Abstract
We previously reported that PU.1 was down-regulated in myeloma cell lines and myeloma cells in a subset of myeloma patients, and that conditional PU.1 expression in PU.1-negative myeloma cell lines, U266 and KMS12PE, induced growth arrest and apoptosis. To elucidate the molecular mechanisms of the growth arrest and apoptosis, we performed DNA microarray analyses to compare the difference in gene expression before and after PU.1 induction in U266 cells. Among cell cycle-related genes, cyclin A2, cyclin B1, CDK2 and CDK4 were down-regulated and p21 was up-regulated, while among apoptosis-related genes, TRAIL was found highly up-regulated. When TRAIL was knocked down by siRNAs, apoptosis of PU-1-expressing cells was inhibited, suggesting that TRAIL plays a critical role in PU.1-induced apoptosis in both U266 and KMS12PE myeloma cells. In both U266 and KMS12PE cells expressing PU.1, PU.1 directly bound to a region 30 bp downstream of the transcription start site of the TRAIL gene. Up-regulation of PU.1 induced transactivation of the TRAIL promoter in reporter assays, and disruption of the PU.1-binding site in the TRAIL promoter eliminated this transactivation. Therefore, we conclude that PU.1 is capable of inducing apoptosis in certain myeloma cells by direct transactivation of TRAIL.
Keywords: myeloma, PU.1, TRAIL, apoptosis, p21
Introduction
Multiple myeloma is an incurable hematological malignancy that is resistant to several chemotherapeutic agents as well as hematopoietic stem cell transplantation (Attal et al., 2003). Recently, several new types of agents for myeloma, such as thalidomide, lenalidomide and the proteasome inhibitor bortezomib, have been investigated, but these agents do not lead to long-term complete remission of myeloma, even though they are very effective and improve the survival duration of patients (Hideshima et al., 2003; Hideshima et al., 2001; Singhal et al., 1999). In contrast, the new types of molecular target agents for leukemia and lymphoma include imatinib, which targets bcr-abl tyrosine kinase and brings about striking improvement of the remission and survival rates of chronic myeloid leukemia patients, and rituximab, which targets CD20 antigen on the surface of B lymphoma cells and improves the remission and survival rates of lymphoma patients. These observations indicate that understanding the pathogenesis of a disease is very important for designing new types of molecular target agents. Nevertheless, in contrast to leukemia and lymphoma, the pathogenesis of multiple myeloma is still not well understood.
PU.1 is an ETS family transcription factor that is important for both myelopoiesis and lymphopoiesis (Klemsz et al., 1990; McKercher et al., 1996). Gene expression generally requires long-range distal enhancer regions in addition to the promoter region (Loots et al., 2000; Okuno et al., 2002a; Okuno et al., 2002b; Radomska et al., 2002; Yu et al., 1999). In the case of the PU.1 gene, the long-range distal enhancer region is located in a 14-kb 5’ upstream region in mice and a 17-kb 5’ upstream region in humans (Li et al., 2001; Okuno et al., 2005; Tatetsu et al., 2007). In Friend leukemia, FEEV is integrated into the 14-kb 5’ upstream region of the PU.1 gene locus and results in failure of PU.1 down-regulation in erythroblasts, thereby leading to erythroleukemia in mice (Moreau-Gachelin et al., 1988). We previously reported that proper murine PU.1 gene expression requires the 14-kb 5’ upstream regulatory region, which consists of two highly conserved regions among different mammals, and that the FEEV integration site in Friend leukemia is located between these two conserved regions (Okuno et al., 2005). Moreover, conditional knockout of this 14-kb 5’ upstream region led to down-regulation of PU.1 to 20% of wild-type mice, and all of these mice developed acute myeloid leukemia (Rosenbauer et al., 2004). These mice also developed T cell lymphoma with up-regulation of PU.1 in T lymphoma cells, since one of the two highly conserved regions in the 14-kb 5’ upstream region has suppressor activity for PU.1 expression in the T cell lineage through wnt signaling (Rosenbauer et al., 2006). These data indicate that inappropriate regulation of the PU.1 gene, including failure of down-regulation or up-regulation in proper differentiation stages, leads to hematological malignancies in different hematological lineages (Tenen, 2003).
We recently reported that PU.1 is down-regulated in the majority of myeloma cell lines and freshly isolated myeloma cells from a subset of multiple myeloma patients (PU.1 low-to-negative subset), whereas normal plasma cells express relatively high levels of PU.1 (Tatetsu et al., 2007). PU.1 is down-regulated by methylation of the 17-kb 5’ enhancer region and the promoter region in myeloma cell lines, whereas only the promoter region is methylated in T cell lines (Amaravadi and Klemsz, 1999). Myeloma patients in the PU.1 low-to-negative subset may have a poor prognosis. In addition, conditional expression of PU.1 in PU.1-negative myeloma cell lines, U266 and KMS12PE, was found to induce cell growth arrest and apoptosis (Tatetsu et al., 2007). These data suggest that down-regulation of PU.1 may be an important genetic change for oncogenesis or growth maintenance of multiple myeloma cells. To elucidate the effects of PU.1 expression in U266 cells, we performed DNA microarray analyses and compared the differences in gene expression before and after PU.1 expression.
Results
Interferon-stimulated genes (ISGs) are mainly up-regulated in the U266 myeloma cell line after PU.1 induction
As previously reported (Tatetsu et al., 2007), we generated a stable U266tetPU.1 cell line derived from the PU.1-negative myeloma cell line U266 with a tet-off conditional expression system of PU.1. RNA was purified from U266tetPU.1 cells before (day 0) and at days 1 and 3 after PU.1 induction and subjected to DNA microarray analyses (Illumina:Sentrix® Human-6 Expression BeadChip). The expression levels of genes were analyzed using the GeneSpring7.2 software, and compared among days 0, 1 and 3. A total of 47,296 human genes were analyzed, among which 21,565 genes were found to be expressed and 25,731 genes were not expressed. Among the 21,565 genes expressed, 479 genes were up-regulated by more than 2-fold and 1,697 genes were down-regulated by more than 50% on either day 1 or day 3 after PU.1 induction. The 30 genes exhibiting the highest up-regulation after PU.1 induction are shown in Table 1 (day 1) and Supplementary Table S1 (day 3). IFIT1, IFITM1, IFIT2, IFIT4, IFI27, G1P2, C1orf29, LY6E, LAMP3 and G1P3, which are known as IFN-stimulated genes (ISGs) (de Veer et al., 1998), were up-regulated by more than 20-fold at day 1 and more than 5-fold at day 3. These up-regulations were confirmed by semi-quantitative PCR of IFIT4 and IFI27 (data not shown). These data indicate that PU.1 induction activated the expression of many ISGs in U266 myeloma cells. In contrast, the 30 most down-regulated genes at day 1 and day 3 after PU.1 induction are shown in Supplementary Table S2 and Supplementary Table S3. Among these down-regulated genes, Syndecan 1, which is a marker of plasma cells and known as CD138, was the most down-regulated gene 3 days after PU.1 induction.
Table 1.
