Abstract
The Ca2+-binding proteins (CBPs) calbindin D28k, calretinin and parvalbumin are phenotypic markers of functionally diverse subclasses of neurons in the adult brain. The developmental dynamics of CBP expression are precisely timed: calbindin and calretinin are present in prospective cortical interneurons from mid-gestation, while parvalbumin only becomes expressed during the early postnatal period in rodents. Secretagogin (scgn) is a CBP cloned from pancreatic β and neuroendocrine cells. We hypothesized that scgn may be expressed by particular neuronal contingents during prenatal development of the mammalian telencephalon. We find that scgn is expressed in neurons transiting in the subpallial differentiation zone by embryonic day (E) 11 in mouse. From E12, scgn+ cells commute towards the extended amygdala and colonize the bed nucleus of stria terminalis, interstitial nucleus of the posterior limb of the anterior commissure, dorsal substantia innominata (SI), and the central and medial amygdaloid nuclei. Scgn+ neurons can acquire a cholinergic phenotype in the SI or differentiate into GABA cells in the central amygdala. We also uncover phylogenetic differences in scgn expression since this CBP defines not only neurons destined to the extended amygdala but also cholinergic projection cells and cortical pyramidal cells in the fetal non-human primate and human brains, respectively. Overall, our findings emphasize the developmentally shared origins of neurons populating the extended amygdala, and suggest that secretagogin can be relevant to the generation of functional modalities in specific neuronal circuitries.
Keywords: Ca2+-binding protein, extended amygdala, forebrain, mouse, primate
Introduction
Temporal and spatial coordination of intracellular Ca2+ signalling is essential to a cell’s ability for continuous dynamic adaptation to microenvironmental stimuli. Ca2+ signalling in neurons is particularly important in controlling presynaptic Ca2+ dynamics that underpin neurotransmitter release, transducing the activity of synaptic inputs (Burnashev & Rozov, 2005), and orchestrating multimodal intracellular signalling tuning neuronal excitability (Wang et al., 2004; Cohen & Greenberg, 2008). Both the homeostatic maintenance of intracellular [Ca2+] and the precise temporal control of its activity-dependent transients require effective mechanisms including Ca2+ extrusion by plasmalemmal Ca2+-ATPases (Strehler et al., 2007), dissipating Ca2+ oscillations via Ca2+ uptake by intracellular stores (Nicholls, 2009), and chelation of free, cytosolic Ca2+ by Ca2+-binding proteins (CBPs) (Andressen et al., 1993).
CBPs are generally viewed as ‘buffers’ to attenuate stochastic Ca2+ peaks in neurons (Andressen et al., 1993). Members of the EF-hand family of CBPs invariably contain a 3-D motif to bind Ca2+ (Heizmann, 1986). Ancestral representatives of the CBP family, e.g. calmodulin, are ubiquitously expressed with a high degree of evolutionary conservation, and control fundamental cellular functions ranging from the cell cycle, cell motility, axon polarization to synaptic signalling (Andressen et al., 1993). In contrast, the parvalbumin (PV) and calbindin subfamilies, the latter including the vitamin D-dependent 28 kDa isoform of calbindin (CB) and calretinin (CR), exhibit phylogenetically preserved tissue-specific expression patterns in vertebrates (Freund & Buzsaki, 1996; Klausberger & Somogyi, 2008), and are restricted to morphologically distinct subpopulations of GABAergic interneurons and local projection cells in rodent, primate and human corticolimbic circuits and extended amygdala (EA) with the exception of CB that is also expressed by cortical pyramidal and dentate granule cells (Celio, 1990).
The consensus exists that, although their developmental dynamics are different, CBPs are late markers of postmitotic GABA cells in both cortical and striatal territories (Flames & Marin, 2005; Wonders & Anderson, 2006): CB+ pioneer neurons populate the cerebral cortex by embryonic day (E) 14 in mouse (Sanchez et al., 1992), and are also present in human fetal brain by week 14 of pregnancy (Brun et al., 1987). CR+ neurons invade the developing cerebrum by mid-gestation in both rodents and human (Verney & Derer, 1995; Meyer et al., 1998). While PV first appears at E13 in the spinal sensory system, the onset of PV expression in forebrain GABAergic neurons is restricted to the first postnatal week (Solbach & Celio, 1991), except in human telencephalon where PV+ Cajal-Retzius cells were noted by gestational weeks 20–24 (Verney & Derer, 1995).
Secretagogin (scgn) is a recently discovered CBP harbouring six putative EF-hand motifs (Rogstam et al., 2007) that was cloned from β cells of the pancreatic islands of Langerhans and endocrine cells of the gastrointestinal tract (Wagner et al., 2000). Although the distribution and neurochemical specificity of scgn+ neurons in the adult mouse, primate (Mulder et al., 2009b) and human brain (Attems et al., 2007) has recently been explored, scgn expression in the developing nervous system remains elusive. We report that scgn is expressed in mouse telencephalon from E11, and identifies neurons inhabiting the EA in mouse, primate and human brains.
Materials and methods
Animals and tissue preparation
Mouse embryos at E10.5–E18.5 (n = 4–6/time point, n ≥ 2 pregnancies/analysis) were obtained from time-mated C57Bl6/N, glutamate decarboxylase 67 (GAD67)-GFP (Δneo) (Tamamaki et al., 2003) or choline-acetyltransferase (ChAT)(BAC)-EGFP mice (Tallini et al., 2006). We considered the day when vaginal smear was found in females as E0.5. Neonates were killed by decapitation on postnatal day (P) 0, P1 or P2. Whole brains were immersion fixed in 4% paraformaldehyde (PFA) in Na-phosphate buffer (PB, 0.1M, pH7.4) overnight. Tissue samples were cryoprotected in an ascending gradient of sucrose in PB (up to 30%) for at least 48h prior to cryostat sectioning (14-μm thickness). Sections were thaw-mounted on SuperFrost+ glass slides.
Adult GAD67-GFP (n = 3) and ChAT(BAC)-EGFP (n = 2) reporter mice were anesthetized by isoflurane (5%, 1L/min flow rate) and transcardially perfused with 4% PFA in PB (100 ml, 3–4 ml/min flow rate) that was preceded by a pre-rinse with ice-cold physiological saline. Whole brains were removed from the skull, divided into fore- and hindbrain regions and post-fixed in the same fixative overnight. Tissue blocks were cryoprotected in 30% sucrose as above and serial 40 μm-think coronal sections were cut on a cryostat microtome.
Grey mouse lemur (Microcebus murinus, Primates) embryos and fotuses were obtained from a colony bred in captivity for the past ~35 years with stock originating from the dry forest of the South-western coast of Madagascar (Perret & Aujard, 2001). Pregnant female mouse lemurs were maintained in isolation in appropriate cages, mimicking wild breeding conditions (Perret, 1992), with constant temperature and humidity. Food and water were provided ad libitum. The mean duration of gestation in mouse lemurs is 61 ± 0.2 days (Perret, 1990). The embryos (n = 2) used in this report were harvested freshly from a spontaneous abortion at day 33 of intrauterine development. Fetal brain (n = 1) was collected by hysterectomy under ketamine anesthesia that was indicated because of abnormal bleeding of the female on day 50 of pregnancy. None of the offspring were viable. Whole embryos and fetal brain samples were fixed in 4% PFA in PB for 48h, rinsed in phosphate-buffered saline (PBS; 0.01M, pH 7.4) and kept in PBS also containing 0.1% NaN3 until processing. Primate tissues were cryoprotected and sectioned (14-μm thickness) onto SuperFrost+ glass slides. Brains of adult grey mouse lemurs were processed as described (Mulder et al., 2009b).
All experiments on animals conformed to the European Communities Council Directive (86/609/EEC) and were approved by the Home Office of the United Kingdom (mice) or the Ministry of Agriculture, France (#A91.114.1; lemurs). Particular care was taken to minimize the number of animals and their suffering throughout the experiments.
Expression profiling
Total RNA isolated from tissues microdissected from C57Bl6/N embryos at E12 – P2 was subjected to scgn expression analysis after confirming RNA integrity (Supporting Fig. 1A). Quantitative real-time PCR (qPCR) reactions were validated by preliminary testing of amplification efficacy and by excluding the possibility of genomic DNA contamination in the presence (+) or absence (−) of reverse transcriptase in parallel and running the samples on 1.5% agarose gel (Supporting Fig. 1A1). qPCR reactions were performed with custom designed primers for scgn (Supporting Fig. 1A2–A4) (Mulder et al., 2009b). TATA binding protein (forward primer: 5′-ACCCTTCACCAATGACTCCTATG-3′; reverse primer: 5′-ATGACTGCAGCAAATCGCTTGG-3′) was used to normalize scgn expression.
Biochemistry
Protein samples were analyzed under denaturing conditions. After electrophoresis, proteins were transferred onto Immobilon-FL PVDF membranes (Millipore, Billerica, MA, USA) and probed with rabbit anti-scgn (1:2,000) and mouse anti-β-actin (1:4,000) primary antibodies (Mulder et al., 2009b). Immunoreactivities were revealed using IRDye680 and IRDye800 secondary antibodies (Invitrogen/Molecular Probes, Paisley, United Kingdom). Blots were scanned on a Li-Cor Odyssey-IR imager (Li-Cor Biosciences, Lincoln, NA, USA).
