the tailing resulting from the asymmetrical mCherry emission. The areas under the QD peaks were normalized to the area under the QD spectrum in the absence of mCherry. The resulting normalized QD emissions were plotted relative to the acceptor concentrations. The FRET efficiencies were calculated using the equation:
| (3) |
where FDA is the fluorescence of the donor in the presence of the acceptor and FD is the fluorescence of the donor in the absence of the acceptor. The method of estimating the donor-acceptor distance (R) has been described before (11, 16, 32) and is outlined in the supplemental information.
RESULTS AND DISCUSSION
As shown in Table 2, QDs with eight different organic coatings from three distinct sources were evaluated using a FRET assay to determine their capacity for self-assembly to biomolecules via polyhistidine (metal-affinity) coordination. These three major categories of QDs consist of: (1) QDs coated using ligand exchange with DHLA (QD type 1, or QD1 in Table 2); DHLA-PEG600-OH (QD2); DHLA-PEG), or DHLA-PEG750-OCH3 (QD3); DHLA-mPEG); (2) Invitrogen Qdots, which are coated with an amphiphilic block co-polymer, including carboxyl Qdots (QD4) and amino-PEG Qdots (QD5); and (3) lipid-PEG coated QDs, specifically carboxyl-functionalized (QD6), amine-functionalized (QD7), and non-functionalized (QD8) T2-MP EviTags from Evident Technologies. In order to relate the results of the FRET assay to the fundamentals of the interaction between the his-tagged fluorescent protein and the QD, basic QD properties, such as photoluminescence emission, hydrodynamic diameter, zeta potential, and quantum yield, were first characterized and compared.
Table 2.
Summary of FRET Pair Specifications and Key Parameters
| FRET Donor (Acceptor: His6-mCherry) | J (10−15 M−1 cm3) | R0 (Å) | E at 1:1QD:FP | max E |
|---|---|---|---|---|
| (QD1) DHLA 550 nm QDs | 4.73 | 44.9 | 0.15 | 0.48 |
| (QD2) DHLA-PEG 550 nm QDs | 4.57 | 43.8 | 0.02 | 0.18 |
| (QD3) DHLA-mPEG 550 nm QDs | 4.76 | 45.2 | 0.01 | 0.17 |
| (QD4) Qdot® 545 ITK™ carboxyl quantum dots | 4.56 | 61.1 | <0.01 (0.09)a | 0.21 (0.47)a |
| (QD5) Qdot® 545 ITK™ amino (PEG) quantum dots | 5.10 | 62.1 | N/A | N/A |
| (QD6) Carboxyl-functionalized 540 nm T2-MP EviTag | 4.77 | 52.6 | 0.51 | 0.76 |
| (QD7) Amine-functionalized 540 nm T2-MP EviTag | 3.82 | 59.6 | 0.37 | 0.64 |
| (QD8) Non-functionalized 540 nm T2-MP EviTag | 4.20 | 53.9 | N/A | N/A |
Ni2+ added to enhance QD-FP interaction.
Quantum Dot Properties
All eight of the QDs tested exhibit emission peaks between 540 and 550 nm (Table 1 and Figure S1 of the Supplementary Information). These similar emission spectra translate into comparable spectral overlap integrals with the mCherry acceptor (J; Table 1), but significant differences are seen in the Förster distances, R0, calculated for each of the FRET pairs because of variations in the quantum yields of the distinct QDs. At identical donor-acceptor (QD-protein) distances, QDs with higher QYs, and therefore larger Förster distances, will exhibit increased FRET efficiencies. Thus, when determining conjugate conformation, it is important to utilize donor-acceptor distances that can be extracted from the empirical FRET results as well as the relative FRET efficiency.
Table 1.
Summary of Quantum Dot Properties
| QD Type | Water-soluble Coating | Peak Emission(nm) | QYa | Hydrodynamic Diameter(nm)b | Zeta Potential at pH 7 (mV)c | Zeta Potential at pH 9.5 (mV)c | |
|---|---|---|---|---|---|---|---|
| Dihydrolipoic acid (DHLA) coated QDs | DHLA | (QD1) | 550 | 0.11 | 9.8 ± 0.3 | −29.8 ± 1.8 | −23.3 ± 2.6 |
| DHLA-PEG600-OH | (QD2) | 548 | 0.10 | 12.2 ± 0.9 | −12.9 ± 1.3 | −34.6 ± 2.0 | |
| DHLA-PEG750-OCH3 | (QD3) | 550 | 0.12 | 12.9 ± 0.5 | −2.8 ± 0.5 | −18.2 ± 0.6 | |
| Invitrogen: Qdot 545 ITK | Polymer (-COOH) | (QD4) | 544 | 0.75 | 13.0 ± 0.3 | −3.4 ± 0.8 | −31.0 ± 1.3 |
| Polymer (-PEG2000-NH2) | (QD5) | 550 | 0.74 | 15.9 ± 0.8 | −2.3 ± 0.7 | −4.8 ± 1.2 | |
| Evident Technologies: T2-MP EviTags | Lipid-PEG2000-COOH | (QD6) | 548 | 0.29 | 14.9 ± 1.7 | −0.6 ± 0.2 | −26.5 ± 0.8 |
| Lipid-PEG2000-NH2 | (QD7) | 540 | 0.77 | 20.8 ± 0.5 | −4.0 ± 0.6 | −31.8 ± 0.5 | |
| Lipid-PEG2000-OCH3 | (QD8) | 542 | 0.39 | 23.2 ± 0.7 | −10.0 ± 0.5 | −15.5 ± 1.3 | |
QY measured relative to Rhodamine 6G in water.
Hydrodynamic diameter values are the mean ± standard deviation of the volume-weighted size distribution.
Zeta potential values are means ± standard deviations of n = 3 or n = 4 measurements.