Thirty genes showing the highest up-regulation at 1 day after PU.1 induction
| Gene | Fold Expression |
|---|---|
| IFIT1 | 167.8 |
| IFITM1 | 155.2 |
| IFIT2 | 126.8 |
| IFIT4 | 98.2 |
| IFI27 | 83.3 |
| TNFSF10 | 76.5 |
| G1P2 | 71.0 |
| IFI44L | 55.3 |
| LY6E | 44.7 |
| ISG20 | 43.5 |
| TRIM22 | 41.6 |
| IFI44 | 37.5 |
| IFITM3 | 36.3 |
| OASL | 35.7 |
| RSAD2 | 35.1 |
| PRIC285 | 28.0 |
| MX1 | 27.8 |
| LAMP3 | 25.6 |
| CMPK2 | 25.5 |
| IRF7 | 24.8 |
| MT2A | 24.7 |
| PARP12 | 24.7 |
| CXCL10 | 22.8 |
| USP18 | 20.8 |
| LOC285510 | 19.2 |
| IL1RN | 18.7 |
| SLC24A1 | 18.2 |
| SP110 | 17.6 |
| G1P3 | 17.2 |
| SP110 | 16.9 |
Cell cycle arrest induced by PU.1 may be partially related to up-regulation of p21
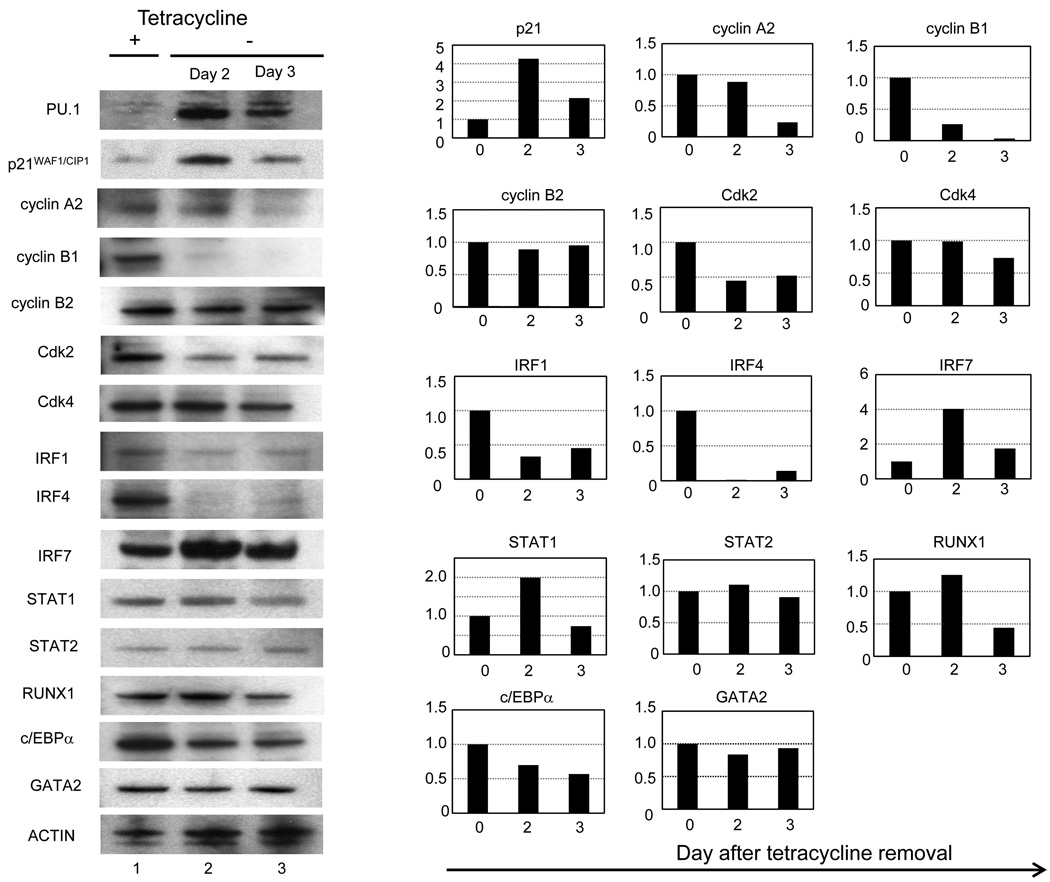
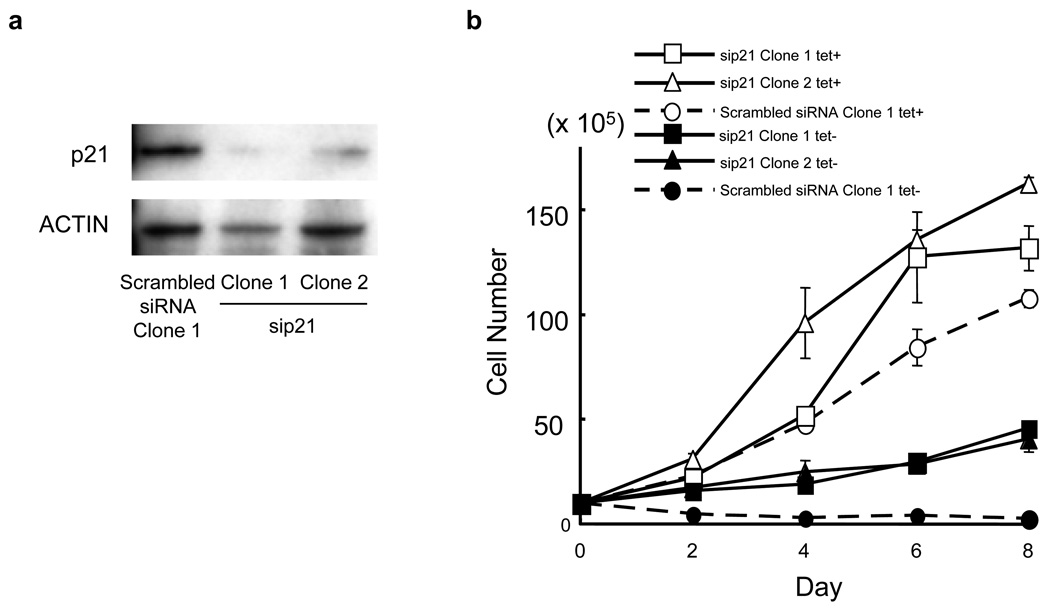
Next, we investigated the mechanisms of the cell growth arrest of U266 cells induced by PU.1 by comparing the gene expression profiles of cell cycle-related genes. Most of the cyclin genes, particularly cyclin A2, B1, B2, D1, D2 and E2, were all continuously down-regulated from day 1 to day 3 after PU.1 induction, and E2F1 and E2F2 were also down-regulated (Table 2). Regarding the CDK families, CDK2 and CDK4 were down-regulated to 77% and 40%, respectively. We previously confirmed down-regulation of cyclin D1 at the protein level (Tatetsu et al., 2007). In addition, cyclins, CDKs, and E2F were analyzed by western blot. We confirmed that cyclin A2 and B1, CDK2 and 4 were also down-regulated at the protein levels (Figure 1), whereas E2F2 protein level was not changed (data not shown). The down-regulation of these cyclins and CDK family members was consistent with the growth arrest of U266tetPU.1 cells after PU.1 induction. We further evaluated the expression profiles of growth inhibitory genes, including the tumor suppressor genes p53, p15 and p16, and found that only p21 was up-regulated by 3.4-fold at day 1 and 2.2-fold at day 3 after PU.1 induction (Table 2). The protein level of p21 was also increased by 4.3-fold (Figure 1). Therefore, we next evaluated the effect of up-regulated p21 on the growth arrest of U266tetPU.1 cells expressing PU.1 by knocking p21 using stably expressed siRNAs (Figure 2a). Suppression of p21 partially restored growth of U266tetPU.1 cells expressing PU.1, suggesting that p21 may be partially involved in the growth arrest of U266tetPU.1 cells induced by PU.1 (Figure 2b).