Generation of anti-secretagogin antibodies and histochemistry
Within the framework of the Human Protein Atlas program (www.proteinatlas.org), a rabbit antibody against a recombinant fragment of human scgn (amino acids (AA) 135–273) was generated (Mulder et al., 2009a). The specificity of the ensuing anti-scgn antibody has been extensively evaluated (Mulder et al., 2009b) in accordance with existing guidelines on the application of primary antibodies (Fritschy, 2008). We have further validated our novel anti-scgn antibody by comparing its labelling pattern with that of a commercial polyclonal anti-scgn antibody raised in goat against scgn’s AA164-276 fragment (R & D Systems, Minneapolis, MN, USA; Supporting Fig. 2A) by both Western blotting (Supporting Fig. 2B) and histochemistry (Supporting Fig. 2C). We find that these antibodies unequivocally recognize a major protein band corresponding to scgn’s calculated molecular weight in Western applications (Supporting Fig. 2B), and reveal the same neuron populations in E15 mouse forebrain (Fig. 3, Supporting Fig. 2C). Furthermore, our anti-scgn antibody produces a staining pattern in the olfactory bulb that is indistinguishable from that of a polyclonal anti-scgn antibody generated against the complete human scgn sequence (Wagner et al., 2000) (J. Attems & L. Wagner, personal communication).
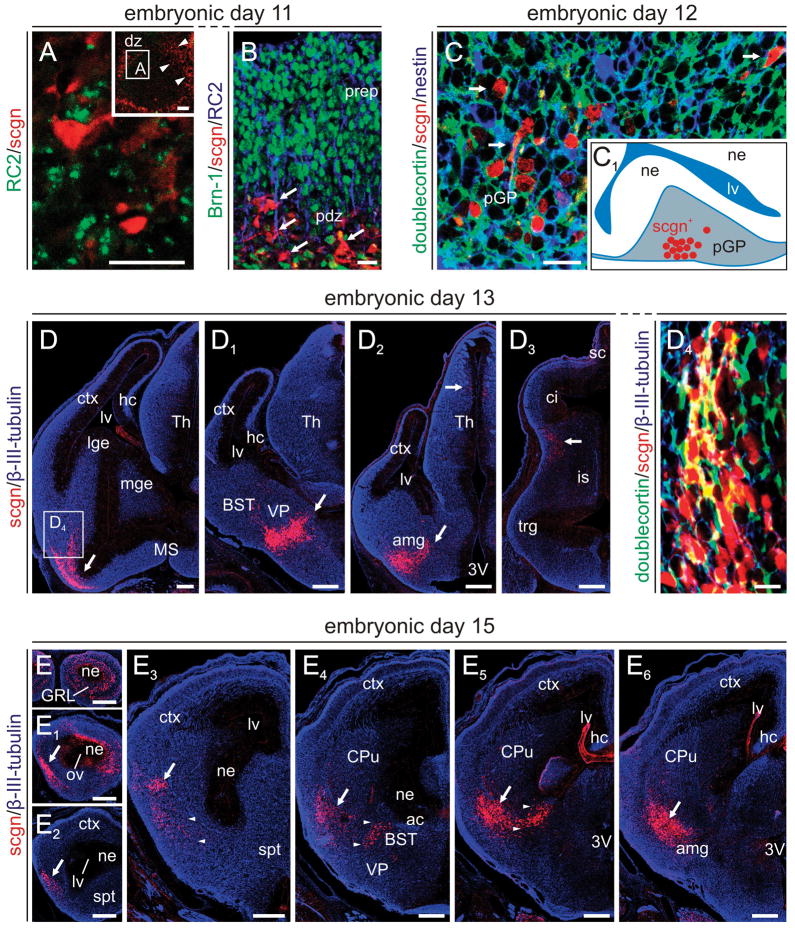
Fig. 3. Scgn+ neurons in the embryonic mouse telencephalon.
(A) Scgn marks early postmitotic neurons intermingled in a fine meshwork of radial glia fibers exiting the olfactory differentiation zone (arrowheads, inset). (B) Scgn+ cells concentrate in the pallidal differentiation zone (pdz) at the basal surface of the telencephalic vesicle (arrows) by E11. Scgn+ cells lack both Brn-1 and RC2, labelling neocortical neurons (Sugitani et al., 2002) and radial glia (Fishell & Kriegstein, 2003), respectively. (C) By E12, scgn+ cells transit in the differentiation zone committing neurons to the prospective globus pallidus (pGP). Inset denotes the anatomical localization of scgn+ neurons in relation to neighbouring neurogenic territories. (D-D3) Scgn+ neurons (arrows) concentrate in subnuclei of the extended amygdala. Open rectangle in (D) identifies the location of migrating scgn+ neurons, a subset apparently co-expressing doublecortin, shown at high-resolution (D4). (E-E5) Distribution of scgn+ neurons in the mouse forebrain, including olfactory bulb, at E15. Arrowheads point to neurons that are destined to the interstitial nucleus of the posterior limb of the anterior commissure (IPAC) and substantia innominata, while arrow indicates neurons populating the prospective amygdala. List of abbreviations is available in Supporting Table 1 on-line. Scale bars = 20 μm (A–B,D1), 200 μm (D–D3, E–E6).
Multiple immunofluorescence histochemistry with cocktails of primary antibodies (Table 1) was performed in both species studied (Mulder et al., 2009b). Immunosignals were visualized using combinations of carbocyanine (Cy) 2-, Cy3- and Cy5-conjugated antibodies raised in donkey (1:200–1:400, Jackson ImmunoResearch, West Grove, PA, USA). Hoechst 35,528 (Sigma, St. Louis, MA, USA), a nuclear dye, has been applied to reveal tissue architecture. Tissue autofluorescence in sections from adult mouse and primate brains was quenched by Sudan Black B (Schnell et al., 1999).
Table 1.
| Protein target | Species | Dilution | Source | Cat. No. | Fluorophore | Reference |
|---|---|---|---|---|---|---|
| Brn-1 | goat | 1:1,000 | Santa Cruz | SC-6028 | Cy2 | Keays et al. (2007) |
| Calretinin | goat | 1:2,000 | SWant | CG1 | Cy5 | Schwaller et al. (1993) |
| Calbindin D28k | mouse (IgG) | 1:2,000 | SWant | 300 | Cy5 | Celio (1990) |
| Choline-acetyltransferase | goat | 1:200 | Chemicon/Millipore | AB144P | Cy2, Cy3 | Härtig et al. (1998) |
| Doublecortin | goat | 1:100 | Santa Cruz | SC-8066 | Cy2, Cy3 | Nagatsuka et al. (2003) |
| GABA | guinea pig | 1:1,000 | Chemicon/Millipore | AB175 | Cy3 | McDonald & Pearson (1989) |
| GAD65/67 | mouse (IgG) | 1:500 | Nordic BioSite | MSA-225 | Cy5 | Karlsen et al. (1991) |
| Lhx8 | goat | 1:100 | Santa Cruz | SC-2216X | Cy3, Cy5 | Kitanaka et al. (1998) |
| Nestin | mouse (IgG) | 1:1,000 | Chemicon/Millipore | MAB353 | Cy5 | Bossolasco et al. (2005) |
| PSA-NCAM | mouse (IgM) | 1:200 | Chemicon/Millipore | MAB5324 | Cy5 | Bernier et al. (2002) |
| RC2 | mouse (IgM) | 1:200 | Chemicon/Millipore | MAB5740 | Cy5 | Aguado et al. (2006) |
| Secretagogin | goat | 1:1,000 | R & D Systems | AF4878 | Cy3 | This report |
| β-III-tubulin (TUJ1) | mouse (IgG) | 1:2,000 | Promega | G7121 | Cy5 | Berghuis et al. (2007) |
| Tyrosine hydroxylase | mouse (IgG) | 1:1,000 | R & D Systems | MAB1423 | Cy2 | Arita et al. (2002) |
Confocal laser-scanning microscopy
Sections were inspected and representative images of immunoreactivity were acquired on a Zeiss 710LSM confocal laser-scanning microscope (Zeiss, Jena, Germany) equipped to separate emission signals through spectral detection and unmixing. Emission spectra for each dye was limited as follows: Hoechst (420–485 nm), Cy2 (505–530 nm), Cy3 (560–610 nm), and Cy5 (640–720 nm). Image surveys were generated using the tile scan function with optical zoom varied from 0.6x–1.5x at 10× primary magnification (objective: EC Plan-Neofluar 10×/0.30). Co-localization was defined as immunosignals being preset without physical signal separation in ≤1.0-μm optical slices at 40× (Plan-Neofluar 40×/1.30) or 63× (Plan-Apochromat 63×/1.40) primary magnification (Mulder et al., 2009b). Images were processed using the ZEN2009 software (Zeiss). Multi-panel figures were assembled in CorelDraw X3 (Corel Corp., Ottawa, ON, Canada).
Quantitative morphometry, 3-D rendering and statistics
The diameter of scgn+ neurons was measured after capturing images of scgn+ cell assemblies in pallidal and EA territories at 40× primary magnification. The somatic diameter of individual neurons was measured on the premise that scgn is a cytosolic protein (Attems et al., 2007) and is homogenously distributed throughout the neuronal perikarya. Only neurons with smooth surfaces and processes were included in our analysis to avoid bias due to partial profiles of cell fragments.
Serial coronal sections (sampling interval: 140 μm) spanning the entire forebrain from an E15 mouse were double-labelled to reveal scgn+ neurons and cell nuclei (Hoechst 35,528). Single x–y plane images were acquired (Zeiss 710LSM), and 3-D reconstructed using the BioVis3D software (BioVis3D, Montevideo, Uruguay).