Because emission wavelength is a property of the QD core size (33), based on their similar emission wavelengths, the eight QDs examined here have similar nanoparticle dimensions (~4 nm according to technical support from Evident and Invitrogen). Significant differences in the hydrodynamic diameter were, however, apparent in the DLS measurements (Table 1), which reflect subtle differences in the contribution of the hydrated organic coatings to the hydrodynamic interactions.
Ligand-exchange coated QDs
DHLA-coated QDs (QD1) were the only nanoparticles that exhibited sub-10 nm hydrodynamic diameters, which can be attributed to the small DHLA-based moieties and removal of the native TOP/TOPO surfactant during the aqueous phase transfer (10, 34). DHLA, among the smallest molecules capable of conferring water solubility to QDs (3), binds the ZnS capping layer via bidentete thiols and contains a carboxylic acid that holds the QD in suspension as a colloid when deprotonated. Although this thin organic coating is ideal for minimizing the donor-acceptor distance, which is critical for achieving high FRET efficiencies, colloidal stabilization based on electrostatic repulsion is vulnerable to changes in the environment and the protonation state of the carboxyl groups, i.e. pH. For example, compared to the other seven QDs used in this study, we measured a progressive decrease in photoluminescence over time for the DHLA-coated QDs (QD1) in the presence of 1 M NaCl (Figure S2), which can be attributed in aggregation build up in the solution; DHLA-coated QDs are also known to be more stable in alkaline solutions than at acidic pHs (34).
The other two QD varieties coated using ligand-exchange, the DHLA-PEG (QD2; coated with DHLA-PEG600-OH) and DHLA-mPEG (QD3; coated with DHLA-PEG750-OCH3) QDs, exhibit 25% and 32% increases in hydrodynamic diameter over the DHLA coating, respectively (Table 1, see (35)). The increase in size is accompanied by enhanced colloidal stability since solubility is mediated by the ethylene glycol repeats. These QDs also exhibited less variation in the photoluminescent output over time, including that under high salt conditions (Figure S2).
Ligand exchange with these molecular scale caps decreases the QY of the QDs relative to the starting QDs in organic solvents. For example, all three of the ligand-exchanged QDs had quantum yields between 0.10 and 0.12. The exact value depends on the nature of the starting TOP/TOPO-capped nanocrystals; higher yields (0.2–0.3) have been reported for these materials (34).
QDs coated with amphiphilic block co-polymers
The amphiphilic block co-polymer coated carboxyl Qdots from Invitrogen (QD4) exhibited the smallest hydrodynamic diameter (13 nm) of the commercially available QDs (Table 1), a value in good agreement with previously reported measurements (35). The hydrophobic region of the amphiphilic poly(acrylic) acid polymer interdigitates with the TOP/TOPO layer and presents a high density of carboxylic acids at the QD-solvent interface. While the hydrophilic carboxylic acids confer water solubility to the QDs similarly to those on the small molecule DHLA, the larger, more neutral polymer does appear to improve the QD passivation as well, providing the Qdot with relatively reliable photoluminescence over time, even in high salt conditions (Figure S3). This coating type preserves the high QY of the QDs even in aqueous solution, consistently producing batches with QYs greater than 0.70. The amino-PEG Qdots (QD5) add another layer to the same coating scheme used for the carboxyl Qdots. The amine-terminated PEG2000 chains that are conjugated to the carboxyl groups on the surface of the carboxyl Qdots add almost 3 nm to the hydrodynamic diameter of the Qdots (Table 1), but the hydration of the PEG molecules results in a QD surface that significantly reduces non-specific interactions (36).
Lipid-PEG coated QDs
The three EviTags exhibited rather different hydrodynamic diameters although they used the same core-shell structure and lipid-PEG coating scheme (Table 1). Each coating consists of a micelle-like shell of amphiphilic 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carboxy(polyethylene glycol)-2000], i.e. DSPE-PEG2000, from Avanti Polar Lipids, Inc. (Alabaster, Alabama), terminated with a carboxyl-, amine-, or methoxy-group (Table 2). The reduction in hydrodynamic diameter as the PEG terminal group changes from –OCH3 to –NH2 to –COOH does not correlate with a decrease in photoluminescence (PL) stability, even under high salt conditions (Figure S4). These differences in the hydrodynamic diameter may indicate variations in the density and orientation of the lipid-PEG coating. A previous report on thickness measurements of DSPE-PEG2000 Langmuir-Blodgett monolayer films, using neutron reflectivity, indicated that a densely packed monolayer of DSPE-PEG2000 could reach a thickness of 10.7 nm, but smaller thicknesses were measured for less densely packed monolayers. In that case, the decrease in the monolayer thickness was found to be related to the brush height of the PEG polymer; the lipid demonstrated no density-dependent variation in its geometry (37). This effect may be even more pronounced on a nanoparticle, where surface curvature inherently increases the lateral space available for PEG molecules. The difference in hydrodynamic sizes measured for these various nanocrystals may thus be attributed to differences in the contributions of the end-functionalized PEG (-COOH, -NH2, and -OCH3) to the coating quality and hydrodynamic structure. The zeta potential measurement results also signal surprising differences between the three coating types that are not consistent with the surface charge that would be expected based on the lipid-PEG terminal group. This is a further indication of the differences in the structure of the coatings beyond the functional moiety. We should also note that while the variation in coating thickness does not appear to affect PL stability of QDs, it does seem to influence polyhistidine self-assembly, as discussed below.