Table 2.
Changes in cell cycle-related genes after PU.1 induction
| Gene symbol | Day1/Day0 (fold) | Day3/Day0 (fold) |
|---|---|---|
| CCNA1 | 1.35 | 1.07 |
| CCNA2 | 0.47 | 0.36 |
| CCNB1 | 0.65 | 0.86 |
| CCNB2 | 0.54 | 0.48 |
| CCNB3 | 0.93 | 0.97 |
| CCND1 | 0.60 | 0.57 |
| CCND2 | 0.64 | 0.34 |
| CCND3 | 1.94 | 0.71 |
| CCNE1 | 0.91 | 0.84 |
| CCNE2 | 0.39 | 0.37 |
| CCNH | 1.14 | 0.70 |
| E2F1 | 0.49 | 0.61 |
| E2F2 | 0.22 | 0.26 |
| RB1 | 1.08 | 1.07 |
| CDK2 | 0.77 | 0.81 |
| CDK4 | 0.40 | 0.77 |
| CDK6 | 0.95 | 0.99 |
| CDK7 | 0.62 | 1.10 |
| P15 | 1.31 | 1.59 |
| p16 | 1.09 | 1.01 |
| p18 | 0.77 | 0.64 |
| p19 | 0.92 | 0.93 |
| p21 | 3.40 | 2.16 |
| p27 | 0.90 | 0.80 |
| P53 | 0.82 | 1.04 |
| P57 | 0.79 | 1.80 |
Figure 1.
IRF7 and p21WAF1/CIP were up-regulated and IRF4 was down-regulated in U266tetPU.1 cells after PU.1 induction. Western blotting analyses of cell cycle related genes, IRFs, STATs, and transcription factor essential for hematopoiesis were performed on cell extracts of U266tetPU.1 cells before (lane 1) and at 2 day (lane 2) and 3 days (lane 3) after PU.1 induction by removal of tetracycline. Right panels are based on the densitometry of the left panel. Each expression level of day 0 is set as 1, and relative expression levels at day 2 and 3 were shown in each right panel.
Figure 2.
Growth arrest of U266tetPU.1 cells expressing PU.1 may be partially induced by up-regulated p21. (a) The protein levels of p21 were strongly suppressed by stably expressed siRNA for p21. Western blot analysis of p21 2 day after PU.1 induction in U266tetPU.1 derivative cell lines expressing siRNA for p21 or its scramble was shown. (b) Growth curve of two U266tetPU.1 derivative cell lines expressing siRNA for p21 with (open triangle and open square) or without tetracycline (closed triangle and closed square) and one U266tetPU.1 derivative cell line expressing scrambled siRNA with (open circle) or without tetracycline (closed circle). Error bars represent the standard deviation derived from three independent experiments.
IRF7 shows the highest up-regulation and IRF4 does strong down-regulation among transcription factors after PU.1 induction
Among the genes up-regulated after PU.1 induction, IRF7 was the only transcription factor that was highly up-regulated at both day 1 (24.8-fold) and day 3 (6.0-fold) after PU.1 induction (Table 1 and Supplementary Table S1). The protein level of IRF7 was also highly up-regulated, as evaluated by western blotting analysis (Figure 1). Therefore, we speculated that IRF7 may be a key molecule for activating the expression of ISGs. In contrast, IRF4, which is also known as MUM1 or Pip and is highly expressed in almost all myeloma cells (Iida et al., 1997), was down-regulated to 44% at day 1 and 36% at day 3. Protein level of IRF4 was also strongly downregulated to 1.8% at day 2 and 18% at day 3 after PU.1 induction (Figure 1). Other up-regulated transcription factor in both mRNA and protein levels was STAT1, which binds to IRFs and translocates to the nucleus during interferon signal transduction. In conclusion, both the most up-regulated transcription factor, IRF7, and the strongly down-regulated transcription factor, IRF4, belong to the interferon signal transduction pathway.
We next evaluated the expression levels of genes involved in lineage commitment and differentiation. The critical transcription factors for lineage commitments were evaluated with the DNA microarray data, and RUNX1, which is essential for definitive hematopoiesis and directly regulates PU.1 (Huang et al., 2008), was found to be down-regulated to about 50% at days 1 and 3 after PU.1 induction, while no changes in expression were detected for GATA-2 and C/EBPα. To confirm protein expression levels of these transcription factors, we performed western blot analyses and found that protein levels of RUNX1 and C/EBPα were down-regulated to 44% and 57% 3 day after PU.1 induction, respectively (Figure 1). In contrast, protein level of GATA-2 was not changed and there was no expression of GATA-1 in U266tetPU.1 cells both before and after PU.1 induction by western blot analyses.
TRAIL may be a key molecule for apoptosis of U266tetPU.1 and KMS12PEtetPU.1 cells induced by PU.1
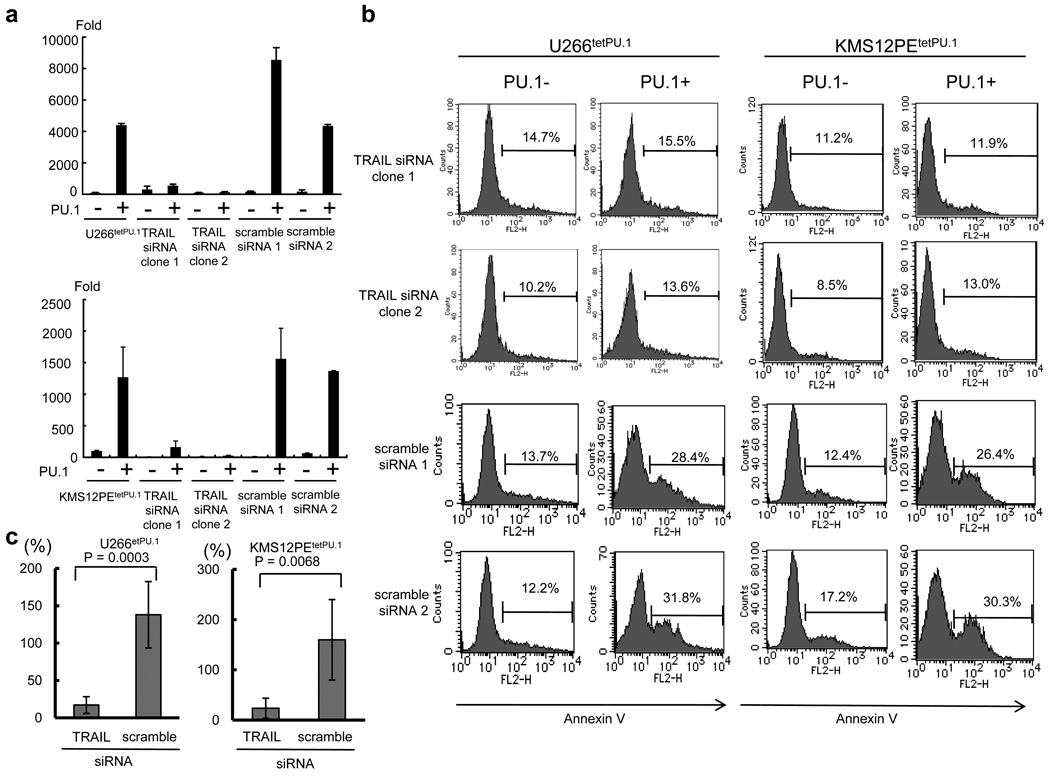
Among the apoptosis-related genes, TRAIL (TNFSF10), a ligand for death receptors DR4 and DR5, was up-regulated by 76.5-fold at day 1 (Table 1) and 1.7-fold at day 3 after PU.1 induction in U266tetPU.1 cells, and these data were confirmed by Real Time PCR (44.0-fold at day 2) (Figure 3a). TRAIL was also strongly up-regulated (12.1-fold at day 2) after PU.1 induction in KMS12PE tetPU.1 cells (Figure 3a). It is well known that TRAIL can induce apoptosis in myeloma cells (Mariani et al., 1997; Mitsiades et al., 2002). Therefore, to clarify whether TRAIL is a key molecule for the apoptosis of U266tetPU.1 and KMS12PE tetPU.1 cells induced by PU.1, we stably introduced siRNAs for TRAIL into both cell lines and obtained stable transformants exhibiting strong knockdown of TRAIL (Figure 3a). Stable expression of siRNA targeting TRAIL inhibited apoptosis of U266tetPU.1 and KMS12PE tetPU.1 cells induced by PU.1, whereas scrambled siRNAs did not (Figure 3b), and the difference in apoptosis between siRNA for TRAIL and its scrambled counterparts was statistically significant (Figure 3c). Taken together, TRAIL may mainly induce apoptosis in U266tetPU.1 and KMS12PE tetPU.1 cells after PU.1 expression.