Data were expressed as means ± sem and analyzed using Student’s t-test (SPSS v.16.0, SPSS Inc., Chicago, IL, USA). A p value of < 0.05 was considered statistically significant.
In situ hybridization in human fetal brain
Human fetal specimens at mid-gestation (21–22 weeks of gestation; n = 3) were obtained from saline-induced abortions (Wang et al., 2004; Hurd et al., 2005). Protocols were approved by the local institutional review board as part of a large-scale study to evaluate the molecular effects of prenatal drug exposure on human neurodevelopment (Hurd et al., 2005). Specimens were fixed in 1% PFA and frozen at −80 °C. Coronal cryosections (20 μm) spanning corticolimbic areas including the amygdaloid complex were cut. In situ hybridization was conducted as described (Wang et al., 2004): cDNA fragments of human scgn (NM_006998) were obtained from human brain total RNA by reverse transcription P C R u sing the primer pairs: 5′-GGTTGATTTTCAGCCCAAGA-3′ (sense), 5′-TTTGCACAAGGTGAAACAGC-3′ (anti-sense). The RNA probe was transcribed in the presence of [35S]-uridine 5′-[α-thio]triphosphate (specific activity 1000–1500 Ci/mmol; New England Nuclear, Boston, MA). In situ hybridization was carried out as described (Hurd, 2003) by applying the labelled probe to the brain sections at a concentration of 2 × 103 cpm/mm2 of the coverslip area overnight at 55°C in a humidified chamber. Two adjacent sections from each subject were studied. The slides were then apposed to Imaging Plates (FUJIFILM Corporation, Tokyo, Japan) along with 14C-standards (American Radiolabelled Chemicals, St Louis, MO, USA). Films were developed with a phosphoimaging analyzer (FLA-7000), and images were analyzed by the MultiGauge software (FUJIFILM Corporation).
Nomenclature
We have adopted the nomenclature of (Paxinos & Franklin, 2001) to describe the organization of the developing mouse brain. In addition, we have relied on the nomenclature introduced by Bons et al. (1998) for the adult mouse lemur to identify brain areas in the developing grey mouse lemur brain. A comprehensive list of abbreviations of neuroanatomical structures can be found in Supporting Table 1.
Results
Expression profiling in developing brain
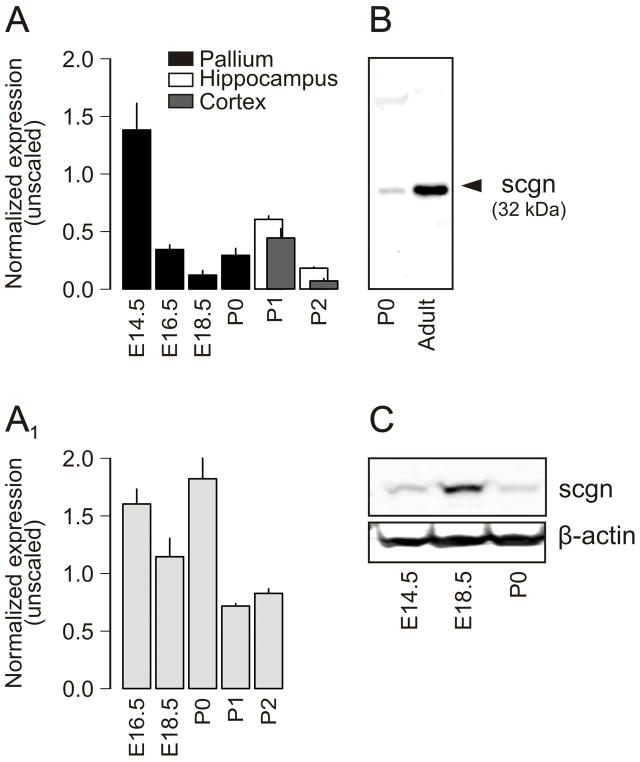
Recent findings demonstrate that scgn is a CBP identifying neurochemically heterogeneous subsets of neurons in adult rodent, primate (Mulder et al., 2009b) and human forebrain (Attems et al., 2007). However, it remains unknown whether scgn is expressed during neurodevelopment. We assessed scgn mRNA levels in the mouse cerebral cortex (including hippocampus; Fig. 1A) and amygdaloid complex (Fig. 1A1) by qPCR analysis (Supporting Fig. 1) during mid- and late-gestation, and in neonates. We establish that pallial scgn mRNA expression is robust by E14.5, whilst moderate to low between E16 - P2 in the developing mouse neocortex and hippocampus (Fig. 1A). In contrast, scgn mRNA levels in the amygdaloid complex remain largely stable until birth with a marked decline being apparent after P1 (Fig. 1A1).
Fig. 1. Scgn is expressed in embryonic mouse forebrain.
(A,A1) Scgn mRNA expression profiling of microdissected mouse brain tissues as measured by qPCR relative to TATA binding protein (TBP). The dorsal telencephalon (pallium) has been subdivided into the hippocampus and neocortex in neonates. (A1) Scgn mRNA expression in the fetal amygdaloid complex. (B) A novel anti-scgn antibody (HPA006641) recognizes a single protein band corresponding to scgn’s calculated molecular weight in both neonatal and adult brain samples. Forebrain (P0; 40 μg protein/lane) and olfactory bulb (adult; 20 μg protein/lane) samples were processed simultaneously. (C) Temporal presence of scgn in developing mouse forebrain as determined by Western analysis. β-Actin served as loading control.
Within the framework of the Human Protein Atlas program (Uhlen et al., 2005; Mulder et al., 2009a), we have generated antibodies to >8,000 proteins, including a polyclonal antibody recognizing a phylogenetically conserved scgn epitope (Mulder et al., 2009b). Here, we confirmed that this antibody recognizes a single protein target in samples prepared from neonatal mouse forebrain that is identical in size to that seen in adult brain (Fig. 1B; Supporting Fig. 2), and corresponds to scgn’s calculated molecular weight of 32 kDa (www.ensembl.org). We explored whether scgn is expressed in the developing central nervous system at the protein level by detecting scgn protein upon loading fetal and neonatal forebrain lysates (40 μg/lane) on denaturing SDS-PAGE (Fig. 1C).
The developmental dynamics of scgn mRNA expression suggest that this CBP may be transiently expressed by select cell populations in the fetal brain. Alternatively, scgn+ cells may be born by ~E14.5 with their population size remaining stable over time. Thus, the gradual decrease in scgn mRNA expression may merely reflect a proportional reduction in the prevalence of scgn+ cells during the progressive expansion of the embryonic forebrain until birth.
Scgn expression during organogenesis
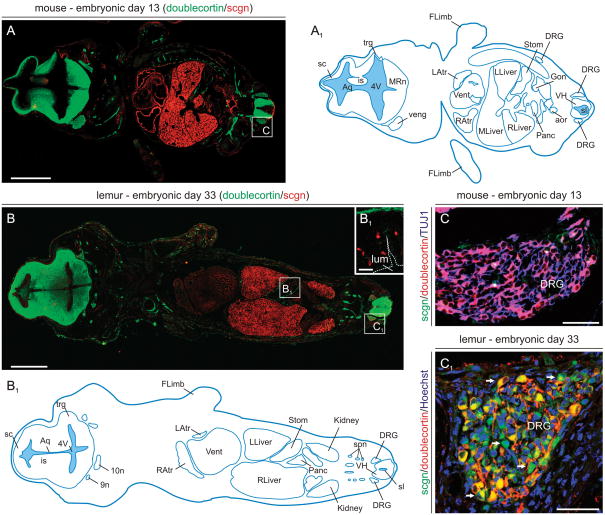
We have tested scgn’s expression sites at mid-gestation by analyzing horizontal sections spanning the whole body of mouse (E13) and grey mouse lemur (E33) embryos. We used grey mouse lemurs because detailed information is available on both the intrauterine development of this prosimian primate (Perret, 1990), and the neurochemical specificity of scgn+ neurons in the adult lemur brain (Mulder et al., 2009b). Since the distinct timelines of rodent and primate embryogenesis may be a potential confounding factor in comparative analyses, we have chosen developmental stages in either species at which the general (Supporting Fig. 3) and organ systems anatomy (Fig. 2) of the embryos are similar. We find significant scgn immunolabeling in the heart, pancreas, kidney and gonads of both mouse (Fig. 2A) and lemur embryos (Fig. 2B), corroborating prior findings in human tissues (Wagner et al., 2000; Lai et al., 2006). We also show that scgn+ putative enteroendocrine cells (Lai et al., 2006; Gartner et al., 2007) populate the developing stomach in both species (Fig. 2B1). Whilst we failed to detect scgn immunosignal in the mouse dorsal root ganglion (DRG; Fig. 2C) at E13, scgn+ neurons co-expressing doublecortin (Fig. 2C1) were present in the lemur DRG. Scgn is not expressed in the liver during adulthood (Mulder et al., 2009b). Therefore, scgn immunoreactivity in embryonic liver may either indicate transient expression of this CBP or represent a methodological artifact due to unexpected tissue immunogenicity. Overall, our results suggest that scgn is expressed in several organ systems of mid-gestation mammalian embryos.
Fig. 2. Scgn expression in mouse and primate embryos at mid-gestation.