His-tag-mediated self-assembly
FRET assays were performed to determine under what conditions and to what extent the his-tag mediates self-assembly of fluorescent proteins to QDs. The FRET-based assay is particularly useful here because it both confirms the proximity of the nanoparticle and biomolecule and allows the derivation of a separation distance due to its strong distance dependence. This provides insight into both self-assembly and the overall conjugate structure. In all cases, a non-his-tagged fluorescent protein mCherry was used as a negative control in order to ascertain that his-tag was necessary to induce the interaction between the donor (QD) and acceptor (FP). Self-assembly was also attempted under high salt conditions (1 M NaCl) in cases where the QD PL was stable under these buffer conditions (see supplemental information). The extreme salt concentration is disruptive to electrostatic interactions and therefore can be used to test whether the interaction is metal chelation-based or mediated electrostatically. His-based metal chelation, for example to Ni-NTA, can tolerate extreme conditions and the presence of denaturing agents including 5% SDS, 6 M guanidine, 8 M urea, and greater than 1 M NaCl (7, 15, 20). In addition, supplementation with NiCl2 was included to determine if the presence of additional chelating ions would affect QD-protein binding.
Ligand-exchange coated QDs
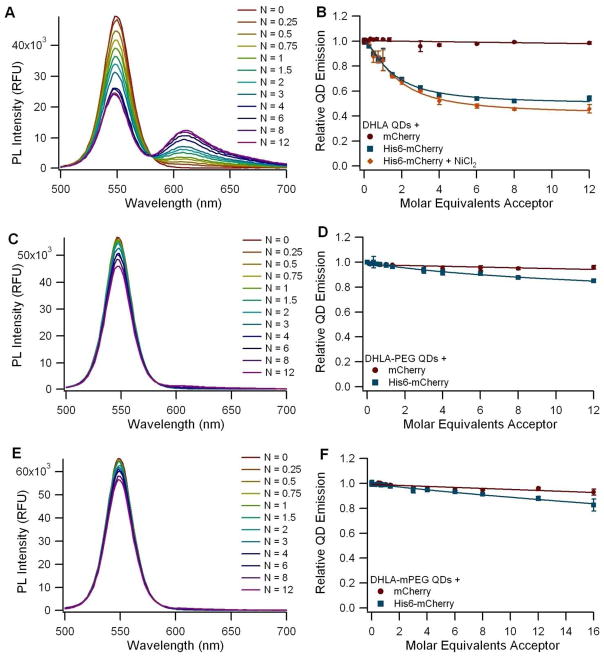
Our FRET assays revealed that, of the three DHLA-based organic coatings, only QDs coated with DHLA (QD1) (with the smallest size of the three) demonstrated a good capacity for self-assembly via polyhistidine coordination (Figure 2). The spectra collected from the DHLA-QDs incubated with His6-mCherry show a clear and progressive decrease in the QD emission with a concomitant dose-dependent increase in the sensitized emission of the fluorescent protein, with an isosbestic point around 580 nm (Figure 2A). It was not possible to test self-assembly with these QDs under high salt conditions because DHLA QDs exhibit reduced stability in those conditions (Figure S2), but the addition of the divalent cation (Ni2+) did not appear to enhance binding (Figure 2B). The approximately 6 nm donor-acceptor distance extracted for the DHLA-coated QD and His6-mCherry implies that the FP barrel structure is indeed very close to the QD surface. These findings are consistent with previously reported kinetic data on the binding of his-tagged biomolecules to DHLA-coated QDs, which indicated that the his-tag binds directly to the ZnS capping layer of the QD (15).
Figure 2.
FRET assay using 550 nm QDs coated with (A-B) DHLA (QD1), (C-D) DHLA-PEG600-OH (QD2), or (E-F) DHLA-PEG750-OCH3 (QD3). A, C, and E: Spectra of QDs incubated with varying ratios (N) of His6-mCherry monomers per QD. B, D, and F: Graph of the normalized QD emission relative to the number of acceptor molecules per QD. Circles (●) represent QDs incubated with a non-his-tag control mCherry, squares (■) represent experiments with His6-mCherry as the acceptor, and the diamonds (◆) represent experiments with His6-mCherry following supplementation with Ni2+. All points are mean ± standard deviation with n = 3.
It has been previously documented that the polyhistidine-mediated coordination of a non-fluorescent protein such as maltose-binding protein (MBP) to the DHLA-coated can QDs result in a photoluminescence (PL) increase of up to 20% (38). This was attributed to the passivation of surface defects in the DHLA coating by his-tags, thereby reducing the emission loss in aqueous media. This effect could be problematic for the analysis of the FRET assays described above, since his-tag binding increases the QD emission concomitantly as the QD emission is being reduced by energy transfer to the His6-mCherry proteins. The effect of his-tag binding on QD PL was examined for all eight QDs and the two that seemed to be affected by the presence of the his-tagged protein, the DHLA QDs and the carboxyl-functionalized EviTags (Figure S8), were tested in an alternative FRET assay format. The modified FRET assay, in which the total number of proteins was fixed while the ratio of fluorescent to non-fluorescent proteins was varied, was explored to see if the inclusion of the non-fluorescent protein would increase the overall FRET efficiency of the system. In this particular experimental protocol, the modified conjugate configuration did not improve the conjugate FRET efficiency (see supplemental information and Figure S9).
Both PEG-conjugated DHLA moieties (QD2 and QD3) exhibited marginal QD PL quenching when mixed with His6-mCherry (Figure 2C and E), demonstrating an inability of the his-tag to stably interact with the nanoparticles. DHLA-PEG-capped QDs, though not amenable to his-driven conjugation with proteins (due to the steric hinderance from the PEG coating), have been shown to tightly bind his-terminated peptides, as these are able to penetrate the PEG brush and interact directly with the QD Zn-rich surface. QD-peptide assemblies made using these QDs have been employed in enzymatic assays and conjugate uptake by live cells, where the improvements in colloidal stability were beneficial (39).