Figure 3.
Apoptosis of U266tetPU.1 and KMS12PEtetPU.1 cells expressing PU.1 may be highly induced by up-regulated TRAIL. (a) TRAIL is up-regulated in U266tetPU.1 (upper panel) and KMS12PEtetPU.1 cells (lower panel) after PU.1 induction. Real-time PCR of TRAIL was performed for U266tetPU.1 and KMS12PEtetPU.1 cells and their transformants stably expressing siRNAs for TRAIL or its scrambled sequences. Bar graphs represent the mean and s.d. of three experiments. (b) The siRNAs for TRAIL inhibit apoptosis of U266tetPU.1 and KMS12PEtetPU.1 cells expressing PU.1. After conditional expression of PU.1, Annexin V-positive cells of U266tetPU.1 and KMS12PEtetPU.1 cells stably expressing siRNA for TRAIL or its scrambles were determined as apoptotic cells. (c) Suppression of apoptosis by siRNA for TRAIL is statistically significant. Based on the results of Annexin V staining as shown in Figure 3b, percent increases of apoptotic cells in U266tetPU.1 and KMS12PEtetPU.1 cells stably expressing siRNA for TRAIL or its scrambles after PU.1 induction are shown compared to those before PU.1 induction (set as 100%). Means ± s.d. for three independent clones expressing siRNA for TRAIL or its scrambled siRNA are shown.
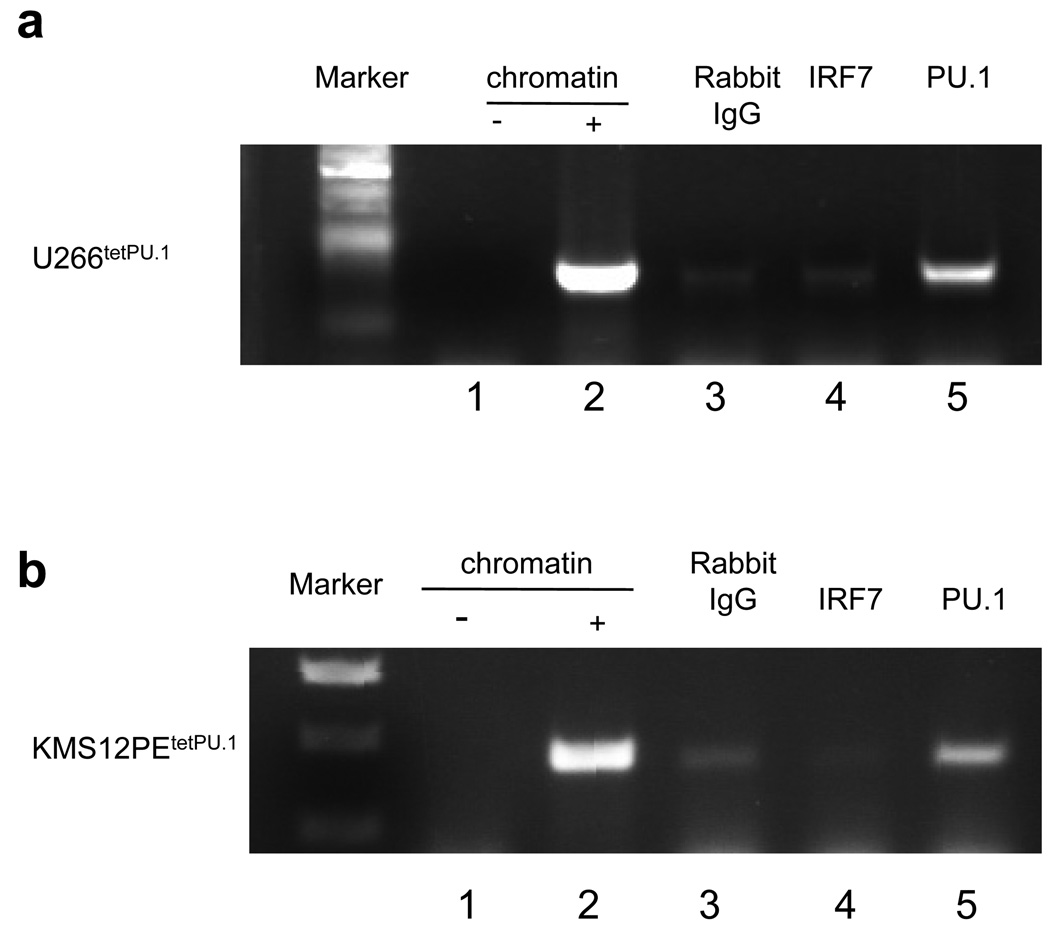
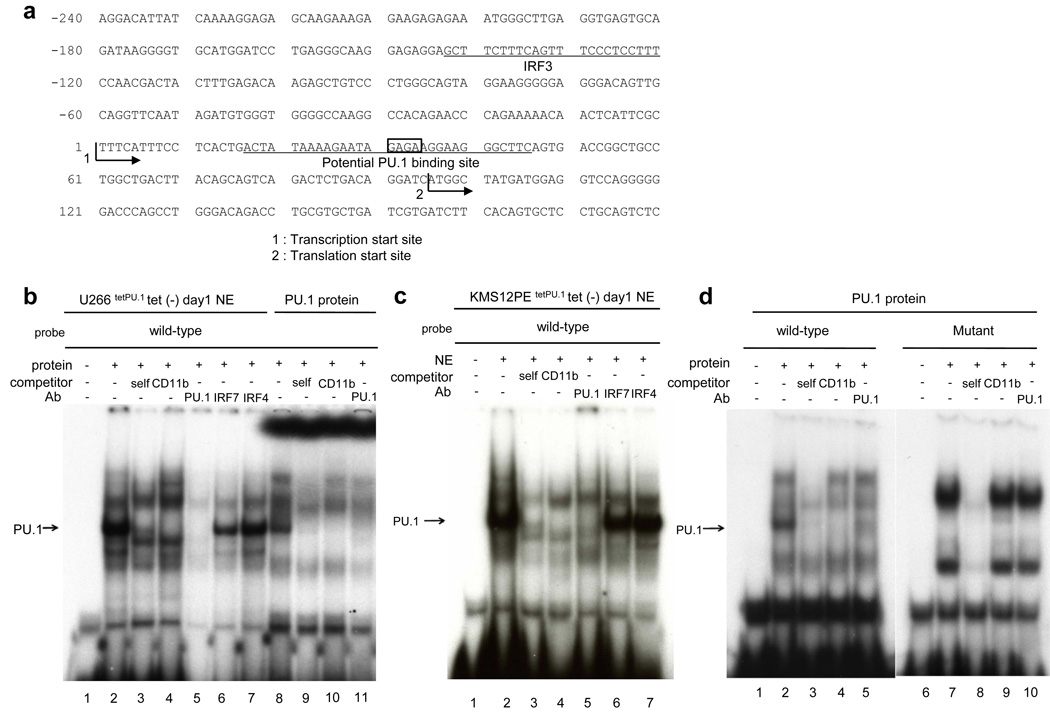
PU.1 directly binds to the TRAIL promoter in U266tetPU.1 and KMS12PEtetPU.1 cells
Next, we explored how TRAIL was induced after PU.1 expression. It was previously reported that IRF3 up-regulated the TRAIL promoter after paramyxovirus infection (Kirshner et al., 2005). In our microarray data, IRF3 was not up-regulated, whereas IRF7 was highly up-regulated, after PU.1 induction in U266tetPU.1 cells. Therefore, we performed chromatin immunoprecipitation (ChIP) assays of the TRAIL promoter with anti-IRF7 and anti-PU.1 antibodies, and unexpectedly found that PU.1 itself, but not IRF7, directly bound to the TRAIL promoter (Figure 4a). In case of KMS12PEtetPU.1 cells expressing PU.1, PU.1 also bound to the TRAIL promoter (Figure 4b). Therefore, we evaluated the TRAIL promoter to search for transcription binding sites and found one potential PU.1-binding site located in 30-bp 3’ downstream of the transcription start site (Figure 5a). We performed EMSAs using oligonucleotides harboring the potential PU.1-binding site and nuclear extracts of U266tetPU.1 cells and identified several bands for protein binding (Figure 5b). Competition with CD11b promoter oligonucleotides including the PU.1-binding site and addition of the anti-PU.1 antibody eliminated one band for protein binding (Figure 5b, lanes 4 and 5) of the oligonucleotides for the 30-bp 3’ downstream region of the transcription start site (Figure 5a), indicating that the binding to the oligonucleotides was PU.1-specific. We also identified the same PU.1 binding complex with the same oligonucleotides and nuclear extracts of KMS12PEtetPU.1 cells expressing PU.1 (Figure 5c). In addition, in vitro-translated PU.1 protein bound to the same oligonucleotides and CD11b oligonucleotides and the anti-PU.1 antibody eliminated the binding (Figure 5d, lane 1–5), indicating that PU.1 binds to the oligonucleotides. Next we introduced mutations into the PU.1-binding site (GAGA to TCGC) in the oligonucleotides and performed EMSAs. We detected two major shifted bands, but these did not disappear after competition with the CD11b oligonucleotides or addition of the anti-PU.1 antibody (Figure 5d, lane 6–10), indicating that the mutations in the PU.1-binding motif completely abolished PU.1 binding to the TRAIL promoter region. Therefore, these data indicate that PU.1 binds to the 30-bp 3’ downstream region of the transcription start site of the TRAIL promoter.
