Abstract
Ubiquitin is a versatile cellular signaling molecule that can form polymers of eight different linkages, and individual linkage-types have been associated with distinct cellular functions. Though little is currently known about Lys11-linked ubiquitin chains, recent data indicate that they may be as abundant as Lys48-linkages and involved in vital cellular processes. Here we report the generation of Lys11-linked polyubiquitin in vitro, for which the Lys11-specific E2 enzyme UBE2S was fused to a ubiquitin binding domain. Crystallography and NMR analyses of Lys11-linked diubiquitin reveal that Lys11-linked chains adopt compact conformations in which Ile44 is solvent exposed. Furthermore, we identify the OTU family deubiquitinase Cezanne as the first deubiquitinase with Lys11-linkage preference. Our data highlight the intrinsic specificity of the ubiquitin system that extends to Lys11-linked chains, and emphasize that differentially linked polyubiquitin chains must be regarded as independent posttranslational modifications.
Introduction
Protein ubiquitination is a versatile posttranslational modification with roles in protein degradation, cell signaling, intracellular trafficking and the DNA damage response1,2. Ubiquitin polymers are linked through one of seven internal lysine (K) residues or through the N-terminal amino group. Importantly, the type of ubiquitin linkage determines the functional outcome of the modification1. The best-studied ubiquitin polymers, Lys48- and Lys63-linked chains, have degradative and non-degradative roles, respectively2,3. However, recent data revealed an unexpected high abundance of so-called atypical ubiquitin chains, which account for more than half of all ubiquitin linkages in S. cerevisiae4,5.
Polyubiquitin chains are assembled on substrates through the concerted action of a three-step enzymatic cascade, involving an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and E3 ubiquitin ligases6. While E3 ligases confer substrate specificity, E2 enzymes commonly determine the type of chain linkage7. Lys48- and Lys63-specific E2 enzymes that generate large amounts of these linkage types in vitro have been identified8,9, allowing structural analysis of these chain types as well as a detailed understanding of specificity of ubiquitin binding domains (UBDs) and deubiquitinases (DUBs) (reviewed in1). This information is currently lacking for atypical ubiquitin chains.
Lys11-linkages between ubiquitin molecules are as abundant as Lys48-linkages, each accounting for roughly 30% of all linkages in yeast4. Early data indicated that Lys11-linked chains are, like Lys48-linked chains, efficient proteasomal degradation signals10, but only few reports have implicated Lys11-linked ubiquitin chains in distinct biological processes. An E2 enzyme, UBE2S/E2-EPF, was identified that assembled Lys11 linkages in vitro10, and the human anaphase promoting complex (APC/C) was recently found to use a combination of UBE2C/UbcH10 and UBE2S to assemble Lys11 linkages on its substrates11-14. This places Lys11-linked ubiquitin chains at the heart of the mammalian cell cycle, suggesting that DUBs and ubiquitin binding proteins with Lys11-specificity are candidate cell cycle regulators. S. cerevisiae lacks homologs of UBE2C and UBE2S, and its APC/C assembles Lys48-linkages on cell cycle substrates15. However, S. cerevisiae contain large amounts of Lys11-linked ubiquitin chains, and these have been implicated in endoplasmic reticulum-associated degradation (ERAD)4. In mammalian cells, Lys11 linkages were also found to be enriched in UBA/UBX protein complexes, which interact with the key ERAD regulator p97/cdc4816. Hence, Lys11-linked chains seem to regulate important cellular processes, including ERAD and the cell cycle, and possibly others. This poses the questions whether Lys11-linked polyubiquitin act merely as a redundant proteasomal targeting signal, or whether these chains, and hence degradation of their targets, is regulated independently.
Here we analyze the Lys11-specific E2 enzyme UBE2S, which assembles Lys11-linked ubiquitin chains on its own C-terminal tail in vitro, and also generates limited amounts of free, i.e. unattached, Lys11-linked diubiquitin. We engineer a UBE2S fusion protein that synthesizes free Lys11-linked polymers with markedly increased efficiency, allowing high-level purification of Lys11-linked ubiquitin dimers, trimers and tetramers, facilitating detailed structural and biochemical studies. Lys11-linked ubiquitin chains adopt a unique conformation, distinct from Lys48-, Lys63-linked and linear chains. We further identify Cezanne as the first DUB with preference for Lys11-linkages, suggesting that Lys11-linked ubiquitin chains are independently regulated degradation signals within the ubiquitin system.