QDs coated with amphiphilic block co-polymers
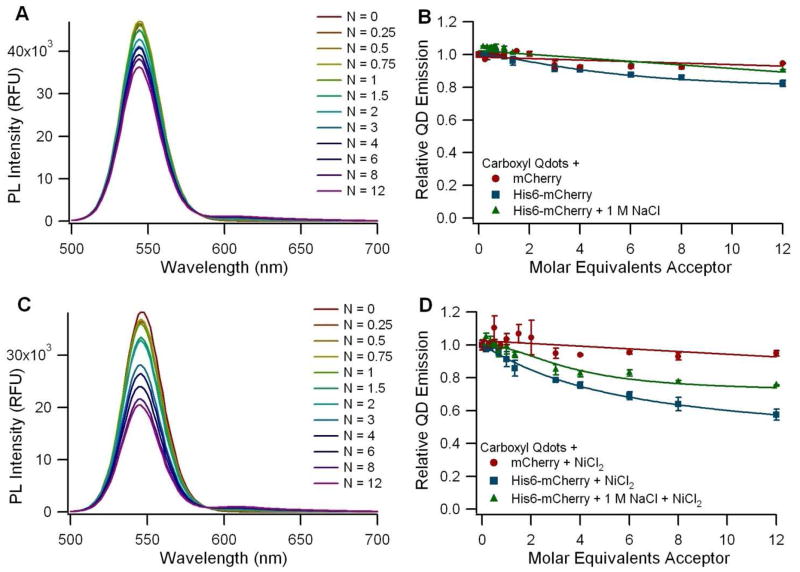
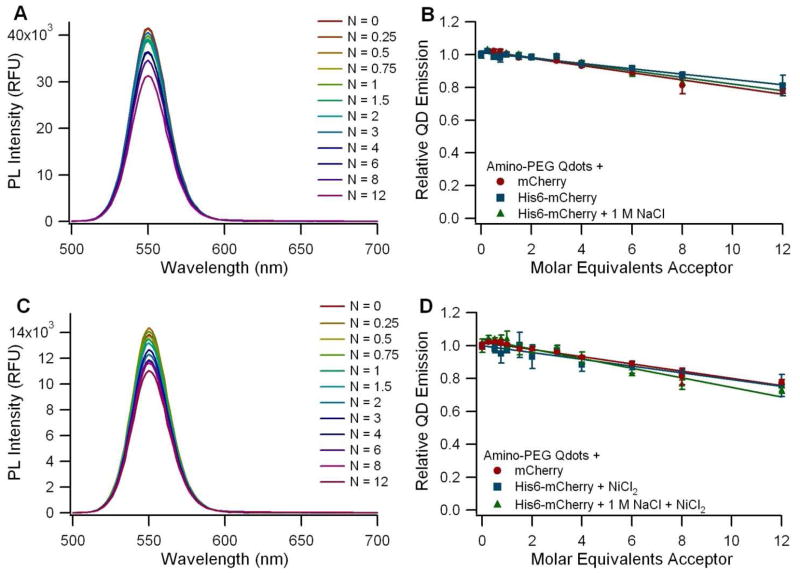
The carboxyl Qdots from Invitrogen (QD4) showed only a limited capacity to self-assemble with the His6-tagged mCherry in borate buffered saline, and this subtle interaction was disrupted in the presence of 1 M NaCl (Figure 3A and B). However, a stronger FRET signal was observed following supplementation with NiCl2. The Ni2+-dependent FRET signal was also reduced in the presence of 1 M NaCl, but not eliminated as in the non-Ni2+-dependent case (Figure 3C and D). In the absence of the supplemented Ni2+, we found that the limited interaction between the fluorescent protein and the carboxyl Qdot is non-specific, electrostatically-mediated, and can be disrupted with high salt concentrations. The supplemented Ni2+, however, appears to complex with the high density of carboxyl groups on the Qdot surface, enabling the interaction with the protein via Ni2+ chelation. The same interaction was observed in another study when his-tagged luciferase molecules were incubated with carboxyl Qdots in the presence of Ni2+ (40). Other divalent ions including Ca2+, Mg2+, and Mn2+ were tested in that study as well, but Ni2+ was the most effective one in inducing the interaction between the his-tagged protein and the Qdot, while maintaining protein functionality (40). Calculation of the corrected donor-acceptor distance yielded a median separation of 94 Å between the QD core and the FP fluorophore (see Table S1 in the supplementary information). This distance is significantly larger than the hydrodynamic radius of 65 Å measured for the QDs (Table 1), and is consistent with the model in which the FPs bind to the outer surface of a solid polymer coating that surrounds the QD. In contrast, the amino-PEG ITK Qdots (QD5) showed no difference in QD emission regardless of whether his-tagged or control mCherry was used (Figure 4B), and addition of NiCl2 did not increase the level of interaction (Figure 4D). We attribute this to the presence of the amino-PEG moieties on the Qdot surface, which shield the nanoparticles from interacting with the protein (even non-specifically, as seen with the carboxyl Qdots) and prevent the immobilization of Ni2+ ions on the nanoparticle surface. The zeta potential measurements showed that the carboxyl Qdots are highly negatively charged while the amino-PEG Qdots have a near neutral zeta potential, supporting the finding that the carboxyl Qdots are primed for both electrostatic interactions and metal chelation whereas the amino-PEG Qdots are not.
Figure 3.
FRET assay using 545 nm Invitrogen ITK Carboxyl Qdots (QD4) before (A-B) and after (C-D) supplementation with Ni2+. A and C: Spectra of carboxyl Qdots (QD4) incubated with varying ratios (N) of His6-mCherry per QD. B and D: Graphs of the normalized QD emission with respect to the ratio of acceptor molecules per QD. Circles (●) represent a non-his-tagged mCherry control, squares (■) represent the His6-mCherry acceptor, and triangles (▲) represent the addition of His6-mCherry in the presence of 1 M NaCl to diminish the effect of electrostatic interactions.
Figure 4.