Figure 4.
PU.1 binds to the TRAIL promoter region in both U266tetPU.1 and KMS12PEtetPU.1 cells in vivo. (a) and (b) Chromatin immunoprecipitation (ChIP) assays reveal that PU.1, but not IRF7, binds to the TRAIL promoter region. ChIP assays were performed on U266tetPU.1 (a) and KMS12PEtetPU.1 cells (b) at 1 day after PU.1 induction. Chromatin DNA was immunoprecipitated with anti-IRF7 or anti-PU.1 antibodies, or rabbit IgG as a control. After extraction of genomic DNA, PCR amplification of the TRAIL promoter and exon 1 region was performed with water (lane 1), input chromatin DNA (lane 2) or genomic DNA immunoprecipitated by rabbit IgG (lane 3), anti-IRF7 antibody (lane 4) or anti-PU.1 antibody (lane 5).
Figure 5.
PU.1 binds to a 30-bp 3’ downstream region of the transcription start site of the TRAIL gene. (a) A potential PU.1-binding site is located in a 30-bp 3’ downstream region of the transcription start site of the TRAIL gene. The sequence of the TRAIL promoter region and 180 bp of exon 1 is shown. The potential PU.1-binding site is located in a 30-bp 3’ downstream region of the transcription start site in the 5’ non-coding region of exon 1. The sequence for oligonucleotides utilized for EMSA in Figure 5b–d is underlined (wild-type). A previously described binding site for IRF3 is also shown (Kirshner et al., 2005). (b)(c) PU.1 binds to the 30-bp 3’ downstream region of the transcription start site. An EMSA was performed with nuclear extracts of U266tetPU.1 (b) and KMS12PEtetPU.1 cells (c) at 1 day after PU.1 induction and the oligonucleotides shown in Figure 4a (wild-type). “Self” refers to competition with itself. “CD11b” refers to competition with oligonucleotides complementary to the CD11b promoter region including the PU.1-binding site (CD11b oligonucleotides). The relative position of the PU.1 complex is shown on the left side of the panels (arrow). PU.1, IRF7, and IRF4 refer to antibodies used for supershift assays. The anti-PU.1 antibody does not create a supershift but instead eliminates the PU.1 complex (lane 5). In vitro translated PU.1 generated PU.1 specific band as a positive control as shown in (d) (b, lane 8–11). (d) In vitro-translated PU.1 protein specifically binds to the 30-bp 3’ downstream region of the transcription start site. EMSAs were performed with in vitro-translated PU.1 and wild-type oligonucleotides or mutant oligonucleotides of the PU.1-binding site (GAGA to TCGC). The relative position of the PU.1 complex is shown on the left side of the panels (arrow). The CD11b oligonucleotides and anti-PU.1 antibody eliminate the PU.1 complex (lanes 4 and 5). In contrast to the PU.1 complex with the wild-type oligonucleotides, there is no PU.1 complex with the mutant oligonucleotides that can be diminished by competition with the CD11b oligonucleotides or anti-PU.1 antibody (lanes 7–10).