Results
Analysis of E2 enzymes involved in Lys11 chain formation
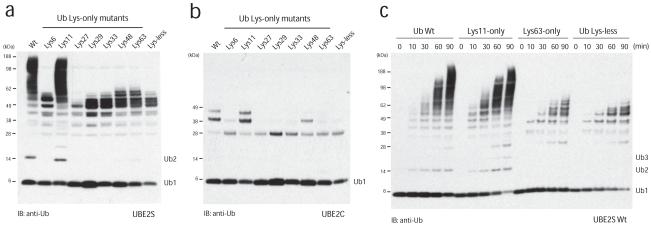
Specific E2 enzymes have been described for the assembly of free Lys48- and Lys63-linked ubiquitin chains, and the biology of these posttranslational modifications is now known in great detail. In order to study the elusive Lys11 linkage, we analyzed two human E2 conjugating enzymes that have been associated with this chain type, namely UBE2C/UbcH1011 and UBE2S/E2-EPF10, for their ability to assemble unattached polyubiquitin chains in vitro in absence of an E3 ligase. UBE2S generated small amounts of free diubiquitin (Fig. 1a), while UBE2C did not assemble unattached ubiquitin chains (Fig. 1b). UBE2S, but not UBE2C, also underwent autoubiquitination, resulting in the appearance of high molecular weight species of UBE2S (Fig. 1a). Linkage type analysis using single-Lys ubiquitin mutants (Lys6-only, Lys11-only etc.) revealed that UBE2S assembled Lys11 linkages specifically (Fig. 1a). UBE2S autoubiquitinated several of its 17 Lys residues, however with ubiquitin mutants lacking Lys11, these monoubiquitin modifications were not extended (Fig. 1a, c), and autoubiquitination with Lys-less and Lys63-only ubiquitin followed similar kinetics resulting in 6-7 distinct bands of UBE2S, most likely corresponding to multi-monoubiquitinated species (Fig. 1c). Lys11-specificity of UBE2S was verified by tryptic analysis of wild-type diubiquitin by LC-MS/MS (Supp. Fig. 1), in which we detected peptides derived from Lys11-linked diubiquitin, and with substantially lower intensity also from Lys48- and Lys63-linked diubiquitin. Other linkages were not detected (Supp. Fig. 1).
Figure 1.
UBE2S is a Lys11-specific E2 enzyme
(a) UBE2S and (b) UBE2C were analyzed in autoubiquitination assays in the presence of E1, ubiquitin and Mg•ATP. The panel of single-Lys ubiquitin mutants reveals the intrinsic linkage specificity. Autoubiquitination is visualized with a polyclonal anti-ubiquitin antibody. UBE2S, but not UBE2C, autoubiquitinates and also assembles unattached Lys11-linked ubiquitin chains. (c) Time course assay for autoubiquitination by UBE2S. The reaction for wild-type (wt) and Lys11-only ubiquitin leads to similar high-molecular weight conjugates, while for the Lys-less (K0) and Lys63-only ubiquitin an equivalent pattern of multi-monoubiquitination is observed.
