Abstract
The evolutionary appearance of p53 likely preceded its role in tumor suppression, suggesting that there may be unappreciated functions for this protein. Using genetic reporters as proxies to follow in vivo activation of the p53 network in Drosophila, we discovered that the process of meiotic recombination instigates programmed activation of p53 in the germline. Specifically, double-stranded breaks in DNA generated by the topoisomerase Spo11 provoked functional p53 activity, which was prolonged in cells defective for meiotic DNA repair. This intrinsic stimulus for the p53 regulatory network is highly conserved because Spo11-dependent activation of p53 also occurred in mice. Our findings establish a physiological role for p53 in meiosis and suggest that tumor suppressive functions may have been co-opted from primordial activities linked to recombination.
The p53 gene family mediates adaptive responses to genotoxic stress (1–3) and is broadly conserved (4, 5). It is widely accepted that the p53 regulatory network is generally compromised in human cancers but several lines of evidence indicate that during evolution, animals rarely lived long enough to experience cancer (6, 7). Therefore, tumor suppressive functions associated with p53 were likely derived from primordial activities that are poorly understood. We used Drosophila to investigate physiological properties of this network because, in this organism, a single p53 member exists and, like its mammalian counterparts, it coordinates stress responses and promotes genome stability (8–12).
We constructed transgenes (fig. S1) that place green fluorescence protein (GFP) under the control of an enhancer taken from sequences upstream of the Drosophila reaper (rpr) locus, which includes a p53 consensus binding site (13). Two transgenic lines were produced and designated as “p53Rps” for p53 reporter strains. One strain produces nuclear localized GFP (p53R-GFPnls) and the other does not (p53R-GFPcyt).
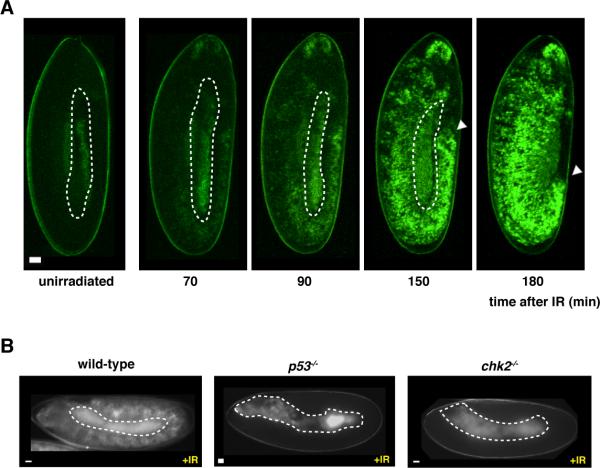
Ionizing radiation (IR) leads to DNA damage and activates p53 in various model systems. Therefore, we exposed p53Rps transgenic embryos to IR and followed GFP expression by time-lapse live imaging. GFP was observed as early as 70min after exposure to IR, and was prominent at 180min in virtually all embryos (Fig. 1A and movie S1). p53Rps expression was not observed in unirradiated embryos nor was it observed in flies lacking p53 or chk2 (Fig. 1B), its upstream activating kinase (14). To confirm that damaged DNA is responsible for activation, we also observed induction of GFP in response to UV radiation and injected DNA fragments (fig. S2). Thus, the p53Rps reporters serve as authentic proxies that enable us to monitor p53 activation in live animals in real time.
Fig. 1. Transgenic reporters as in vivo surrogates for p53 activation.
A) Live imaging of p53R-GFPnls expression after radiation (IR) challenge at indicated time points (also see movie S1). Autofluorescent yolk material is marked with dotted lines. Note that IR challenge did not interfere with germ band retraction (white arrow). (B) shows stimulus induced p53R-GFPnls expression seen in wild-type, is not seen in p53−/− or chk2−/− animals. Scale bar, 10μm.
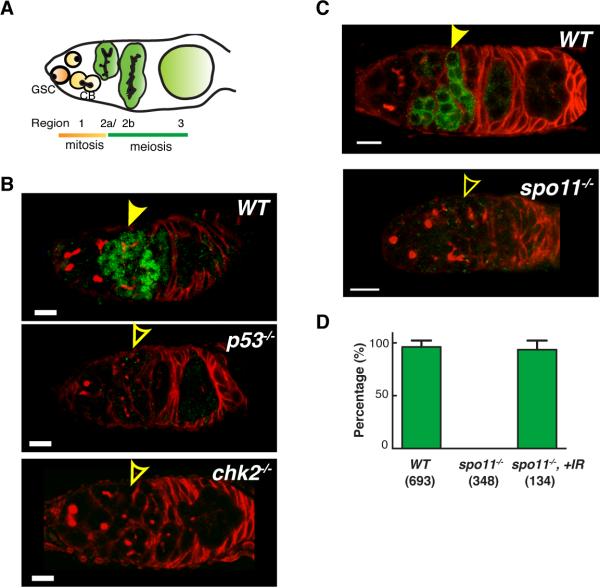
We surveyed reporter activity throughout development and found little or no evidence for unstimulated expression but, surprisingly, transient p53Rps expression was observed in germline precursors of all females. Activity was localized in region 2a and 2b of the germarium and was absent beyond region 3 and in all egg chambers (Fig. 2). As in somatic tissues, p53Rps activity was absent in the germaria of p53−/− or chk2−/− animals (Fig. 2B, fig. S4). Because p53 is present in this tissue, the pulse of p53Rps activity in oocyte precursors reflects the “on” state of this regulatory network, rather than the presence or absence of protein.