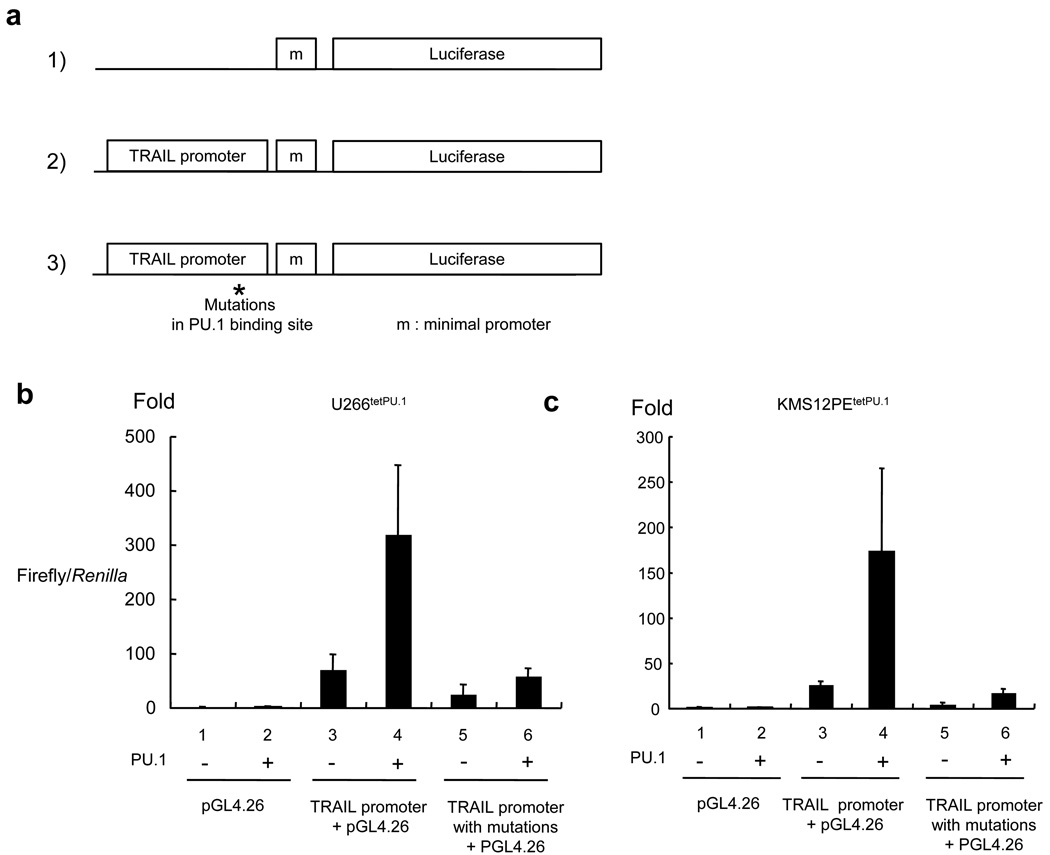
PU.1 directly transactivates the TRAIL promoter in U266tetPU.1 and KMS12PEtetPU.1 cells
To evaluate whether the binding of PU.1 can directly transactivate the TRAIL promoter, we performed luciferase reporter assays with a construct comprised of the TRAIL promoter and a luciferase reporter gene in U266tetPU.1 and KMS12PEtetPU.1 cells before and after PU.1 induction (Figure 6a). With the wild-type TRAIL promoter, the expression level was up-regulated by 70-fold in U266tetPU.1 cells and 26-fold in KMS12PEtetPU.1 cells compared to that of pGL4.26 alone when tetracycline was not removed (without PU.1 expression) (Figure 6b and c). In addition, when PU.1 was up-regulated, reporter gene expression was highly up-regulated by 4.6-fold compared to that with no PU.1 expression in U266tetPU.1 cells, and 6.6-fold in KMS12PEtetPU.1 cells. We further examined whether the up-regulated reporter gene expression was dependent on PU.1 binding to the TRAIL promoter. To achieve this, we introduced the same mutations (GAGA to TCGC) into the PU.1-binding site in the 30-bp 3’ downstream region of the transcription start site of the TRAIL promoter construct that were shown to abolish PU.1 binding in EMSAs (Figure 5d), and found that these mutations completely eliminated the up-regulation of reporter gene expression relative to the control plasmid (pGL4.26) in both U266tetPU.1 and KMS12PEtetPU.1 cells regardless of whether or not PU.1 was induced (Figure 6b and c). Taken together, these data suggest that TRAIL expression in both U266tetPU.1 and KMS12PEtetPU.1 cells expressing PU.1 is highly dependent on direct binding of PU.1 to the TRAIL promoter.
Figure 6.
PU.1 directly transactivates the TRAIL promoter in luciferase reporter assays. (a) U266tetPU.1 and KMS12PEtetPU.1 cells were transfected with 1) pGL4.26, 2) pGL4.26 with the TRAIL promoter and exon 1 5’ non-coding region, or 3) pGL4.26 with the TRAIL promoter and exon 1 5’ non-coding region containing mutations in the PU.1-binding site in the 30-bp 3’ downstream of the transcription start site. pRL-TK was co-transfected to express Renilla activity for standardization of the transfection efficiency. (b) (c) The TRAIL promoter activity is dependent on the PU.1-binding site. The Firefly/Renilla ratio of U266tetPU.1 (b) or KMS12PEtetPU.1 cells (c) before PU.1 induction transfected with pGL4.26 was set as 1 and the transactivation activities are shown as the fold expression. Three independent pools for each plasmid transfection with or without tetracycline were prepared for reporter assays and the mean and the standard deviation for each pool are shown.
Discussion
We previously reported that down-regulation of PU.1 in myeloma cells is necessary for their growth and survival, and that conditional expression of PU.1 in myeloma cells induced growth arrest and apoptosis (Tatetsu et al., 2007). In the present study, we investigated the genes involved in the growth arrest and apoptosis of U266 myeloma cells by performing DNA microarray analyses and comparing the gene expressions before and after PU.1 induction. We found that among cell cycle related genes, p21 was up-regulated and cyclin A2, cyclin B1, CDK2, and CDK4 were down-regulated after PU.1 induction. Among apoptosis-related genes, TRAIL was directly transactivated by PU.1 and induced apoptosis in both U266tetPU.1 and KMS12PEtetPU.1 cells expressing PU.1.
After PU.1 induction, many cyclins and CDKs were down-regulated, and these findings are compatible with the growth arrest of U266 cells. In contrast, among tumor suppressor genes, only p21 was up-regulated after PU.1 induction, and this was confirmed at the protein level. Given the fact that knockdown of p21 by siRNA partially rescue the PU.1 induced growth arrest of U266tetPU.1 cells, p21 may partially mediate the induction of growth arrest of myeloma cells by PU.1. The mechanism of the up-regulation of p21 after PU.1 induction in U266tetPU.1 cells is now being elucidated. It was reported that IRF1 directly transactivates p21 gene expression by binding to the p21 gene promoter (Coccia et al., 1999). Among the IRF family members, IRF7 and IRF9 were highly up-regulated at 24.8-fold and 3.9-fold in mRNA levels, respectively. It is reasonable that these IRFs may induce p21 expression after PU.1 induction.