Biochemical analysis of UBE2S
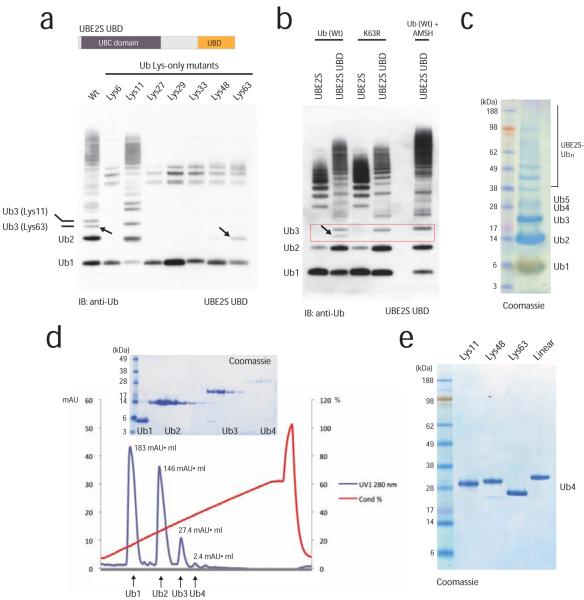
We set out to harness the capability of UBE2S to produce free Lys11-linked ubiquitin chains. Human UBE2S comprises 222 residues with an N-terminal conserved catalytic Ubc domain spanning residues 1-156. The C-terminal 25 residues include nine Lys residues that are conserved in UBE2S homologs (www.ensembl.org), while the remaining 40 residues form a non-conserved Lys-free linker (Fig. 2a). Mutation of the catalytic Cys residue to Ala (UBE2S C95A) rendered UBE2S inactive, while the C95S mutant acted as a ubiquitin-trap, since the Ser residue was still charged with ubiquitin by the E1 enzyme, but failed to discharge efficiently (Fig. 2a), similar to what has been reported for UBE2N/Ubc1317. Autoubiquitination of UBE2S occurred in cis, as wild-type UBE2S was unable to ubiquitinate GST-tagged UBE2S C95A in trans (Fig. 2b). The autoubiquitination of UBE2S appeared to be favored compared to formation of free Lys11-linked chains, and free chain production is inefficient. The Lys-rich tail of UBE2S is a likely target for autoubiquitination, and removal of the last 25 residues (UBE2S ΔC) reduced autoubiquitination (Fig. 2c), increased formation of free diubiquitin (data not shown), and retained specificity for Lys11 linkages (Fig. 2c). From 25 mg input ubiquitin, ~1 mg Lys11-linked diubiquitin could be purified by cation exchange (Fig. 2d).
Figure 2.
Assembly of Lys11-linked diubiquitin
(a) Domain structure of UBE2S, and autoubiquitination reactions with UBE2S wild-type and catalytic mutants. (b) UBE2S autoubiquitination occurs in cis. Wild-type UBE2S was mixed with GST-tagged inactive UBE2SC95A, and after precipitation of the GST-tagged protein, ubiquitination in supernatant (left) and precipitate (right) is analyzed. (c) Removal of the Lys-rich tail of UBE2S decreases autoubiquitination while preserving Lys11 specificity. (d) Purification of Lys11-linked diubiquitin by cation exchange chromatography. The integrated peak area (mAU*ml) is indicated. A gel showing protein-containing fractions is shown as an inset.
Generation of Lys11-linked ubiquitin tetramers
In order to increase the yields of Lys11-linked dimers and to obtain longer polymers, we created an UBE2S variant with increased capability to form free ubiquitin chains. We replaced the Lys-rich tail of UBE2S that is targeted by cis-autoubiquitination (residues 196-222) with the ubiquitin binding ZnF-UBP domain of human USP5/IsoT (residues 173-289)18 (Fig. 3a). This UBD binds ubiquitin with nanomolar affinity and interacts with the free C-terminus of ubiquitin, leaving the Lys11 side chain accessible for chain elongation18. The UBE2S UBD fusion protein was markedly more efficient in producing ubiquitin dimers, trimers, and tetramers. However, UBE2S UBD also incorporated Lys63-linkages in these oligomers (see Lys63-only mutant in Fig. 3a). Two distinct trimer bands were observed in reactions using wild-type ubiquitin, but not with the Lys11-only ubiquitin mutant, indicating alternating or branched linkages with wild-type ubiquitin, since differently linked ubiquitin chains have distinct electrophoretic mobility (Fig. 3a,b). Two linkage types (Lys11 and Lys63) in the wild-type ubiquitin reaction were further confirmed by LC-MS/MS analysis. Formation of Lys63-linkages could be prevented either by using the ubiquitin K63R mutant, or by incubation with the Lys63-specific deubiquitinase AMSH19 (Fig. 3b). Indeed, AMSH removed only the faster migrating of the two triubiquitin bands, showing that a chain with alternate linkages had been created by UBE2S-UBD (Fig. 3b). When we included AMSH directly in the assembly reactions, we were able to remove the contaminating Lys63 linkages in situ (Fig. 3b). This protocol allowed large scale generation and purification of Lys11-linked di-, tri- and tetraubiquitin (Fig. 3c,d,e) with improved yields. Almost 50% of the input ubiquitin was converted into Lys11-linked oligomers using UBE2S UBD, while UBE2S ΔC only assembled 15% of input ubiquitin into Lys11-linked dimers (Fig. 3b, compare integrated peak area in Fig. 2d and 3d). The generation of Lys11-linked ubiquitin chains in large quantities allowed detailed structural analysis of this chain type.