Fig. 2. Meiotic recombination instigates programmed activation of p53 in the Drosophila female germline.
(A) Schematic illustration of female germarium in Drosophila ovary. Region 1 contains germline stem cells (GSC) and their progeny, cystoblasts (CB). Cysts in region 2a and 2b initiate meiosis and further develop into egg chambers in region 3. Regions in the germarium are visualized using immunostaining for hu-li tai shao (HTS) protein. (B) and (C): Genotypes as indicated were stained for GFP (green) and HTS (red). Animals in (B) carry p53R-GFPnls, or p53R-GFPcyt in (C). Stereotyped GFP activation in region 2 is indicated by solid arrows and the absence of staining with open arrows. (D) Percentage of germarium with p53R-GFPcyt expression in regions 2a and 2b. Means ± standard deviations from at least three independent trials are plotted and sample size is denoted within parenthesis. Scale bars, 10μm.
Meiotic recombination is initiated in region 2a and 2b by Spo11 (also known as mei-W68), a topoisomerase that generates DNA double strand breaks (DSBs) needed for strand exchange (15). Therefore, we tested p53Rps activity in germaria lacking Spo11. Activation of p53 in regions 2a and 2b was absent in spo11−/− ovaries (Fig. 2C). Furthermore, DSBs introduced by IR were sufficient to restore p53Rps activity in regions 2a and 2b of spo11−/− germaria (Fig. 2D). Thus, DSBs formed during the initiating steps of meiotic recombination provoke activation of p53.
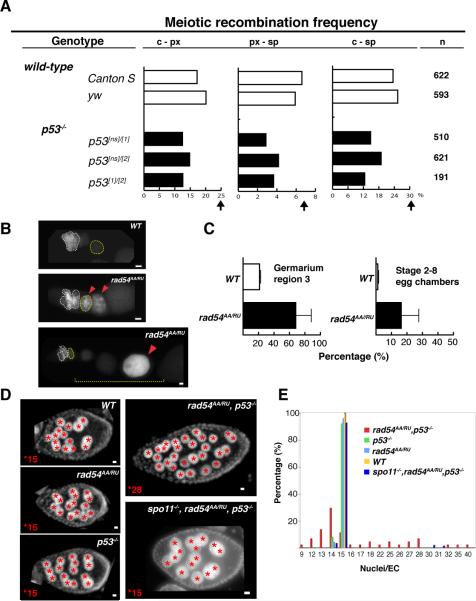
Flies lacking p53 are viable and fertile. Therefore, the gene does not exert essential functions needed for gametogenesis or proper chromosome segregation (table S2). To test whether meiotic recombination frequencies were altered in p53−/− animals, we assayed three distinct p53 alleles in trans-allelic combinations for meiotic exchange frequencies. Across all intervals, reduced crossover rates, ranging from 21% to 54%, were observed (Fig. 3A, table S1).
Fig. 3. Functions of p53 activity during meiosis.
(A) Meiotic recombination frequency in wild-type and p53−/− flies. Recombination frequency is reduced in all p53 transallelic combinations when compared to wild-type (Canton S and yw) at all three intervals. Arrows show expected frequencies (FlyBase). (B) Immunostainings of GFP in p53R-GFPcyt animals that are wild-type (WT) or rad54 mutants. Persisting GFP expression beyond region 2 (white dotted circle) is observed in rad54 mutants (noted by red arrows). (C) The incidence of p53Rps expression was quantified in Region 3 (noted by yellow dotted circle) and in stage 2 to 8 egg chambers (yellow dotted line) as indicated. (D) Images of egg chambers stained with DAPI from indicated genotypes. Red stars mark individual nurse cell nuclei. (E) Distribution of nurse cell nuclei number per egg chamber (nuclei/EC). Single gene mutants show 15 nuclei/EC, but p53−/−,rad54AA/RU ovaries exhibit a broad distribution, ranging from 9 to 40 nuclei/EC, which is restored to normal in spo11−/−,rad54AA/RU,p53−/− animals (blue group). Scale bars, 10μm.