FRET assay using 545 nm Invitrogen ITK Amino-PEG Qdots (QD5) before (A-B) and after (C-D) supplementation with Ni2+. A and C: Spectra of amino-PEG Qdots (QD5) incubated with varying ratios (N) of His6-mCherry per QD. B and D: Graph of the normalized QD emission with respect to the number of acceptor molecules per QD. Circles (●) represent QDs incubated with a non-his-tagged mCherry control, squares (■) represent the His6-mCherry acceptor, and triangles (▲) represent the addition of His6-mCherry in the presence of 1 M NaCl to diminish the effect of electrostatic interactions.
It is worth noting that the carboxyl Qdots (QD4) produced minimal sensitized emission from the his-tagged mCherry even when FRET quenching is significant (Figure 3C). In fact, the relative intensity of the sensitized emission from the protein is the largest in the case of the DHLA QDs (QD1) (Figure 2A), even though there is more FRET quenching observed for the carboxyl-functionalized EviTags (QD2) (Figure 5A). This difference in sensitized emission between the samples is an artifact of the disparity between the relative brightnesses of the three classes of QDs. The Qdots (QD4 and QD5) required dramatically lower fluorimeter gain settings compared to the EviTags (QD6, QD7, QD8), which still had lower gains than applied for the DHLA QDs (QD1, QD2, QD3), because of their significantly enhanced relative brightness. Protein emission intensity may have paled in comparison to the brightness of the Qdots. Because it was necessary to optimize gain settings to the QD-type under examination, the relative intensities of the peaks should only be compared within one type of QD, not from figure to figure
Figure 5.
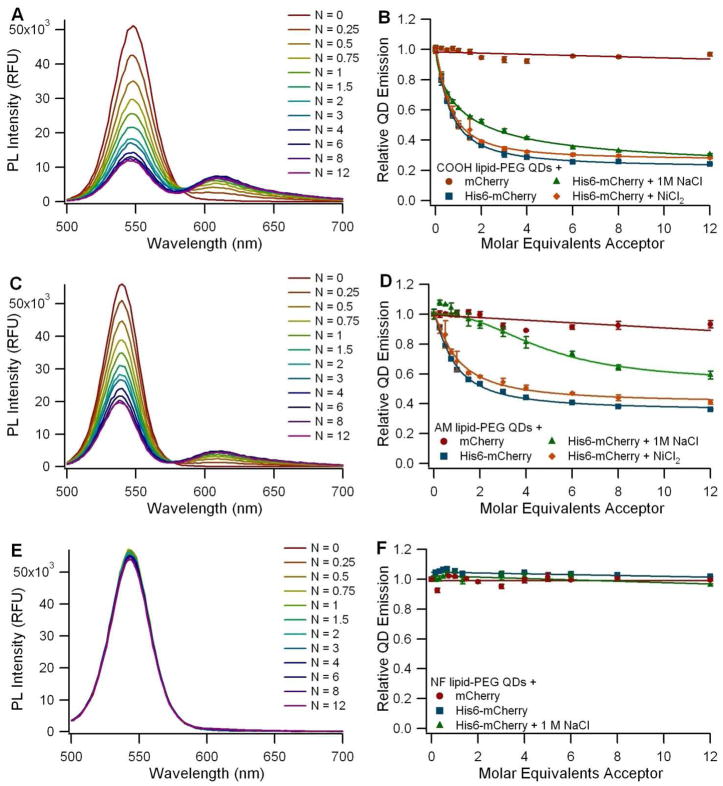
FRET assay using 540 nm (A-B) carboxyl- (QD6), (C-D) amine- (QD7), and (E-F) non-functionalized (QD8) T2-MP EviTags. A, C, and E: Background-subtracted spectra of EviTags incubated with various ratios (N) of His6-mCherry molecules per QD. B, D, and F: Graph of the normalized QD emission relative to the number of acceptor molecules per QD. Circles (●) represent EviTags incubated with a non-his-tagged mCherry control, squares (■) represent QDs incubated with the His6-mCherry acceptor, triangles (▲) represent experiments with His6-mCherry in the presence of 1 M NaCl, and diamonds (◆) represent experiments with His6-mCherry following supplementation with Ni2+.
Lipid-PEG coated QDs
The interaction between the His6-mCherry and the EviTags was found to be dependent on the terminal functional group present in the lipid-PEG coating. The carboxyl-functionalized EviTags (QD6) were shown to be excellent FRET donors with 70% quenching of the QD emission at a donor to acceptor ratio of 1:3 (Figure 5A and B). His-tag binding to carboxyl-functionalized EviTags proceeded in both saline and high salt and was not enhanced by the addition of Ni2+. This indicates that the interaction was neither electrostatically-driven nor dependent on the chelation of free divalent cations by the carboxyl groups, as is the case with the carboxyl Qdots.
The incubation of the carboxyl-functionalized EviTags with non-fluorescent his-tagged proteins resulted in a mild increase in the QD PL (Figure S8), indicating that these EviTags may have surface defects similar to those described for the DHLA QDs. As with the DHLA QDs, the modified conjugate configuration designed to account for this increase in PL due to binding did not yield improved FRET efficiencies (Figure S9).
Amine-functionalized EviTags (QD7) also showed a capacity for the polyhistidine-mediated self-assembly (Figure 5C and D), but this interaction was more susceptible to disruption from high salt concentrations. Not only was the extent of QD quenching lessened in the high salt buffer, but the shape of the dose-dependent quenching curve was altered as well, indicating that the mechanism of the interaction between the amine-functionalized EviTags and His6-mCherry likely contains an electrostatic component, where transient non-specific interactions between the amines in the coating and the fluorescent protein may provide the opportunity for close proximity between the his-tag and the QD surface. Although these non-specific associations do appear to contribute to the probe assembly in this case, the presence of some conjugate formation in the high salt solution implies the incidence of polyhistidine-derived metal chelation as well.