Figure 3.
Assembly of Lys11-linked tetraubiquitin
(a) UBE2S engineering to increase yields of free Lys11-linked ubiquitin chains. The C-terminal tail was replaced with the ZnF-UBP domain of USP5/IsoT. The fusion protein assembles free chains of up to five ubiquitin molecules, yet it is less specific and also incorporates Lys63-linkages with wild-type and Lys63-only ubiquitin (indicated by arrows). (b) Incorporation of Lys63-linkages can be counteracted by using a K63R ubiquitin mutant, or by including the Lys63-specific DUB AMSH in the reaction, as observed by disappearance of the faster migrating Lys63-linkage contamination. (c) 5 μl aliquot of a 1 ml chain assembly reaction using 25 mg ubiquitin shows that di-, tri- and tetraubiquitin is generated in milligram quantities. (d) Cation exchange chromatography was used to purify Lys11-linked ubiquitin chains. The integrated peak area (mAU*ml) is specified. A gel showing protein-containing fractions is shown as an inset. (e) Purified ubiquitin tetramers of Lys11, Lys48, Lys63 and linear linkages have different electrophoretic mobility on 4-12% SDS-PAGE gels.
Solution studies of Lys11-linked diubiquitin
Ubiquitin chains are dynamic entities and can adopt multiple conformations in solution. The solution properties of Lys11-linked ubiquitin chains were studied with Nuclear Magnetic Resonance (NMR) spectroscopy. 1H, 15N-heteronuclear correlation spectra (HSQC) provide a fingerprint of the local environment of individual residues. These so-called chemical shifts report on the resonance frequencies of all backbone amide protons and nitrogens, and chemical shift perturbations as a consequence of e.g. the formation of an interface are highly specific.
We assembled uniformly labeled Lys11 diubiquitin from 13C, 15N-labeled K63R mutant ubiquitin. To subsequently deconvolute the contribution from both parts of the interface, in a second species only the distal moiety of Lys11-linked diubiquitin was 13C, 15N-labeled. To achieve this, assembly reactions with 13C, 15N-labeled K11R and unlabeled wild-type ubiquitin were performed, in which the K11R mutant serves as a distal chain terminator. To minimize buffer effects, the two labeled diubiquitin species, as well as labeled K63R, and K11R monoubiquitin (all at 100 μM) were dialyzed simultaneously against neutral (pH 7.2) or acidic (pH 3.5) buffer also containing 150mM NaCl to mask nonspecific interactions. Relaxation experiments and measurements at different concentrations confirmed monodispersity, and allowed to exclude aggregation effects for all species at the chosen experimental conditions. We assigned and confirmed the chemical shift positions in all species with standard triple-resonance experiments (Supp. Fig. 2a). To generate chemical shift perturbation maps, we compared uniformly labeled Lys11-linked diubiquitin to K63R monoubiquitin, and distally labeled Lys11-linked diubiquitin to K11R monoubiquitin. To assess the effect of K63R and K11R mutations we compared the labeled monoubiquitin species to find perturbation differences of <0.1 p.p.m., with exception of the flexible loop region in ubiquitin near Lys11 that is slightly more perturbed (Supp.Fig. 2b).
Immediately apparent was the doubling of a defined subset of resonances in the spectra of uniformly labeled Lys11-linked diubiquitin in all tested conditions (Fig. 4b,c and Supplementary Data), associated with interface formation. As expected, the resonances for Lys11 and Gly76 involved in the Lys11-linkage were markedly shifted compared to monoubiquitin (Fig. 4b). The chemical shift perturbation map of this species contains contributions of both sides of the interface (Fig. 4b). To deconvolute the individual contributions, we analyzed chemical shift perturbations of distally labeled Lys11-linked diubiquitin in comparison to K11R monoubiquitin (Fig. 4b). This revealed the set of perturbed resonances that correspond to the interface on the distal moiety. Importantly, all resonances that were found to be perturbed in the distally labeled Lys11-linked diubiquitin have equivalent or similar perturbations in the uniformly labeled Lys11-linked diubiquitin. However, we cannot exclude or quantify contributions to these perturbations from the proximal moiety in this case.
Figure 4.
NMR Solution studies of Lys11-linked diubiquitin.