To understand the function of p53 during meiotic recombination, we monitored p53 activity in mutants defective for proper meiosis. Rad54 is required to properly resolve DNA crossovers and in the germaria of rad54 (okra) mutants, unrepaired DNA breaks abnormally persist (16). In such animals, we found persistent activation of p53 beyond region 2 (Fig. 3B,C), as indicated by p53Rps expression in region 3 of the germarium and later-staged egg chambers, where a substantial increase in the incidence of reporter activation was observed (Fig. 3C). To test whether persistent p53 activation is functionally relevant, we examined animals doubly mutated for p53 and rad54. p53−/−,rad54AA/RU females were sterile despite the fact that corresponding trans-allelic combinations of the single gene mutants were fertile (see methods for allele descriptions). Furthermore, severe oogenesis defects, not seen in single mutants, were evident in double mutants, including abnormal numbers of nurse cell nuclei (Fig. 3D) and shortened egg lengths (fig. S5). To test whether genetic interactions between p53 and rad54 were instigated by meiotic recombination, we created spo11,p53,rad54 triple mutants. In these animals, fertility and normal nurse cell numbers was restored (Fig. 3D and E). Defects in egg length were also suppressed (fig. S5). Hence, genetic interactions between p53 and rad54 required the action of spo11. Furthermore, these results indicate that failure to properly resolve meiotic recombination can lead to sustained and functionally relevant p53 activity.
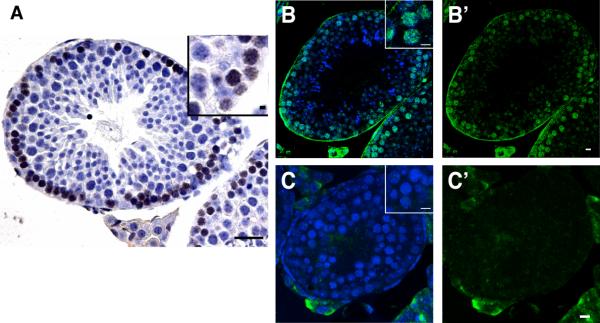
To determine whether activation of p53 by meiotic recombination is conserved, we examined mouse testes with antibodies that specifically detect phosphorylated-p53 at Ser15 (17). In seminiferous tubules from wild-type animals, p53 was transiently activated in early spermatocytes (Fig. 4, A and B). On the basis of nuclear morphology (described in supplemental methods), staining appeared to peak between leptotene and zygotene stages and disappeared after early pachytene stage. This pattern is consistent with earlier studies on Spo11 activity (18) and p53 promoter driven-chloramphenicol acetyltransferase (CAT) transgenes in this tissue (19). Staining for phospho-p53 (Ser15) was absent from testes of Spo11-deficient mice (Fig. 4, B and C), where spermatocytes survive through the leptotene and zygotene stage (20). Therefore, we can exclude cell loss as a reason for the absence of signal and, as in flies, meiotic recombination is necessary to provoke p53 activation.
Fig. 4. Spo11 dependent p53 activation is conserved in the mouse.
(A–C) Immunostainings for phospho-Ser15 p53 in paraffin sections from wild-type (A, B) and Spo11 knockout (C) mouse testes. DAB chromogen (A, brown) and fluorescent (B,C, green) detection methods show transient activation of p53 in meiotic cells is absent in Spo11-deficient testes. DNA staining (hematoxylin or Hoechst) is in blue. B' and C' show signal without Hoechst counterstain. Insets in (B) and (C) show nuclear localized staining of Ser15-p53 under comparable magnifications. Scale bar, 100 μm (A); 10μm (B–C, insets in A).
We demonstrate that meiosis, a defining step in sexual reproduction, signals a programmed burst of p53 activation. This activity is stimulated by the action of Spo11 in Drosophila females (recombination does not occur in males) and in mice. In flies, activation requires the kinase chk2 (which also has roles later in oogenesis (21)) but appears to be independent of the ATM and ATR kinases (fig. S3), suggesting that alternative transducers could mediate Spo11-dependent activation of p53.
In mice (Fig. 4), potential functions of Spo11-mediated activation of p53 are indicated by altered kinetics of gametogenesis in p53-deficient mice (22, 23) as well as giant-cell degenerative syndrome in the testes of p53-deficient males (24), and implantation defects in females (25). Additional layers of complexity or redundant activities conferred by the p63 and p73 paralogs (26, 27), could obscure conserved functions since recombination appeared normal without p53 (28). Nevertheless, these findings raise the possibility that the act of recombination during meiosis may have been an intrinsic primordial stimulus that shaped ancestral features of the p53 regulatory network. Future studies could elucidate whether p53 directly impacts crossover reactions (29) or imposes quality control on selected gametes.