Non-functionalized EviTags (QD8), in comparison, showed no capacity for polyhistidine-mediated self-assembly (Figure 5E and F), perhaps in part due to the lack of electrostatic attraction, but more likely due to hindrance from a dense lipid-PEG coating.
The significant differences in the quenching effect of the three EviTag QDs is surprising given that the only difference between the three QD types is the functional group at the terminal end of the lipid-PEG coating moiety. However it may be explained, in part, by the hydrodynamic diameter measurements. If the large diameter of the non-functionalized EviTags is indeed indicative of a coating with closely packed lipid-PEG molecules that are extended brush-like, then this coating may be impenetrable to the his-tagged FP, similar to what was reported for DHLA-PEG-capped QDs. If the 35% decrease in the hydrodynamic diameter of the carboxyl-functionalized EviTags, in contrast, reflects a less dense, or even patchy, lipid-PEG coating, then some regions of the QD surface may be exposed, thus amenable to the binding of the polyhistidine tag. It is unclear, however, what role the TOP/TOPO layer that passivated the surface following the inorganic synthesis plays in this scenario. The amine-functionalized EviTags exhibited a behavior between the two discussed above, both in hydrodynamic diameter and the extent of FRET. In fact, given the much higher quantum yield of the amine-functionalized EviTag, one would expect a greater quenching than that with the carboxyl-functionalized EviTag, if the fluorescent proteins were bound to the QDs with the same amount and were positioned at the same distance from the QD core. Instead, we found that quenching was reduced with the amine-functionalized EviTags. Calculations of the corrected donor-acceptor distances yielded median R values of 56 Å and 71 Å for the carboxyl- and amine-functionalized EviTags, respectively (Table S1). Both of these donor-acceptor distances are shorter than half of the hydrodynamic diameter of the QDs, as measured with DLS (Table 1), suggesting that in both cases the fluorescent proteins may be embedded in the coating layer rather than outside of the PEG corona. The flexibility of the hydrated PEG chains, particularly when not close-packed into a dense brush-like conformation, could permit this physical arrangement. Furthermore, the difference in the donor-acceptor distance for carboxyl-functionalized or amine-functionalized EviTags could arise if the amino acid linker region between the his-tag and the FP barrel structure is fully extended when the protein is bound to the amine-functionalized EviTag due to the differences in their PEG conformations.
To ensure that the results of the EviTag FRET assays are not dependent on these particular batches of QDs, self-assembly experiments were repeated many times with different coating batches and QD emission wavelengths (paired with alternative GFP-like FPs to ensure appropriate spectral overlap where necessary) utilizing QDs purchased during the past few years, making it likely that multiple batches of Avanti’s lipid-PEG were involved for each coating type as well. Our general observation of strong quenching of the carboxyl-functionalized EviTags (QD6), moderate quenching of the amine-functionalized EviTags (QD7), and no quenching of the non-functionalized EviTags (QD8) was consistent for all experiments conducted (data not shown). Our previous results indicated that carboxyl-functionalized EviTags with three different coating batches all showed considerable quenching when bound to FPs via polyhistidine-mediated coordination (11). Thus, these results demonstrate the utility of polyhistidine coordination for the bioconjugation of biomolecules to QDs coated with carboxyl-functionalized lipid-PEG molecules even though the technique does not extend to all lipid-PEG-based coatings.
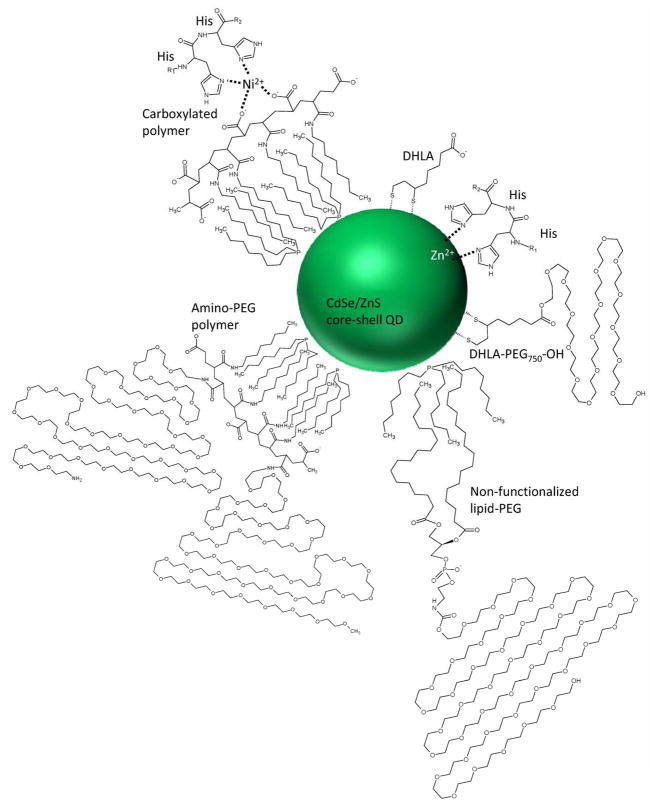
In this study, we compared QDs with eight different coatings using three different coating schemes and determined their capacity for polyhistidine coordination as a means to conjugate biomolecules to nanoparticles by self-assembly. This is highly desirable for the assembly of nanoparticle based imaging probes and delivery systems because of its technical simplicity and has even been extended to self-assembling his-modified DNA to QDs (16). The aggregate of these results demonstrates the importance of accessibility for his-tag-mediated coordination. Successful self-assembly depends on enabling access either with small ligand coatings like DHLA or with porous polymeric coatings like the carboxyl-functionalized lipid-PEG (Figure 6). DHLA-coated QDs (QD1) exhibited effective his-tag mediated binding and short donor-acceptor distances, but exhibit some environmental sensitivity and pH-dependent colloidal instability. Using QDs coated with PEGylated DHLA (QD2 and QD3) improved the stability and environmental sensitivity, making them advantageous for certain applications including intracellular imaging, but steric hindrance precluded his-tagged proteins from penetrating to the QD surface, even in cases where his-tagged peptides had been able to bind. The ZnS surface of the carboxyl- (QD4) and amino-PEG (QD5)Qdots was similarly inaccessible for his-tag binding, but chelation of supplemented Ni2+ to the high density of carboxyl groups on the carboxyl Qdot polymer coating did allow for Ni2+-mediated association of the protein with the Qdot, similar to the way in which a his-tagged protein binds to a Ni2+-containing metal affinity chromatography column. While the added distance between the donor and acceptor resulting from this binding scheme may make the strategy suboptimal for FRET-based assays, it may be useful for labeling Qdots with proteins for applications where the protein should be completely exposed at the QD surface to maintain functionality. Finally, the lipid-PEG coated EviTags demonstrated a range of his-tag binding permissiveness, depending on the specific functional group at the PEG terminus. Lipid-PEGs terminated with a methoxy-group (QD6) generated a thick, impenetrable coating, while coatings with terminal carboxyl-groups (QD7) were much thinner and susceptible to his-tag binding. Amine-functionalized EviTags (QD8) performed in between the two.