(a) Overlay of 15N,1H HSQC spectra of ubiquitin K63R (red) onto Lys11-linked diubiquitin K63R (blue). The expansion illustrates the doubling of peaks observed for Lys29, Ile30, Asp32 and Lys33. The signal for Asp52 is unperturbed. (b, c) Weighted chemical shift perturbation according to residue number for Lys11-linked diubiquitin with both molecules 13C, 15N–labeled (blue, K63R ubiquitin mutant) or only labeled distally (orange, K11R ubiquitin mutant). Shown are chemical shift perturbations observed for doubled peaks calculated as the weighted difference between the chemical shift position in the Lys11-linked diubiquitin mutants and their respective monoubiquitin counterparts at pH7.2 (b) and pH 3.5 (c). Stars (*) indicate exchange broadened residues, and arrows indicate K29 and K33. (d) Combined chemical shift perturbation differences for Lys48- and Lys63-linked diubiquitin. This data was kindly provided by Takeshi Tenno 22.
This analysis shows that Lys11-linked dimers have a defined pattern of perturbed resonances in solution, which is distinct from the pattern observed for Lys48-, or Lys63-linked diubiquitin20-22 (Fig. 4d), reflecting (a) distinct conformation(s) of Lys11-linked diubiquitin. Strikingly, the backbone resonances corresponding to residues 41-51 of ubiquitin (including Ile44) are not perturbed, suggesting that this region, which is involved in the Lys48 diubiquitin interface21,22 (Fig. 4d) and in most ubiquitin-UBD interactions23-25, is not involved in the dimer interface in Lys11-linkages. Instead, the chemical shift perturbations indicate three regions of the ubiquitin surface that contribute to the interface and/or are affected by the Lys11 isopeptide bond: The flexible β-hairpin loop spanning residues 5-15; residues 29-36 that include the C-terminal part of the ubiquitin helix; and the C-terminal residues from 69-76 (Fig. 4b).
To further distinguish between interface residues and residues perturbed as a result of forming the isopeptide linkage, we analyzed chemical shift perturbations also at low pH. It has previously been shown for Lys48-linked diubiquitin that low pH ‘opens’ the compact conformation of this chain type resulting in a more transient interface21. If a similar interface ‘opening’ also occurred for Lys11-linkages, this may allow a more confident definition of interface residues. Although at pH 3.5 we observe fewer perturbations compared to pH 7.2, several residues remain perturbed (Fig. 4c). On the other hand, Lys29 and Lys33 are perturbed only at pH 7.2 but not at pH 3.5 (indicated by arrows in Fig. 4b,c), suggesting that these residues are located at an interface at pH 7.2.
Crystal structure of Lys11-linked diubiquitin
Lys11-linked ubiquitin dimers at 5 mg ml−1 were crystallized, and diffraction data were collected at the ESRF (Grenoble) to 2.2 Å resolution. The structure was solved by molecular replacement, and refined to final statistics found in Table 1. Six Lys11-linked diubiquitin molecules are present in the asymmetric unit.
Table 1.
Data collection and refinement statistics
| Lys11-linked diubiquitin | |
|---|---|
| Data collection | |
| Space group | P2221 |
| Cell dimensions | |
| a, b, c (Å) | 79.23, 79.96, 221.23 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 24.92 – 2.20 (2.32 – 2.20)* |
| R merge | 0.106 (0.485) |
| I / σI | 6.0 (2.0) |
| Completeness (%) | 98.3 (99.7) |
| Redundancy | 3.1 (3.0) |
| Refinement | |
| Resolution (Å) | 24.92 – 2.20 |
| No. reflections | 65986 |
| Rwork / Rfree | 0.205 / 0.252 |
| No. atoms | |
| Protein | 7255 (12 ubiquitin molecules) |
| Ligand/ion | 111 |
| Water | 654 |
| B-factors | |
| Protein | 30.1 |
| Ligand/ion | 41.8 |
| Water | 34.4 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.978 |
Values in parentheses are for highest-resolution shell.