(A,A1) Scgn+ organ systems in mouse embryo. Doublecortin, a microtubule-associated protein widely expressed by migrating cells in the developing nervous system (Gleeson et al., 1999), was used to reveal anatomical boundaries of various organs. (B,B1) Scgn+ organs in the grey mouse lemur embryo. Open rectangles in A and B denote the general location of high-resolution photomicrographs in B1-C1. (B1) Scgn immunoreactivity decorates putative enteroendocrine cells (Mulder et al., 2009b) at the base of gastric pits formed in the stomach of lemur embryos. (C,C1) Prospective sensory neurons in the dorsal root ganglion (DRG) of the lemur (arrows, C1) but not mouse (C) express scgn. Abbreviations are listed in Supporting Table 1. Scale bars = 30 μm (B1), 150 μm (C,C1), 0.5 mm (A,B).
Emergence of scgn in postmitotic neurons in the developing mouse forebrain
We find scgn+ cells at E11 in the mouse telencephalon (Fig. 3A,B). Clusters of scgn+ cells could be observed at least at two locations in the wall of the cerebral vesicle: in its anterior wall forming the olfactory bulb (OB; Fig. 3A) and in the subpial area of the ganglionic eminence (GE). At E12, scgn+ cells transit in the differentiation zone that commits neurons to the prospective globus pallidus (GP; Fig. 3C). Scgn+ cells were immunoreactive for β-III-tubulin, but not nestin (neural progenitor), RC2 (radial glia) or Brn-1 (neocortical pyramidal cell) during the period of E11-12 suggesting that scgn marks postmitotic, non-pyramidal neurons at the subpial surface of the telencephalic vesicle. The scgn+ cell pool expands by E13 with cells traversing the palliosubpallial boundary in two directions: a contingent of cells adopts scgn+/GABA+ phenotype upon entering the OB (Fig. 5C,C1). In the present study, we focus on scgn+ cells that migrate in the subpallium caudally (Fig. 3D-D4) and commit neurons to the EA (Fig. 3D1,D2). Although we did not engage in performing detailed analysis of migratory routes, the gradual colonization of the bed nucleus of the stria terminalis (BST) and amygdaloid territories by scgn+ neurons between E13 and E15 (Fig. 3E-E6) suggests that these cells first migrate caudally in the lateral subpallium before turning, and migrating in the lateral-to-medial direction within the EA. In sum, our analysis reveals that scgn+ cells cytoarchitecturally resembling migrating neurons form a continuous stream along the palliosubpallial boundary before reaching their final destinations in the OB or EA (Fig. 4).
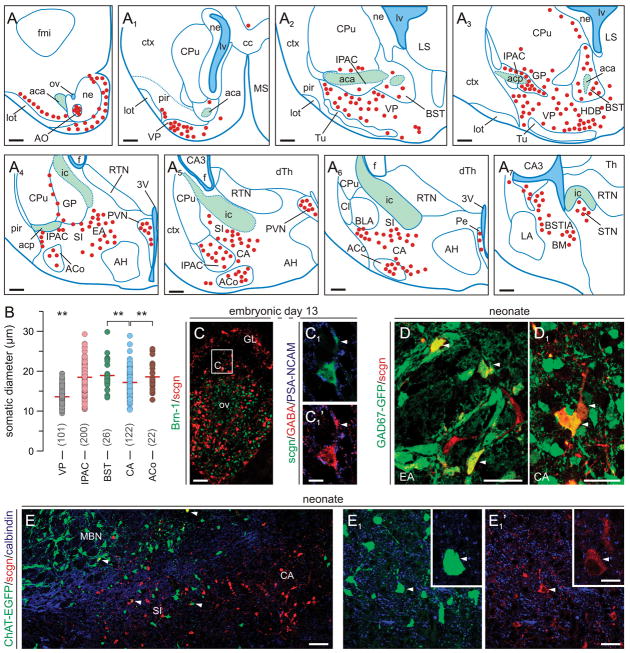
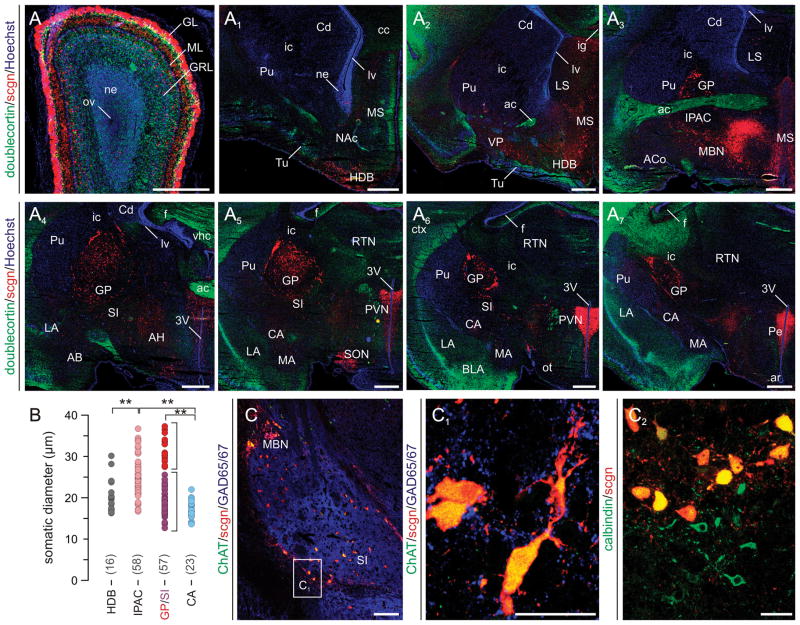
Fig. 5. Scgn in the neonatal mouse forebrain.
(A–A7) Schematic presentation of the regional distribution of scgn+ neurons in coronal sections along the anterior-posterior axis of the new-born mouse brain with a sampling interval of 560 μm. Red circles correspond to scgn+ cells, blue lines demarcate particular nuclei, and green areas identify major axonal pathways. (B) Somatic diameters of scgn+ neurons in the VP and subdivisions of the amygdaloid complex (IPAC, BST, CA, ACo). **p < 0.01 (Student’s t-test). (C-C1) Scgn+ neurons in the olfactory bulb contain GABA by E13. Arrowheads point to the leading edge. (D,D1) Scgn immunoreactivity decorates GABAergic neurons in the extended (EA) and central amygdala (CA). (E-E1′) A subset of GFP-tagged cholinergic neurons inhabiting the prospective substantia innominata (SI) are scgn+ (arrowheads). Abbreviations are listed in Supporting Table 1. Scale bars = 500 μm (A-A7), 200 μm (C), 20 μm (C1,C2).
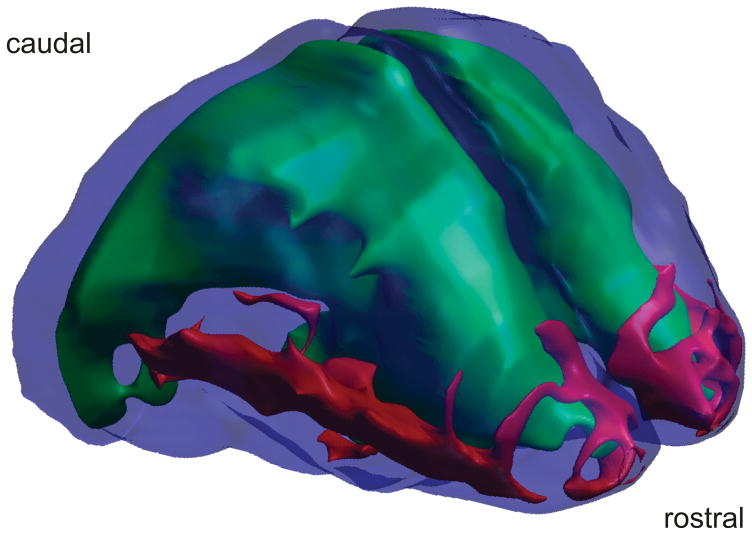
Fig. 4. Spatial spread of scgn+ neurons in the embryonic mouse brain at E15. The distribution of scgn+ neurons migrating towards their final positions along the anterior-posterior axis of the palliosubpallial boundary in the basolateral telencephalon is shown in red.
Green color indicates the lateral ventricles, while deep blue depicts the cortical surface. 3-D reconstruction of the embryonic mouse brain was performed by using serial coronal sections.
Next, we analyzed the distribution of scgn+ neurons in neonatal mouse brain. We observe that the migration of scgn+ cells concludes by birth and scgn+ neurons inhabit, in an anterior-to-posterior direction, the spatially interrelated nuclei of the BST, interstitial nucleus of the posterior limb of the anterior commissure (IPAC), ventral pallidum (VP), dorsal substantia innominata (SI), and the central and medial amygdaloid nuclei (CA/MA) (Fig. 5A-A7). Morphometric analysis revealed that scgn identifies divergent neuron subpopulations with different somatic diameters in the VP and EA (Fig. 5B). By using genetically tagged reporter mice we demonstrate that scgn+ neurons either adopt a GABA phenotype along the longitudinal axis of the EA (Fig. 5D,D1), similar to scgn+/GABA+ neurons in the embryonic OB (Fig. 5C,C1), or co-express ChAT when found in small diameter cholinergic neurons of the dorsal SI (Fig. 5E-E1′). Collectively, our data suggest that by E18 scgn+ neurons can acquire a distribution pattern resembling that in the adult brain, and differentiate into neurochemically distinct subtypes of EA neurons.