Figure 6.
Schematic of various QD coatings and his-tag interactions. The organic coating moieties and their interaction with the polyhisitidine sequence are depicted. Where the his-tag can access the QD surface, coordination of the Zn2+ ions in the ZnS capping layer facilitates self-assembly. In the case of the carboxylated polymer, a divalent cation is chelated by both the carboxyl groups on the polymer surface and the histidine.
In summary, this study systematically compares the capacity of eight QD types for self-assembly with his-tagged proteins. The FRET assay employed was particularly useful for studying the conjugate properties as both the presence of binding and an estimation of the distance between the QD and the protein can be extracted from the experimental results. We found that the accessibility of the ZnS surface is paramount to metal-coordination and that this nanoparticle characteristic can vary dramatically with subtle changes to the colloid’s organic coating. While the results of this screen for materials compatible with his-tag binding is of general interest for QD-biomolecule hybrid device design, it is particularly important for biosensing applications as this self-assembly method produces compact conjugates that are very suitable for FRET-based assays.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge laboratory support from Andrea Fernandez and Annie Zheng as well as helpful discussions with Drs. Steven Dublin and Charles Glaus. This work was supported by NIH through a Nanomedicine Development Center award (1PN2EY018244 to GB), a Program of Excellence in Nanotechnology award (HL80711 to GB), a Center of Cancer Nanotechnology Excellence award (CA119338 to GB), and a National Defense Science and Engineering Graduate (NDSEG) Fellowship (to AMD). IM acknowledges support from DTRA, ONR, and the NRL-NSI.
Footnotes
Supporting Information Available: Results of the calculation of the donor-acceptor distance, assessment of QD stability, characterization of His6-mCherry-NF, evaluation of the QD PL change due to his6 binding, and application of the modified FRET assay are discussed in the supporting information. This material is available free of charge via the internet at http://pubs.acs.org/BC.
LITERATURE CITED
- 1.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 2.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 3.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nature Materials. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 4.Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Luminescent quantum dots for multiplexed biological detection and imaging. Current Opinion in Biotechnology. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 5.Medintz IL, Mattoussi H. Quantum dot-based resonance energy transfer and its growing application in biology. Physical Chemistry Chemical Physics. 2009;11:17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 6.Sapsford KE, Pons T, Medintz IL, Mattoussi H. Biosensing with luminescent semiconductor quantum dots. Sensors. 2006;6:925–953. [Google Scholar]
- 7.Hermanson GT. Bioconjugate Techniques. 2. Academic Press; San Diego: 2008. [Google Scholar]
- 8.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proceedings of the National Academy of Sciences U S A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingerfelt BM, Mattoussi H, Goldman ER, Mauro JM, Anderson GP. Preparation of quantum dot-biotin conjugates and their use in immunochromatography assays. Analytical Chemistry. 2003;75:4043–4049. doi: 10.1021/ac034139e. [DOI] [PubMed] [Google Scholar]
- 10.Mattoussi H, Mauro JM, Goldman ER, Green TM, Anderson GP, Sundar VC, Bawendi MG. Bioconjugation of highly luminescent colloidal CdSe-ZnS quantum dots with an engineered two-domain recombinant protein. Physica Status Solidi B-Basic Research. 2001;224:277–283. [Google Scholar]
- 11.Dennis AM, Bao G. Quantum dot-fluorescent protein pairs as novel fluorescence resonance energy transfer probes. Nano Letters. 2008;8:1439–1445. doi: 10.1021/nl080358+. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. Compact biocompatible quantum dots functionalized for cellular imaging. Journal of the American Chemical Society. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu H, Schops O, Woggon U, Niemeyer CM. Self-assembled donor comprising quantum dots and fluorescent proteins for long-range fluorescence resonance energy transfer. Journal of the American Chemical Society. 2008;130:4815–4827. doi: 10.1021/ja078243f. [DOI] [PubMed] [Google Scholar]
- 14.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nature Materials. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 15.Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. Kinetics of metal-affinity driven self-assembly between proteins or peptides and CdSe-ZnS quantum dots. Journal of Physical Chemistry C. 2007;111:11528–11538. [Google Scholar]
- 16.Medintz IL, Berti L, Pons T, Grimes AF, English DS, Alessandrini A, Facci P, Mattoussi H. A reactive peptidic linker for self-assembling hybrid quantum dot-DNA bioconjugates. Nano Letters. 2007;7:1741–1748. doi: 10.1021/nl070782v. [DOI] [PubMed] [Google Scholar]
- 17.Green NM. Avidin and streptavidin. Methods in Enzymology. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 18.Howarth M, Chinnapen DJF, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, Ting AY. A monovalent streptavidin with a single femtomolar biotin binding site. Nature Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. [Google Scholar]