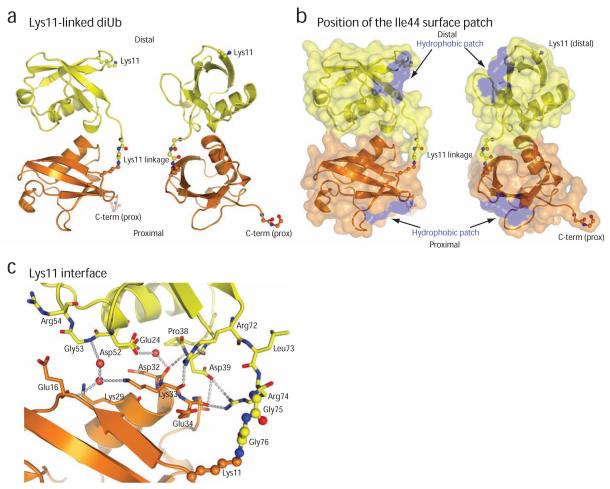
Lys11-linked diubiquitin adopts a compact conformation in the crystals, distinct from any other ubiquitin chain structure observed to date (Fig. 5a, Supp. Fig. 3). Contacts between isopeptide-linked ubiquitin moieties in the crystal structure are entirely polar and do not involve the hydrophobic ubiquitin surface patch (Ile44, Leu8, Val70)(Fig. 5b). The interface instead forms between a surface centered on Glu24 of the distal ubiquitin, and a surface around Lys29 and Lys33 of the proximal ubiquitin. Several direct and water-mediated interactions are formed, including Arg74dist - Glu34prox, backbone (bb), Arg72dist – Glu34prox, bb, Asp39dist, bb- Asp32prox, bb (Fig. 5c). Crystals were obtained at low pH (3.5-5.5) from high salt or PEG precipitants. Low pH conditions may mask additional charged interactions, for example Lys33prox – Asp52dist, which are in close proximity but do not seem to interact in any of the dimer interfaces.
Figure 5.
Crystal structure of Lys11-linked diubiquitin
(a) The crystal structure of Lys11-linked diubiquitin in two orientations. The proximal (orange) and distal (yellow) molecules interact through the ubiquitin helix, and the isopeptide linkage (shown in ball-and-stick representation, with red oxygen and blue nitrogen atoms) is at the surface of the dimer. (b) A semi-transparent surface colored blue for residues Ile44, Leu8 and Val70 shows that the hydrophobic patch is not involved in the interface. (c) Residues at the interface are shown in stick representation, and polar interactions of <3.5Å are shown with dotted lines. Water molecules are shown as purple spheres.
A striking feature of the crystal structure, which is consistent with solution measurements, is the exposed location of Ile44 of the ubiquitin hydrophobic patch (blue in Fig. 5b). Eight of the twelve Ile44 residues are not involved in crystal contacts but point towards solvent channels. The remaining hydrophobic patch residues Val70 and Leu8 are involved in symmetric crystal contacts with non-covalently bound molecules. Interestingly, the additional crystal contacts are compatible with a putative second interface for a Lys11-linked diubiquitin (see Supplementary Discussion and Supp. Fig. 4, 5). Since Val70 and Leu8 are perturbed in the solution measurements (Fig. 4b,c) it is a possibility that the crystal structure has captured two distinct compact conformations that Lys11-linkages can adopt (Supp. Fig. 5). However, it is also possible that the perturbations observed for Val70 and Leu8 in the solution studies originate from indirect effects of the isopeptide linkage (Supp. Fig. 6).
Further comparison of residues located at the asymmetric crystallographic interface (Fig. 5a) with perturbed residues in the solution measurements (Fig. 4) reveals an almost equivalent interface on the proximal ubiquitin moiety, involving Lys29 and Lys33 (Supp. Fig. 7a). A corresponding distal interface however appears to be less well-defined in solution when compared to the crystal structure (Supp. Fig. 7b). This indicates that in solution, the distal ubiquitin may rotate or move slightly, resulting in differences between solution measurements and crystal structure. Nevertheless, the solution measurements and crystallographic analysis of Lys11-linked diubiquitin suggests compact conformations of Lys11-linked ubiquitin chains distinct from Lys48- and Lys63-linked polyubiquitin.