Scgn expression in fetal primate brain
Systematic analysis along the longitudinal axis of the fetal primate brain revealed the first contingent of scgn+ neurons in the granular and glomerular layers of the OB (Fig. 6A). However, unlike in the adult primate brain (Mulder et al., 2009b), neuroblasts migrating in the prenatal rostral migratory stream (Pencea & Luskin, 2003) do not harbour scgn expression (Fig. 6A). Pallial areas are devoid of discernible scgn immunoreactivity. In the basal forebrain, scgn+ neurons were seen in the horizontal diagonal band, nucleus accumbens, medial septum, VP, GP and SI (Fig. 6A1–A7). In contrast to scgn distribution in the neonatal rodent brain, scgn+ cells were only infrequently found in either the CA or MA. In the hypothalamus, substantial scgn+ neuron populations occupied the paraventricular and periverticular nuclei and the supraoptic nucleus (Fig. 6A5–A7). A morphological dichotomy of scgn+ neurons was evident in the basal telencephalon (Fig. 6B): small-to-medium-sized scgn+ neurons populated the horizontal diagonal band, SI and CA. In contrast, large diameter scgn+ neurons were found in the IPAC and GP. Clusters of large diameter neurons enveloping the dorsal and ventral tips of the GP as well as medium-sized neurons heterogeneously invading the ventral subdivision of this territory were ChAT+/CB+ cholinergic neurons (Fig. 6C–C2) (Geula et al., 1993). These data demonstrate that the cholinergic identity of scgn+ cells is acquired by the third trimester of pregnancy.
Fig. 6. Scgn distribution in fetal mouse lemur forebrain.
(A–A7) Scgn expression was identified in coronal sections with a sampling interval of 700 μm along the anterior-posterior axis of the fetal mouse lemur brain at day 50 of gestation. Doublecortin immunoreactivity together with nuclear counterstaining was used to identify anatomical boundaries. Brain regions were identified and termed according to the nomenclature introduced by Bons et al. (1998). (B) Somatic diameters of scgn+ neurons in amygdaloid (IPAC, SI, CA) and pallidal (HDB, GP) nuclei. **p < 0.01 (Student’s t-test). Note the apparent volumetric separation of scgn+ neurons in the GP and SI. (C,C1) Cholinergic cells populating the magnocellular basal nucleus (MBN) as well as SI harbour scgn expression. (C2) The co-localization of scgn and CB, a putative cholinergic marker in Primates (Geula et al., 1993), supports that large diameter cholinergic projection cells are the preferred sites of scgn expression in basal forebrain. Abbreviations are listed in Supporting Table 1. Scale bars = 500 μm (A–A7), 200 μm (C), 20 μm (C1,C2).
Scgn in the adult amygdaloid complex
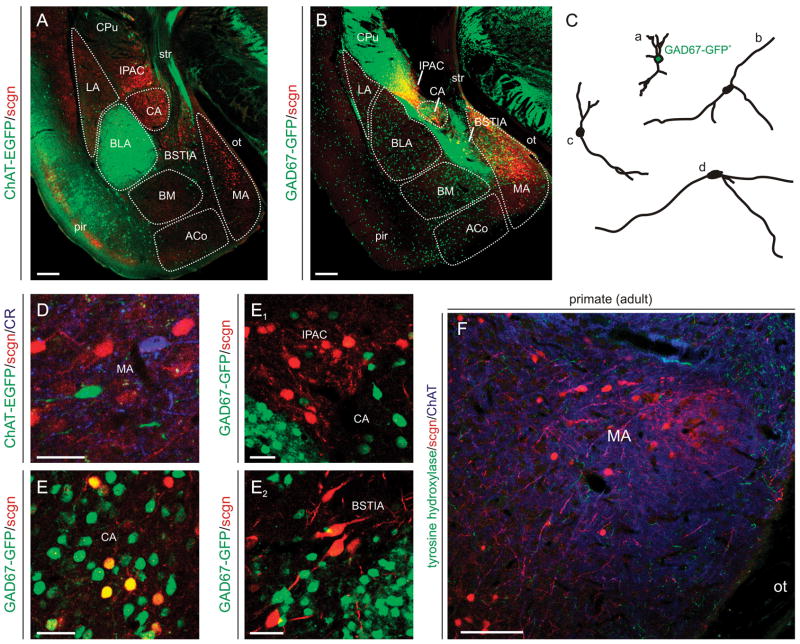
To evaluate the fate of scgn+ neurons in the EA, we have analyzed sections from ChAT-EGFP and GAD67-GFP reporter mice allowing precise delineation of the anatomical boundaries of individual amygdaloid nuclei (Fig. 7A,B). Scgn expression was limited to two morphologically distinct types of neurons in the EA (Fig. 7C): scgn+ neurons with oval perikarya and short ramifying dendrites (Fig. 7C/a) predominate in the CA and IPAC. In contrast, scgn+ neurons as above are intermingled with stellate-like cells with fusiform perikarya and long, smooth primary dendrites in the MA (Fig. 7C/b–d). ChAT+/scgn+ neurons were exclusively identified in the VP and dorsal segment of the SI (Mulder et al., 2009b) but not amygdaloid nuclei (Fig. 7D). GAD67+/scgn+ small-diameter neurons were frequently encountered in the CA and MA (Fig. 7E) but not the IPAC (Fig. 7E1) or the intraamygdaloid segment of the BST (Fig. 7E2).
Fig. 7. Scgn+ neurons in the adult amygdaloid complex.
(A,B) Scgn’s distribution was revealed by using ChAT(BAC)-EGFP+ and GAD67-GFP+ structures as landmarks outlining the anatomical boundaries (dashed lines) of particular subnuclei of the amygdaloid complex. (C) Camera lucida drawings of scgn+ neurons positioned in the dorsal part of the medial amygdaloid nucleus (MA) reveal scgn+/GAD67-GFP+ neurons (a) with short and complex dendritic tafts, and scgn+/GAD67-GFP- stellate-like cells. (D) ChAT(BAC)-EGFP+ neurons lack scgn in the MA. (E–E2) While scgn marks a subset of GABAergic neurons in the central amygdaloid nucleus (CA) (E), multipolar scgn+ cells in the interstitial nucleus of the posteror limb of the anterior commissure (IPAC) and large diameter, bifurcating scgn+ neurons in the intraamygdaloid division of the bed nucleus of the stria terminalis (E2) lack GAD67-GFP expression. (F) Scgn+ neurons in the MA of the adult mouse lemur. Abbreviations are listed in Supporting Table 1. Scale bars = 200 μm (A,B,F), 20 μm (E–E2).
A clear phylogenetic difference in the distribution of scgn+ neurons is the complete absence of scgn immunoreactivity in small-diameter GABAergic neurons of the primate CA and MA. Instead, fusiform scgn+ neurons populate the MA (Fig. 7F).
Based on their cellular origins and connectivity maps, the lateral, basolateral, basomedial and cortical amygdaloid nuclei are classified as pallial structures. In contrast, the BST, CA, MA and SI are of subpallial origin. BST and SI neurons are thought to originate in the pallidal ridge, while neurons inhabiting the CA and MA share their birthplace with striatal neurons (Swanson & Petrovich, 1998). Therefore, the selective expression of scgn in pallidal amygdaloid territories further illustrates the above developmental dichotomy.
Scgn expression in fetal human brain
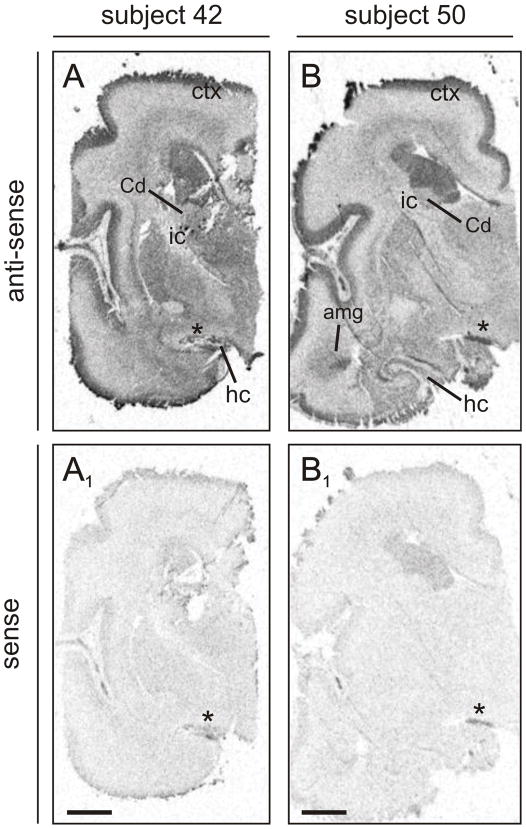
Distinct anatomical organization of scgn mRNA expression with heterogeneous signal in a number of structures was evident in the fetal human brain (Fig. 8). Scgn mRNA distribution patterns were similar in all subjects studied. Invariably strong scgn anti-sense probe hybridization signal was observed throughout the cortical plate of the cerebral cortex (Fig. 8A,B), and in the amygdaloid complex (Fig. 8B). Moderate scgn mRNA expression was observed in the hippocampus, subiculum, thalamic territories, and germinal layers, whereas low signal intensity was seen in the caudate nucleus. These data demonstrate that scgn expression in the mammalian amygdaloid complex is phylogenetically conserved. In addition, our results highlight that scgn expression in pyramidal cells is developmentally regulated and can endure into adulthood in this cell type (Attems et al., 2007).