Supplementary Material
Acknowledgments
We thank S. Keeney for helpful discussions and reading of the manuscript, and the following individuals, laboratories or facilities for helpful discussions, access to instruments, or providing reagents: S. Barolo, M. Buszczak, D. Castrillon, O. Cleaver, K. Hamra, J. Maines, D. McKearin, T. Schupbach, Y. Rong, J. Shelton, T. Wilkie, R. Bachoo, T. Mashimo, B. Thaden, Live Cell Imaging Facility (UTSW), Bloomington Stock Center (Indiana University), Developmental Studies Hybridoma Bank (University of Iowa); A. Ali, A. D'Brot and A. Olivo for assisting experiments. This work was supported by NIGMS (R01GM072124) and NIAAA (R01AA017328). I. Roig was supported by NIH grant R01 HD040916 (to M. Jasin and S. Keeney).
Footnotes
Supporting Online Materials Materials and Methods fig. S1, S2, S3, S4, S5 table S1, S2 movie S1
References
- 1.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Nat Rev Cancer. 2009 Dec;9:862. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Oren M. Nat Rev Cancer. 2009 Oct;9:749. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Cell. 2009 May;137:413. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Lu W-J, Amatruda JF, Abrams JM. Nat Rev Cancer. 2009 Oct;9:758. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 5.Nedelcu AM, Tan C. Dev Genes Evol. 2007 Dec;217:801. doi: 10.1007/s00427-007-0185-9. [DOI] [PubMed] [Google Scholar]
- 6.Aranda-Anzaldo A, Dent MAR. Mech Ageing Dev. 2007 Apr;128:293. doi: 10.1016/j.mad.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Junttila MR, Evan GI. Nat Rev Cancer. 2009 Nov;9:821. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 8.Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM. Oncogene. 2007 Aug;26:5184. doi: 10.1038/sj.onc.1210328. [DOI] [PubMed] [Google Scholar]
- 9.Brodsky MH, et al. Mol Cell Biol. 2004 Feb;24:1219. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin S, et al. Proc Natl Acad Sci USA. 2000 Jun;97:7301. [Google Scholar]
- 11.Lee JH, et al. FEBS Lett. 2003 Aug;550:5. doi: 10.1016/s0014-5793(03)00771-3. [DOI] [PubMed] [Google Scholar]
- 12.Sogame N, Kim M, Abrams JM. Proc Natl Acad Sci USA. 2003 Apr;100:4696. doi: 10.1073/pnas.0736384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodsky MH, et al. Cell. 2000 Mar;101:103. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 14.Peters M, et al. Proc Natl Acad Sci USA. 2002 Aug;99:11305. [Google Scholar]
- 15.McKim KS, Hayashi-Hagihara A. Genes Dev. 1998 Sep;12:2932. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klovstad M, Abdu U, Schupbach T. PLoS Genet. 2008 Feb;4:e31. doi: 10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shieh SY, Ikeda M, Taya Y, Prives C. Cell. 1997 Oct 31;91:325. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 18.Mahadevaiah SK, et al. Nat Genet. 2001 Mar;27:271. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 19.Almon E, et al. Dev Biol. 1993 Mar;156:107. doi: 10.1006/dbio.1993.1062. [DOI] [PubMed] [Google Scholar]
- 20.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Mol Cell. 2000 Nov;6:989. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 21.Abdu U, Brodsky M, Schupbach T. Curr Biol. 2002 Oct 1;12:1645. doi: 10.1016/s0960-9822(02)01165-x. [DOI] [PubMed] [Google Scholar]
- 22.Beumer TL, et al. Cell Death Differ. 1998 Aug;5:669. doi: 10.1038/sj.cdd.4400396. [DOI] [PubMed] [Google Scholar]
- 23.Ghafari F, Pelengaris S, Walters E, Hartshorne G. Hum Reprod. 2009 Feb; doi: 10.1093/humrep/dep022. [DOI] [PubMed] [Google Scholar]
- 24.Rotter V, et al. Proc Natl Acad Sci U S A. 1993 Oct;90:9075. doi: 10.1073/pnas.90.19.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W, Feng Z, Teresky AK, Levine AJ. Nature. 2007 Nov;450:721. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 26.Suh E-K, et al. Nature. 2006 Nov;444:624. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- 27.Tomasini R, et al. Genes Dev. 2008 Oct;22:2677. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gersten KM, Kemp CJ. Nat Genet. 1997 Dec;17:378. doi: 10.1038/ng1297-378. [DOI] [PubMed] [Google Scholar]
- 29.Habu T, et al. Carcinogenesis. 2004 Jun;25:889. doi: 10.1093/carcin/bgh099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.