We also evaluated the gene expression levels of apoptosis-related genes in U266 cells expressing PU.1 and found that TRAIL was highly up-regulated after PU.1 induction and stably expressed siRNAs for TRAIL suppressed the apoptosis of not only U266tetPU.1 but also KMS12PEtetPU.1 cells expressing PU.1. It is likely that TRAIL is a key molecule for inducing apoptosis in U266tetPU.1 and KMS12PEtetPU.1 cells after PU.1 expression, since it was found to effectively induce apoptosis in multiple myeloma cells in vitro (Gazitt, 1999; Mariani et al., 1997; Mitsiades et al., 2001; Mitsiades et al., 2002). We further found that PU.1 itself directly transactivates the TRAIL promoter in both myeloma cell lines. This is a first report to show that PU.1 is a direct transactivator of TRAIL gene expression. From this aspect, PU.1 may be a regulator of unlimited proliferation of hematopoietic cells including plasma cells through up-regulation of the cell death inducer TRAIL.
As shown in Figure 3b and c, siRNA for TRAIL may not completely suppress apoptosis of U266tetPU.1 and KMS12PEtetPU.1 cells after PU.1 expression, suggesting that other factors may be involved in the apoptosis of these cells. IRF4 is an essential transcription factor for generation of plasma cells, since transgenic mice with a conditional knockout of Irf4 in germinal center B cells were unable to differentiate memory B cells into plasma cells (Klein et al., 2006). In addition, it was reported that knockdown of IRF4 induced apoptosis in many genetic types of myeloma cells (Shaffer et al., 2008). Therefore, because protein levels of IRF4 were strongly down-regulated to 0.02-fold at 2 days and 0.18-fold 3 days after PU.1 induction (Figure 1), it is possible that this down-regulation of IRF4 may also induce apoptosis in myeloma cells following PU.1 expression.
In the present study, we found that PU.1 can directly transactivate TRAIL gene expression in myeloma cells. This finding may be universal for all hematopoietic cells, even though we still need to investigate whether PU.1 also induces TRAIL gene expression in other hematopoietic lineages. If this proves to be the case, up-regulation of PU.1 may be a universal therapeutic molecular target for hematopoietic malignancies. Indeed, up-regulation of TRAIL may represent a major target for HDAC inhibitors (Insinga et al., 2005; Nebbioso et al., 2005), and this could be mediated by up-regulation of PU.1. PU.1 also induced down-regulation of IRF4 that may result in apoptosis of myeloma cells (Shaffer et al., 2008). Therefore, up-regulation of PU.1 may represent a possible treatment of multiple myeloma by inducing the combination of up-regulation of TRAIL and down-regulation of IRF4.
Materials and Methods
DNA microarray analysis
We previously generated U266tetPU.1 and KMS12PE tetPU.1 cells that express PU.1 by tet-off system (Tatetsu et al., 2007). RNA was extracted from U266tetPU.1 cells at different three time points, before, and 1 day or 3 days after tetracycline removal. To compare gene expression differences between before and after tetracycline removal, the microarray analysis was performed using Illumina:Sentrix® Human-6 Expression BeadChip as recommended by the manufacturer. Expression levels of genes were analyzed by GeneSpring 7.2 software.
Cell culture
Human myeloma cell line U266tetPU.1 and KMS12PE tetPU.1 cells, and their derivatives were grown in RPMI 1640 containing 10% fetal bovine serum at 37°C.
Western blot analysis
Cell lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. The membranes were incubated with anti-PU.1, anti-p21WAF/CIP1, anti-cyclin A2, anti-cyclin B2, anti-CDK2, anti-CDK4, anti-IRF1, anti-IRF4, anti-IRF7, anti-STAT1, anti-STAT2, anti-RUNX1, anti-c/EBPα, anti-GATA1, anti-GATA2, anti-Actin primary antibody (Santa Cruz Biotechnology) or anti-cyclin B1 (Abcam) for 3–16 h. Finally the membranes were incubated with peroxidase-labeled secondary antibodies for 1 h, and developed using an enhanced chemiluminescence system (Amersham Life Science Inc.).
Generation of U266tetPU.1 and KMS12PEtetPU.1 cells stably expressing siRNA for TRAIL or p21 and those scrambled siRNAs
The siRNA expression vectors were generated by insertion of annealed oligonucleotides for TRAIL or p21 and those scrambled siRNAs into from BamHI to HindIII site of pRNA-U6.1/Zeo or pRNA-U6.1/Hygro (GenScript) (Supplementary Information). These siRNA expression vectors were transfected to U266tetPU.1 or KMS12PEtetPU.1 cells by elecrotoporation and stable transformants were obtained by zeocin or hygromycin selection.
Real-time PCR
Quantitative Taqman PCR was performed with commercially available assay-on demand probe primer sets for TRAIL and β-actin (Applied Biosystems) and Taqman Universal PCR Master Mix reagent using an ABI Prism 7700 Sequence Detection System. The expression levels of β-actin were used to standardize the relative expression levels of TRAIL. The expression level of TRAIL in U266tetPU.1 and KMS12PE tetPU.1 cells before tetracycline removal was set as 100.
Detection of apoptosis
To detect apoptosis of myeloma cells, the cells were stained with an Annexin V Phycoerythrin Apoptosis Detection kit (Medical and Biological Laboratories) and analyzed for Annexin V expression with FACS caliber (Becton Dickinson).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) were performed as previously described (Okuno et al., 2005). Briefly, 1×107 of U266tetPU.1 or KMS12PE tetPU.1 cells 1 day after tetracycline removal were treated with 0.5% formaldehyde and genomic DNA was extracted and sonicated. Small amount of genomic DNA was saved as input DNA. The total input sample was diluted 1/100 before being used as a template for PCR. Soluble chromatin was treated with anti-PU.1 antibody, anti-IRF7 antibody, and rabbit IgG, and immunoprecipitated by protein A agarose beads (Santa Cruz Biotechnology). Each precipitated DNA was eluted and extracted as previously described and subjected for PCR. PCR was performed with TRAIL promoter sense primers, 5’-TGAGGATATGTTAGGGAAAAGCA-3’ located 284-bp upstream of transcription start site, and TRAIL exon 1 antisense primer 5’-GATCACGATCAGCACGCAGGTCT-3’ located 140-bp downstream of transcription start site.
Electromobility shift assay
Nuclear extracts of 5 × 106 U266tetPU. 1 or KMS12PE tetPU.1 cells 1 day after removal of tetracycline were prepared with NE-PER®Nuclear and Cytoplasmic Extraction Reagents (PIERCE). PU.1 cDNA was subcloned into pTNT™Vector (Promega) and subjected to TnT® Quick Coupled Transcription/Translation Systems (Promega) to prepare in vitro translated PU.1 protein. Oligonucleotides, 5’-ACTATAAAAGAATAGAGAAGGAAGGGCTTC-3’ and 5’-GAAGCCCTTCCTTCTCTATTCTTTTATAGT-3’, were annealed and subjected to electromobility shift assay (EMSA) for PU.1 binding site in a 30-bp 3’ downstream region of the transcription start site of the TRAIL gene, whereas oligonucleotides, 5’-ACTATAAAAGAATATCGCAGGAAGGGCTTC-3’ and 5’-GAAGCCCTTCCTGCGATATTCTTTTATAGT -3’ were annealed and utilized as mutant oligonucleotides for the PU.1 binding site of the TRAIL promoter. Annealed oligonucleotides were labeled with [γ-P32]ATP using T4 polynucleotide kinase and incubated with nuclear extracts or in vitro-translated PU.1 protein in 10 mM HEPES (pH 7.8), 50 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, and 5% glycerol for 15 min at 0°C. Reaction mixtures were separated with 6% polyacrylamide gels in 0.5× TBE buffer at 4°C. Gels were dried prior to autoradiography. Anti-PU.1, anti-IRF4, anti-IRF7 antibodies (Santa Cruz) were used for supershift assays. The CD11b promoter oligonucleotides harboring PU.1 binding site 5'-CTACTTCTCCTTTTCTGCCCTTCTTTG-3' and 5'-CAAAGAAGGGCAGAAAAGGAGAAGTAG-3', which have been described previously (Pahl et al., 1993), were annealed and utilized for competition assays.