Fig. 8. Scgn mRNA expression in human fetal telencephalon.
Distribution of scgn mRNA expression in whole sections of the fetal forebrain as revealed by an anti-sense riboprobe. (A,B) The autoradiograms show specific hybridization signal obtained from two fetal subjects. Specific signals were only obtained with hybridizations carried out using the scgn anti-sense but not sense (A1,B1) probe. Abbreviations are referred to in Supporting Table 1. Asterisks indicate folds in the tissue. Scale bars = 1 cm.
Discussion
Our report identifies the developmental dynamics of scgn expression including the migratory routes and final positions of subpallial neurons expressing this CBP in rodent, primate and human fetal brain. Distinct temporal expression patterns of the three ‘classical’ CBPs – CB, CR and PV – have generated broad interest because of the importance of GABAergic neurons in refining the physiological output of neuronal networks (Freund & Buzsaki, 1996; Klausberger & Somogyi, 2008). Here, we show that scgn is expressed earlier than CB, CR or PV in pioneer neurons exiting the pallidal differentiation zone by E11 in mouse. Histochemically noticeable scgn expression is restricted to postmitotic neurons because scgn+ cells lack the expression of RC2 and nestin, radial glia and neural stem/progenitor cell markers (Carleton et al., 2003), respectively. The majority of scgn+ cells we identified migrate towards the prospective EA, and selectively inhabit its subpallial domain by forming a continuum of scgn+ neurons extending from the anterior tip of the VP towards the CA/MA. The lack of Brn-1, a POU homeodomain protein specifying neocortical pyramidal cells (Sugitani et al., 2002), supports that scgn+ cell contingents are destined towards subpallial territories.
Scgn+ neurons commute in at least two major migratory streams along the palliosubpallial boundary and clearly avoid venturing into neocortical territories during forebrain development. Scgn+ neurons populating the OB travel in the anterior direction and upon reaching the olfactory granular layer frequently (>20%) acquire GAD67+/GABA+ phenotypes. In contrast, scgn+ neurons travelling caudally to colonize the EA exhibit a substantially lower percentage (7–9%) of co-localization with GAD67 en route to their final positions. The diversity of neuronal contingents destined to the EA is first evidenced by the bifurcation of their migratory stream at the level of the IPAC: small-to-medium sized scgn+ neurons, many of which are GABAergic (Fig. 5), invade the CA and MA. Whilst we show that scgn can developmentally co-exist with GAD, our prior (Mulder et al., 2009b) and present analysis in adult mouse and primate forebrain reveal a limited likelihood of co-expression of scgn with the other known neuron-specific CBPs, particularly CR and CB. Alternatively, scgn+ neurons can co-express ChAT, a ubiquitous cholinergic marker (Riedel et al., 2002), upon populating the SI.
Intracellular Ca2+ signalling underpins the responsiveness of developing neurons to extracellular guidance cues. We unexpectedly find that scgn is already enriched in subsets of neurons engaged in long-distance migration with histochemically-detectable levels of this CBP maintained throughout neuronal morphogenesis. This notion may pinpoint that the scgn-mediated control of intracellular Ca2+ signalling can play a role in generating adequate cellular responses to microenvironmental stimuli that are specifically present at the palliosubpallial boundary. Otherwise, scgn may be one of the early molecular determinants required for amygdala neurons to integrate into neuronal networks and to acquire specialized functions therein.
In the postnatal nervous system, CBPs are important cytosolic modulators of Ca2+ signalling in neurons whose presence has been associated with the maintenance of distinct electric discharge patterns. In the basolateral amygdala, PV+ interneurons form a primary local modulatory neuronal subnetwork affecting the integration of polymodal sensory information by excitatory principal cells (Woodruff & Sah, 2007a; Woodruff & Sah, 2007b). Our discovery that scgn+ neurons are only present in circumcised clusters in the EA present a number of intriguing possibilities both at the single cell and neuronal network levels: secretagogin is an EF-hand CBP capable of simultaneously binding 4 Ca2+ ions at physiological intracellular Ca2+ levels (Rogstam et al., 2007), with an affinity similar to those of the classical neuronal CBPs. Therefore, when scgn is present in neurons otherwise lacking PV, CR or CR, this CBP may contribute to the refinement of intracellular Ca2+ signalling with an as yet unknown impact on cellular excitability, and integration of afferent inputs. When scgn is co-expressed with CR or CB, it could account for a substantially enhanced Ca2+-buffering capacity thus sub-diversifying the responsiveness and network contribution of a particular neuron. However, we also entertain the possibility that scgn identifies a hitherto unknown, neurochemically distinct class of GABAergic neurons in the CA. Therefore, subsequent studies aimed to elucidate scgn’s functional significance will undoubtedly advance our understanding of the neurobiological principles that govern the organization and function of amygdaloid neuronal circuitries.
Scgn expression exhibits robust phylogenetic differences across mammalian species. Scant scgn expression is found in the SI in rodent brain. However, virtually all cholinergic basal forebrain projection neurons are scgn+ and/or scgn+/CB+ in primate brain. This difference suggests that cholinergic lineage commitment associates with a selective upregulation of scgn expression in higher-order mammals. This evolutionary transitions can be significant in explaining the differential sensitivity of rodent and primate cholinergic neurons to both physiological and noxious stimuli, and might impact cholinergic neurotransmission both at the presynaptic (neurotransmitter release) and postsynaptic (second messenger signalling) levels. Such changes may be required to accommodate the increased complexity and diversity of information processed upon expansion of isocortical areas, the primary targets of cholinergic basal forebrain afferents (Mesulam et al., 1983). A critical difference between scgn expression in prosimian primate and human brain is the unique scgn expression in pyramidal neurons of the human hippocampus (Attems et al., 2007; Attems et al., 2008). Our in situ hybridization data in mid-gestational human embryos corroborate and extend these findings by demonstrating scgn mRNA expression in the neocortex (cortical plate), hippocampus, and prospective amygdala.
In conclusion, our present report establishes scgn as a fourth developmentally-regulated, neuron-specific CBP, whose phylogenetic preservation and selective association with neurochemically distinct subsets of neurons suggest novel dimensions of functional modalities within the extended amygdala circuitry.
Supplementary Material
(A) Isolation of intact RNA is essential for gene expression profiling. Therefore, we ran an aliquot of total RNA (1 μg) isolated from microdissected mouse brains (RNeasy Mini kit, Qiagen, Crawley, United Kingdom) on a 1.0% agarose gel with GelGreen™ (Biotium, Hayward, CA, USA). Sharp 28S and 18S rRNA bands indicate intact total RNA. (A1) Real-time quantitative PCR (qPCR) reactions were validated by preliminary testing of the amplification efficacy and by excluding the possibility of genomic DNA contamination in the presence (+) or absence (−) of reverse transcriptase in parallel and running the samples on 1.5% agarose gel. Data for both secretagogin (scgn) and TATA binding protein (TBP), a housekeeping gene used as internal standard, are shown. (A2) Exon (Ex, solid squares)/intron (lines) map of the scgn gene. Open squares indicate 5′ and 3′ untranslated regions. Arrows denote the relative position and orientation of primers used to amplify scgn cDNA by qPCR. (A3) Primer sequences designed in Ex10 (forward) and Ex11 (reverse) were used to perform qPCR reactions. (A4) This primer pair amplifies with high efficacy as demonstrated by the amplicon quantity from samples of the adult olfactory bulb (OB) and medial septum (MS). NTC: non-translated control.
(A) Amino acid sequence of mouse secretagogin (scgn). Residues in red are conserved between mouse and human (scgn). Horizontal lines denote the protein sequences used to independently generate polyclonal antibodies. (B) Comparative analysis of scgn antibodies generated in rabbit (HPA006641) or goat (R & D Systems, AF4878). Both antibodies recognize a major protein band at 32 kDa in embryonic (pallium) and adult tissues (olfactory bulb). (C-C4) Distribution of scgn+ neurons in the mouse forebrain (E15), including olfactory bulb, as revealed by a polyclonal anti-scgn antibody generated in goat. Arrowheads point to neurons that are likely destined to the interstitial nucleus of the posterior limb of the anterior commissure (IPAC) and substantia innominata. List of abbreviations is available in Supporting Table 1. Scale bars = 100 μm (C,C1), 200 μm (C2–C4).
(A) Grey mouse lemur embryo on gestational day 33. (B) Mouse embryo by day 13 of gestation. Note the apparent parallels in the developmental organization of major organ systems. Scale bars = 1 mm.
Table S1. List of antibodies used for multiple immunofluorescence labelling. Panel of antibodies applied to study the distribution of scgn+ neurons in the developing rodent and primate nervous systems. Staining methods (Riedel et al., 2002; Mulder et al., 2009b) and antibody specificities were described in detail elsewhere (McDonald & Pearson, 1989; Celio, 1990; Karlsen et al., 1991; Schwaller et al., 1993; Hartig et al., 1998; Kitanaka et al., 1998; Arita et al., 2002; Bernier et al., 2002; Nagatsuka et al., 2003; Bossolasco et al., 2005; Aguado et al., 2006; Berghuis et al., 2007; Keays et al., 2007). In particular images, immunofluorescence signals were color-coded for a better identification of fine structures. Abbreviation: Cy, carbocyanine.
Table S2. Nomenclature of brain regions and their list of abbreviations used in this report. The nomenclature by Paxinos & Franklin (2001) and Bons et al. (1998) have been applied to describe neural structures in mouse and grey mouse lemur brains, respectively.