- 20.Sulkowski E. Purification of Proteins by Imac. Trends in Biotechnology. 1985;3:1–7. [Google Scholar]
- 21.Dennis AM, Bao G. Förster Resonance Energy Transfer (FRET) between a fluorescent protein and commercially available quantum dots: a comparison. Proceedings of SPIE. 2008;6866:1–11. [Google Scholar]
- 22.Mei BC, Susumu K, Medintz IL, Mattoussi H. Polyethylene glycol-based bidentate ligands to enhance quantum dot and gold nanoparticle stability in biological media. Nature Protocols. 2009;4:412–423. doi: 10.1038/nprot.2008.243. [DOI] [PubMed] [Google Scholar]
- 23.Susumu K, Mei BC, Mattoussi H. Multifunctional ligands based on dihydrolipoic acid and polyethylene glycol to promote biocompatibility of quantum dots. Nature Protocols. 2009;4:424–436. doi: 10.1038/nprot.2008.247. [DOI] [PubMed] [Google Scholar]
- 24.Mei BC, Susumu K, Medintz IL, Delehanty JB, Mountziaris TJ, Mattoussi H. Modular poly(ethylene glycol) ligands for biocompatible semiconductor and gold nanocrystals with extended pH and ionic stability. Journal of Materials Chemistry. 2008;18:4949–4958. [Google Scholar]
- 25.Dabbousi BO, RodriguezViejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. (CdSe)ZnS core-shell quantum dots: Synthesis and characterization of a size series of highly luminescent nanocrystallites. Journal of Physical Chemistry B. 1997;101:9463–9475. [Google Scholar]
- 26.Hines MA, Guyot-Sionnest P. Synthesis and characterization of strongly luminescing ZnS-Capped CdSe nanocrystals. Journal of Physical Chemistry. 1996;100:468–471. [Google Scholar]
- 27.Peng ZA, Peng XG. Formation of high-quality CdTe, CdSe, and CdS nanocrystals using CdO as precursor. Journal of the American Chemical Society. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 28.Clapp AR, Goldman ER, Mattoussi H. Capping of CdSe-ZnS quantum dots with DHLA and subsequent conjugation with proteins. Nature Protocols. 2006;1:1258–1267. doi: 10.1038/nprot.2006.184. [DOI] [PubMed] [Google Scholar]
- 29.Susumu K, Uyeda HT, Medintz IL, Pons T, Delehanty JB, Mattoussi H. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. Journal of the American Chemical Society. 2007;129:13987–13996. doi: 10.1021/ja0749744. [DOI] [PubMed] [Google Scholar]
- 30.Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WG, Mehlin C. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallographica Section D: Biological Crystallography. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 31.Hink MA, Visser NV, Borst JW, van Hoek A, Visser AJWG. Practical use of corrected fluorescence excitation and emission spectra of fluorescent proteins in Forster resonance energy transfer (FRET) studies. Journal of Fluorescence. 2003;13 J Fluoresc. [Google Scholar]
- 32.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nature Materials. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 33.Qu L, Peng X. Control of photoluminescence properties of CdSe nanocrystals in growth. Journal of the American Chemical Society. 2002;124:2049–2055. doi: 10.1021/ja017002j. [DOI] [PubMed] [Google Scholar]
- 34.Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. Synthesis of compact multidentate ligands to prepare stable hydrophilic quantum dot fluorophores. Journal of the American Chemical Society. 2005;127:3870–3878. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- 35.Pons T, Uyeda HT, Medintz IL, Mattoussi H. Hydrodynamic dimensions, electrophoretic mobility, and stability of hydrophilic quantum dots. Journal of Physical Chemistry B. 2006;110:20308–20316. doi: 10.1021/jp065041h. [DOI] [PubMed] [Google Scholar]
- 36.Bentzen EL, Tomlinson ID, Mason J, Gresch P, Warnement MR, Wright D, Sanders-Bush E, Blakely R, Rosenthal SJ. Surface modification to reduce nonspecific binding of quantum dots in live cell assays. Bioconjugate Chemistry. 2005;16:1488–1494. doi: 10.1021/bc0502006. [DOI] [PubMed] [Google Scholar]
- 37.Bianco-Peled H, Dori Y, Schneider J, Sung LP, Satija S, Tirrell M. Structural study of langmuir monolayers containing lipidated poly(ethylene glycol) and peptides. Langmuir. 2001;17:6931–6937. [Google Scholar]
- 38.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. Journal of the American Chemical Society. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 39.Medintz IL, Pons T, Delehanty JB, Susumu K, Brunel FM, Dawson PE, Mattoussi H. Intracellular delivery of quantum dot-protein cargos mediated by cell penetrating peptides. Bioconjugate Chemistry. 2008;19:1785–1795. doi: 10.1021/bc800089r. [DOI] [PubMed] [Google Scholar]
- 40.Yao H, Zhang Y, Xiao F, Xia Z, Rao J. Quantum dot/bioluminescence resonance energy transfer based highly sensitive detection of proteases. Angewandte Chemie International Edition. 2007;46:4346–4349. doi: 10.1002/anie.200700280. [DOI] [PubMed] [Google Scholar]
- 41.Pong BK, Trout BL, Lee JY. Modified ligand-exchange for efficient solubilization of CdSe/ZnS quantum dots in water: A procedure guided by computational studies. Langmuir. 2008;24:5270–5276. doi: 10.1021/la703431j. [DOI] [PubMed] [Google Scholar]
- 42.Iyer G, Pinaud F, Tsay J, Weiss S. Solubilization of quantum dots with a recombinant peptide from Escherichia coli. Small. 2007;3:793–798. doi: 10.1002/smll.200600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.