Luciferase assay
1.5 kb DNA fragment consists of the 1480-bp human TRAIL promoter and 55-bp of exon 1 5’ non-coding region was amplified with primers 5’-TATACTCGAGGATAGAAGGCAAGGGCAGGAAGT-3’ and 5’-ATATAAGCTTCCGGTCACTGAAGCCCTTCCTTC-3’, then subcloned into from XhoI site to HindIII site of pGL4.26 [luc2/minP/Hygro] (Promega). Subsequently, to introduce mutations into the PU.1 binding site in the 30-bp downstream of TRAIL transcription start site in the obtained plasmid, self-amplification was performed with primers 5’-CACTGACTATAAAAGAATATCGCAGGAAGGGCTTCAGTGA -3’, and 5’-TCACTGAAGCCCTTCCTGCGATATTCTTTTATAGTCAGTG -3’ that are located on the almost same region but have opposite directions. 8 µg of these plasmids were transfected with 0.8 µg of pRL-TK plasmid to 8 × 105 of U266tetPU. 1 or KMS12PE tetPU.1 cells with Lipofectamine LTX Reagent (Invitrogen). Three independent pools for each plasmid were prepared for cultures with or without tetracycline. After 24 hour culture, luciferase activity was quantified with Dual-Glo Luciferase Assay System (Promega). Luciferase activity was standardized with dividing by Renilla activity.
Supplementary Material
Acknowledgments
This work was supported by The Award in Aki's Memory from The International Myeloma Foundation Japan (Y.O.) and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Amaravadi L, Klemsz MJ. DNA methylation and chromatin structure regulate PU.1 expression. DNA Cell Biol. 1999;18:875–884. doi: 10.1089/104454999314737. [DOI] [PubMed] [Google Scholar]
- Attal M, Harousseau JL, Facon T, Guilhot F, Doyen C, Fuzibet JG, et al. Single versus double autologous stem-cell transplantation for multiple myeloma. N Engl J Med. 2003;349:2495–2502. doi: 10.1056/NEJMoa032290. [DOI] [PubMed] [Google Scholar]
- Coccia EM, Del Russo N, Stellacci E, Orsatti R, Benedetti E, Marziali G, et al. Activation and repression of the 2–5A synthetase and p21 gene promoters by IRF-1 and IRF-2. Oncogene. 1999;18:2129–2137. doi: 10.1038/sj.onc.1202536. [DOI] [PubMed] [Google Scholar]
- de Veer MJ, Sim H, Whisstock JC, Devenish RJ, Ralph SJ. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics. 1998;54:267–277. doi: 10.1006/geno.1998.5555. [DOI] [PubMed] [Google Scholar]
- Gazitt Y. TRAIL is a potent inducer of apoptosis in myeloma cells derived from multiple myeloma patients and is not cytotoxic to hematopoietic stem cells. Leukemia. 1999;13:1817–1824. doi: 10.1038/sj.leu.2401501. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson P, Anderson KC. Novel therapeutic approaches for multiple myeloma. Immunol Rev. 2003;194:164–176. doi: 10.1034/j.1600-065x.2003.00053.x. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B, et al. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11:71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- Kirshner JR, Karpova AY, Kops M, Howley PM. Identification of TRAIL as an interferon regulatory factor 3 transcriptional target. J Virol. 2005;79:9320–9324. doi: 10.1128/JVI.79.14.9320-9324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Okuno Y, Zhang P, Radomska HS, Chen H, Iwasaki H, et al. Regulation of the PU.1 gene by distal elements. Blood. 2001;98:2958–2965. doi: 10.1182/blood.v98.10.2958. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons [see comments] Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Mariani SM, Matiba B, Armandola EA, Krammer PH. Interleukin 1 beta-converting enzyme related proteases/caspases are involved in TRAIL-induced apoptosis of myeloma and leukemia cells. J Cell Biol. 1997;137:221–229. doi: 10.1083/jcb.137.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. Embo J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- Mitsiades CS, Treon SP, Mitsiades N, Shima Y, Richardson P, Schlossman R, et al. TRAIL/Apo2L ligand selectively induces apoptosis and overcomes drug resistance in multiple myeloma: therapeutic applications. Blood. 2001;98:795–804. doi: 10.1182/blood.v98.3.795. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Mitsiades CS, Poulaki V, Anderson KC, Treon SP. Intracellular regulation of tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human multiple myeloma cells. Blood. 2002;99:2162–2171. doi: 10.1182/blood.v99.6.2162. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol. 2005;25:2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Huettner CS, Radomska HS, Petkova V, Iwasaki H, Akashi K, et al. Distal elements are critical for human CD34 expression in vivo. Blood. 2002a;100:4420–4426. doi: 10.1182/blood-2002-03-0788. [DOI] [PubMed] [Google Scholar]
- Okuno Y, Iwasaki H, Huettner CS, Radomska HS, Gonzalez DA, Tenen DG, et al. Differential regulation of the human and murine CD34 genes in hematopoietic stem cells. Proc Natl Acad Sci U S A. 2002b;99:6246–6251. doi: 10.1073/pnas.092027799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl HL, Scheibe RJ, Zhang DE, Chen HM, Galson DL, Maki RA, et al. The proto-oncogene PU.1 regulates expression of the myeloid-specific CD11b promoter. J Biol Chem. 1993;268:5014–5020. [PubMed] [Google Scholar]
- Radomska HS, Gonzalez DA, Okuno Y, Iwasaki H, Nagy A, Akashi K, et al. Transgenic targeting with regulatory elements of the human CD34 gene. Blood. 2002;100:4410–4419. doi: 10.1182/blood-2002-02-0355. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal S, Mehta J, Desikan R, Ayers D, Roberson P, Eddlemon P, et al. Antitumor activity of thalidomide in refractory multiple myeloma. N Engl J Med. 1999;341:1565–1571. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- Tatetsu H, Ueno S, Hata H, Yamada Y, Takeya M, Mitsuya H, et al. Down-regulation of PU.1 by Methylation of Distal Regulatory Elements and the Promoter Is Required for Myeloma Cell Growth. Cancer Res. 2007;67:5328–5336. doi: 10.1158/0008-5472.CAN-06-4265. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Yu W, Misulovin Z, Suh H, Hardy RR, Jankovic M, Yannoutsos N, et al. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5' of RAG2. Science. 1999;285:1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.