Acknowledgments
The authors thank Dr. B. Leete (Zeiss Microscopy UK) for help with image processing and analysis. Drs. K.M. Sousa (University of Michigan) and O. Kiehn (Karolinska Institute) are acknowledged for their critical comments on this manuscript. This work was supported by the Scottish Universities Life Science Alliance (T. Harkany), Alzheimer’s Association (T. Harkany), Alzheimer’s Research Trust (ART) UK (J.M. & T. Harkany), European Molecular Biology Organization Young Investigator Programme (T. Harkany), National Institutes of Health grant DA023214 (T. Harkany, Y.L.H.), Swedish Medical Research Council (T. Hokfelt, T. Harkany), European Commission (HEALTH-F2 2007-201159; T. Harkany), Grants-in-Aid for Scientific Research from the MEXT, Japan (Y.Y.), Takeda Science Foundation (Y.Y.), and Knut and Alice Wallenberg Foundation (M.U.). J.M. is recipient of a postdoctoral fellowship from ART UK.
List of Abbreviations
- BST
bed nucleus of stria terminalis
- CA
central amygdaloid nucleus
- CB
calbindin D28k
- CBP
Ca2+-binding protein
- ChAT
choline-acetyltransferase (EC 3.2.1.6)
- CR
calretinin
- Cy
carbocyanine
- DRG
dorsal root ganglion
- E
embryonic day
- EA
extended amygdala
- GAD
glutamic acid decarboxylase
- GE
ganglionic eminence GP, globus pallidus
- IPAC
interstitial nucleus of the posterior limb of the anterior commissure
- MA
medial amygdaloid nucleus
- OB
olfactory bulb
- qPCR
quantitative (real-time) PCR
- P
postnatal day
- PB
Na-phosphate buffer
- PFA
paraformaldehyde
- PV
parvalbumin
- scgn
secretagogin
- SI
substantia innominata
- TBP
TATA binding protein
- TUJ1
β-III-tubulin
- VP
ventral pallidum
Footnotes
Author contributions. T. Harkany, T. Hokfelt and Y.L.H. designed research; J.M., L.S., G.T., J.A.D. performed research; M.U., Y.Y., B.S., M.K. and F.A. contributed new reagents/analytic tools; and J.M. and T. Harkany wrote the paper.
Supporting Information (for on-line publication)
References
- Aguado T, Palazuelos J, Monory K, Stella N, Cravatt B, Lutz B, Marsicano G, Kokaia Z, Guzman M, Galve-Roperh I. The endocannabinoid system promotes astroglial differentiation by acting on neural progenitor cells. J Neurosci. 2006;26:1551–1561. doi: 10.1523/JNEUROSCI.3101-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andressen C, Blumcke I, Celio MR. Calcium-binding proteins: selective markers of nerve cells. Cell Tissue Res. 1993;271:181–208. doi: 10.1007/BF00318606. [DOI] [PubMed] [Google Scholar]
- Arita DY, Di Marco GS, Schor N, Casarini DE. Purification and characterization of the active form of tyrosine hydroxylase from mesangial cells in culture. J Cell Biochem. 2002;87:58–64. doi: 10.1002/jcb.10277. [DOI] [PubMed] [Google Scholar]
- Attems J, Preusser M, Grosinger-Quass M, Wagner L, Lintner F, Jellinger K. Calcium-binding protein secretagogin-expressing neurones in the human hippocampus are largely resistant to neurodegeneration in Alzheimer’s disease. Neuropathol Appl Neurobiol. 2008;34:23–32. doi: 10.1111/j.1365-2990.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- Attems J, Quass M, Gartner W, Nabokikh A, Wagner L, Steurer S, Arbes S, Lintner F, Jellinger K. Immunoreactivity of calcium binding protein secretagogin in the human hippocampus is restricted to pyramidal neurons. Exp Gerontol. 2007;42:215–222. doi: 10.1016/j.exger.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Berghuis P, Rajnicek AM, Morozov YM, Ross RA, Mulder J, Urban GM, Monory K, Marsicano G, Matteoli M, Canty A, Irving AJ, Katona I, Yanagawa Y, Rakic P, Lutz B, Mackie K, Harkany T. Hardwiring the brain: endocannabinoids shape neuronal connectivity. Science. 2007;316:1212–1216. doi: 10.1126/science.1137406. [DOI] [PubMed] [Google Scholar]
- Bernier PJ, Bedard A, Vinet J, Levesque M, Parent A. Newly generated neurons in the amygdala and adjoining cortex of adult primates. Proc Natl Acad Sci U S A. 2002;99:11464–11469. doi: 10.1073/pnas.172403999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bons N, Silhol S, Barbie V, Mestre-Frances N, Albe-Fessard D. A stereotaxic atlas of the grey lesser mouse lemur brain (Microcebus murinus) Brain Res Bull. 1998;46:1–173. doi: 10.1016/s0361-9230(97)00458-9. [DOI] [PubMed] [Google Scholar]
- Bossolasco P, Cova L, Calzarossa C, Rimoldi SG, Borsotti C, Deliliers GL, Silani V, Soligo D, Polli E. Neuro-glial differentiation of human bone marrow stem cells in vitro. Exp Neurol. 2005;193:312–325. doi: 10.1016/j.expneurol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Brun P, Dupret JM, Perret C, Thomasset M, Mathieu H. Vitamin D-dependent calcium-binding proteins (CaBPs) in human fetuses: comparative distribution of 9K CaBP mRNA and 28K CaBP during development. Pediatr Res. 1987;21:362–367. doi: 10.1203/00006450-198704000-00008. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Rozov A. Presynaptic Ca2+ dynamics, Ca2+ buffers and synaptic efficacy. Cell Calcium. 2005;37:489–495. doi: 10.1016/j.ceca.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Carleton A, Petreanu LT, Lansford R, Alvarez-Buylla A, Lledo PM. Becoming a new neuron in the adult olfactory bulb. Nature Neuroscience. 2003;6:507–518. doi: 10.1038/nn1048. [DOI] [PubMed] [Google Scholar]
- Celio MR. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Curr Opin Neurobiol. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Flames N, Marin O. Developmental mechanisms underlying the generation of cortical interneuron diversity. Neuron. 2005;46:377–381. doi: 10.1016/j.neuron.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Fritschy JM. Is my antibody-staining specific? How to deal with pitfalls of immunohistochemistry. Eur J Neurosci. 2008;28:2365–2370. doi: 10.1111/j.1460-9568.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- Gartner W, Vila G, Daneva T, Nabokikh A, Koc-Saral F, Ilhan A, Majdic O, Luger A, Wagner L. New functional aspects of the neuroendocrine marker secretagogin based on the characterization of its rat homolog. Am J Physiol Endocrinol Metab. 2007;293:E347–E354. doi: 10.1152/ajpendo.00055.2007. [DOI] [PubMed] [Google Scholar]
- Geula C, Schatz CR, Mesulam MM. Differential localization of NADPH-diaphorase and calbindin-D28k within the cholinergic neurons of the basal forebrain, striatum and brainstem in the rat, monkey, baboon and human. Neuroscience. 1993;54:461–476. doi: 10.1016/0306-4522(93)90266-i. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Hartig W, Seeger J, Naumann T, Brauer K, Bruckner G. Selective in vivo fluorescence labelling of cholinergic neurons containing p75(NTR) in the rat basal forebrain. Brain Res. 1998;808:155–165. doi: 10.1016/s0006-8993(98)00792-6. [DOI] [PubMed] [Google Scholar]
- Heizmann CW. Intracellular calcium-binding proteins: structure and possible functions. J Cardiovasc Pharmacol. 1986;8(Suppl 8):S7–12. [PubMed] [Google Scholar]
- Hurd YL. In situ hybridization with isotopic riboprobes for detection of striatal neuropeptide mRNA expression after dopamine stimulant administration. Methods Mol Med. 2003;79:119–135. doi: 10.1385/1-59259-358-5:119. [DOI] [PubMed] [Google Scholar]
- Hurd YL, Wang X, Anderson V, Beck O, Minkoff H, Dow-Edwards D. Marijuana impairs growth in mid-gestation fetuses. Neurotoxicol Teratol. 2005;27:221–229. doi: 10.1016/j.ntt.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Karlsen AE, Hagopian WA, Grubin CE, Dube S, Disteche CM, Adler DA, Barmeier H, Mathewes S, Grant FJ, Foster D. Cloning and primary structure of a human islet isoform of glutamic acid decarboxylase from chromosome 10. Proc Natl Acad Sci U S A. 1991;88:8337–8341. doi: 10.1073/pnas.88.19.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanaka J, Takemura M, Matsumoto K, Mori T, Wanaka A. Structure and chromosomal localization of a murine LIM/homeobox gene, Lhx8. Genomics. 1998;49:307–309. doi: 10.1006/geno.1998.5203. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M, Lu B, Xing X, Xu E, Ren G, Huang Q. Secretagogin, a novel neuroendocrine marker, has a distinct expression pattern from chromogranin A. Virchows Arch. 2006;449:402–409. doi: 10.1007/s00428-006-0263-9. [DOI] [PubMed] [Google Scholar]
- McDonald AJ, Pearson JC. Coexistence of GABA and peptide immunoreactivity in non-pyramidal neurons of the basolateral amygdala. Neurosci Lett. 1989;100:53–58. doi: 10.1016/0304-3940(89)90659-9. [DOI] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Wainer BH, Levey AI. Central cholinergic pathways in the rat: an overview based on an alternative nomenclature (Ch1-Ch6) Neuroscience. 1983;10:1185–1201. doi: 10.1016/0306-4522(83)90108-2. [DOI] [PubMed] [Google Scholar]
- Meyer G, Soria JM, Martinez-Galan JR, Martin-Clemente B, Fairen A. Different origins and developmental histories of transient neurons in the marginal zone of the fetal and neonatal rat cortex. J Comp Neurol. 1998;397:493–518. [PubMed] [Google Scholar]
- Mulder J, Bjorling E, Jonasson K, Wernerus H, Hober S, Hokfelt T, Uhlen M. Tissue profiling of the mammalian central nervous system using human antibody-based proteomics. Mol Cell Proteomics. 2009a;8:1612–1622. doi: 10.1074/mcp.M800539-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Zilberter M, Spence L, Tortoriello G, Uhlen M, Yanagawa Y, Aujard F, Hokfelt T, Harkany T. Secretagogin is a Ca2+-binding protein specifying subpopulations of telencephalic neurons. Proc Natl Acad Sci U S A. 2009b;106:22492–22497. doi: 10.1073/pnas.0912484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatsuka Y, Hara-Yokoyama M, Kasama T, Takekoshi M, Maeda F, Ihara S, Fujiwara S, Ohshima E, Ishii K, Kobayashi T, Shimizu K, Hirabayashi Y. Carbohydrate-dependent signaling from the phosphatidylglucoside-based microdomain induces granulocytic differentiation of HL60 cells. Proc Natl Acad Sci U S A. 2003;100:7454–7459. doi: 10.1073/pnas.1232503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG. Mitochondrial calcium function and dysfunction in the central nervous system. Biochim Biophys Acta. 2009;1787:1416–1424. doi: 10.1016/j.bbabio.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. Academic Press; San Diego: 2001. [Google Scholar]
- Pencea V, Luskin MB. Prenatal development of the rodent rostral migratory stream. J Comp Neurol. 2003;463:402–418. doi: 10.1002/cne.10746. [DOI] [PubMed] [Google Scholar]
- Perret M. Influence of social-factors on sex-ratio at birth, maternal investment and young survival in a prosimian primate. Behavioral Ecology and Sociobiology. 1990;27:447–454. [Google Scholar]
- Perret M. Environmental and social determinants of sexual function in the male lesser mouse lemur (Microcebus murinus) Folia Primatol (Basel) 1992;59:1–25. doi: 10.1159/000156637. [DOI] [PubMed] [Google Scholar]
- Perret M, Aujard F. Regulation by photoperiod of seasonal changes in body weight and reproductive function in the lesser mouse lemur (Microcebus murinus): differential responses by sex. Int J Primatol. 2001;22:5–24. [Google Scholar]
- Riedel A, Hartig W, Seeger G, Gartner U, Brauer K, Arendt T. Principles of rat subcortical forebrain organization: a study using histological techniques and multiple fluorescence labeling. J Chem Neuroanat. 2002;23:75–104. doi: 10.1016/s0891-0618(01)00142-9. [DOI] [PubMed] [Google Scholar]
- Rogstam A, Linse S, Lindqvist A, James P, Wagner L, Berggard T. Binding of calcium ions and SNAP-25 to the hexa EF-hand protein secretagogin. Biochem J. 2007;401:353–363. doi: 10.1042/BJ20060918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MP, Frassoni C, varez-Bolado G, Spreafico R, Fairen A. Distribution of calbindin and parvalbumin in the developing somatosensory cortex and its primordium in the rat: an immunocytochemical study. J Neurocytol. 1992;21:717–736. doi: 10.1007/BF01181587. [DOI] [PubMed] [Google Scholar]
- Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- Schwaller B, Buchwald P, Blumcke I, Celio MR, Hunziker W. Characterization of a polyclonal antiserum against the purified human recombinant calcium binding protein calretinin. Cell Calcium. 1993;14:639–648. doi: 10.1016/0143-4160(93)90089-o. [DOI] [PubMed] [Google Scholar]
- Solbach S, Celio MR. Ontogeny of the calcium binding protein parvalbumin in the rat nervous system. Anat Embryol (Berl) 1991;184:103–124. doi: 10.1007/BF00942742. [DOI] [PubMed] [Google Scholar]
- Strehler EE, Filoteo AG, Penniston JT, Caride AJ. Plasma-membrane Ca(2+) pumps: structural diversity as the basis for functional versatility. Biochem Soc Trans. 2007;35:919–922. doi: 10.1042/BST0350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21:323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- Tallini YN, Shui B, Greene KS, Deng KY, Doran R, Fisher PJ, Zipfel W, Kotlikoff MI. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol Genomics. 2006;27:391–397. doi: 10.1152/physiolgenomics.00092.2006. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- Verney C, Derer P. Cajal-Retzius neurons in human cerebral cortex at midgestation show immunoreactivity for neurofilament and calcium-binding proteins. J Comp Neurol. 1995;359:144–153. doi: 10.1002/cne.903590110. [DOI] [PubMed] [Google Scholar]
- Wagner L, Oliyarnyk O, Gartner W, Nowotny P, Groeger M, Kaserer K, Waldhausl W, Pasternack MS. Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J Biol Chem. 2000;275:24740–24751. doi: 10.1074/jbc.M001974200. [DOI] [PubMed] [Google Scholar]
- Wang X, Dow-Edwards D, Anderson V, Minkoff H, Hurd YL. In utero marijuana exposure associated with abnormal amygdala dopamine D2 gene expression in the human fetus. Biol Psychiatry. 2004;56:909–915. doi: 10.1016/j.biopsych.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Woodruff AR, Sah P. Inhibition and synchronization of basal amygdala principal neuron spiking by parvalbumin-positive interneurons. J Neurophysiol. 2007a;98:2956–2961. doi: 10.1152/jn.00739.2007. [DOI] [PubMed] [Google Scholar]
- Woodruff AR, Sah P. Networks of parvalbumin-positive interneurons in the basolateral amygdala. J Neurosci. 2007b;27:553–563. doi: 10.1523/JNEUROSCI.3686-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Isolation of intact RNA is essential for gene expression profiling. Therefore, we ran an aliquot of total RNA (1 μg) isolated from microdissected mouse brains (RNeasy Mini kit, Qiagen, Crawley, United Kingdom) on a 1.0% agarose gel with GelGreen™ (Biotium, Hayward, CA, USA). Sharp 28S and 18S rRNA bands indicate intact total RNA. (A1) Real-time quantitative PCR (qPCR) reactions were validated by preliminary testing of the amplification efficacy and by excluding the possibility of genomic DNA contamination in the presence (+) or absence (−) of reverse transcriptase in parallel and running the samples on 1.5% agarose gel. Data for both secretagogin (scgn) and TATA binding protein (TBP), a housekeeping gene used as internal standard, are shown. (A2) Exon (Ex, solid squares)/intron (lines) map of the scgn gene. Open squares indicate 5′ and 3′ untranslated regions. Arrows denote the relative position and orientation of primers used to amplify scgn cDNA by qPCR. (A3) Primer sequences designed in Ex10 (forward) and Ex11 (reverse) were used to perform qPCR reactions. (A4) This primer pair amplifies with high efficacy as demonstrated by the amplicon quantity from samples of the adult olfactory bulb (OB) and medial septum (MS). NTC: non-translated control.
(A) Amino acid sequence of mouse secretagogin (scgn). Residues in red are conserved between mouse and human (scgn). Horizontal lines denote the protein sequences used to independently generate polyclonal antibodies. (B) Comparative analysis of scgn antibodies generated in rabbit (HPA006641) or goat (R & D Systems, AF4878). Both antibodies recognize a major protein band at 32 kDa in embryonic (pallium) and adult tissues (olfactory bulb). (C-C4) Distribution of scgn+ neurons in the mouse forebrain (E15), including olfactory bulb, as revealed by a polyclonal anti-scgn antibody generated in goat. Arrowheads point to neurons that are likely destined to the interstitial nucleus of the posterior limb of the anterior commissure (IPAC) and substantia innominata. List of abbreviations is available in Supporting Table 1. Scale bars = 100 μm (C,C1), 200 μm (C2–C4).
(A) Grey mouse lemur embryo on gestational day 33. (B) Mouse embryo by day 13 of gestation. Note the apparent parallels in the developmental organization of major organ systems. Scale bars = 1 mm.
Table S1. List of antibodies used for multiple immunofluorescence labelling. Panel of antibodies applied to study the distribution of scgn+ neurons in the developing rodent and primate nervous systems. Staining methods (Riedel et al., 2002; Mulder et al., 2009b) and antibody specificities were described in detail elsewhere (McDonald & Pearson, 1989; Celio, 1990; Karlsen et al., 1991; Schwaller et al., 1993; Hartig et al., 1998; Kitanaka et al., 1998; Arita et al., 2002; Bernier et al., 2002; Nagatsuka et al., 2003; Bossolasco et al., 2005; Aguado et al., 2006; Berghuis et al., 2007; Keays et al., 2007). In particular images, immunofluorescence signals were color-coded for a better identification of fine structures. Abbreviation: Cy, carbocyanine.
Table S2. Nomenclature of brain regions and their list of abbreviations used in this report. The nomenclature by Paxinos & Franklin (2001) and Bons et al. (1998) have been applied to describe neural structures in mouse and grey mouse lemur brains, respectively.