Panel screening of DUBs against Lys11-linked ubiquitin chains
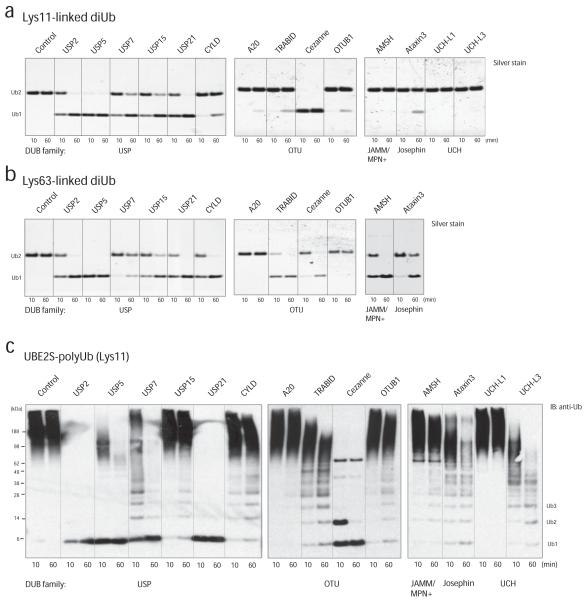
The structural features of Lys11-linkages pose the question whether deubiquitinases exist that act specifically on this chain type. Importantly many DUBs are indeed endowed with intrinsic specificity for particular Lys linkages26. We used 14 representative members of the five human DUB families (Supp. Table 1) and analyzed their ability to hydrolyze Lys11-linked diubiquitin or Lys11-polyubiquitinated UBE2S, in comparison to cleavage of Lys63-linked di- and polyubiquitin (Fig. 6, Supp. Fig. 8). We had previously established conditions at which these DUBs are active and specific against their preferred linkage27 (Supp. Table 1), and used the same enzyme concentration and assay setup with our newly generated Lys11-linked chains.
Figure 6.
Hydrolysis of Lys11 linkages by DUBs.
(a, b) Analysis of a panel of DUBs from all human DUB families against Lys11- (a) and Lys63-linked (b) diubiquitin. Two lanes per DUB in a silver stained gel indicate hydrolysis of ubiquitin dimers after 10 and 60 min, respectively. The Lys63-linked dimers were not tested against UCH family members as these were previously found to be inactive against this chain type 33. (c) The panel of DUBs was analyzed against Lys11-polyubiquitinated UBE2S and ubiquitin was visualized by Western blotting.
The USP domain DUBs USP2, USP5/IsoT, USP7, USP15 and USP21 cleaved Lys11- and Lys63-linked diubiquitin equally well, while the USP domain of CYLD showed Lys63-specificity as reported before28. The catalytic OTU domains of TRABID (Lys63-specific29) and A20 (Lys48-specific30), as well as full-length OTUB1 (Lys48-specific31), showed only weak activity against Lys11-linkages, while cleaving Lys63- or Lys48-linkages, respectively, with high activity (Fig. 6a,b and Supp. Fig. 8a). In contrast, the Cezanne OTU domain had higher activity against Lys11-linkages when compared to Lys63-linked diubiquitin (Fig. 6a,b and see below). AMSH, a JAMM domain metalloprotease with Lys63 specificity19 did not hydrolyze Lys11-linked diubiquitin, allowing us to include this DUB in chain assembly reaction (see above). Ataxin-3, a Josephin DUB with preference for Lys63-linkages in mixed linkage chains32, cleaved Lys11-linked diubiquitin weakly, and the ubiquitin C-terminal hydrolases UCH-L1 and UCH-L3 did not cleave Lys11-linkages, consistent with their inability to hydrolyze free ubiquitin chains33 (Fig. 6a,b). UCH-L3 was highly active against ubiquitin with a C-terminal hexahistidine-tag (Supp. Fig. 8b).
We next analyzed this DUB panel with Lys11-polyubiquitinated UBE2S compared to Lys63-polyubiquitinated TRAF6 (Fig. 6c, Supp. Fig. 8c). The results from this analysis were remarkably similar to those obtained from the dimer cleavage analysis. USP2, USP5, USP7 and USP21 acted on the attached Lys11-linked chains, and USP21 and USP2 were able to hydrolyze all linkages to monoubiquitin. In contrast, USP15 and CYLD had limited activity on the Lys11-polyubiquitinated substrate, and appeared to release longer chains rather than to produce monoubiquitin (Fig. 6c). A20 did not hydrolyze any Lys11-linked chains from polyubiquitinated UBE2S, while TRABID and OTUB1 released small amounts of Lys11-linked polymers. Cezanne appeared to act on Lys11-linkages specifically, as it produced a single ubiquitinated species, suggesting that it had not hydrolyzed the proximal monoubiquitin(s) from UBE2S (Fig. 6c). Also Ataxin-3, AMSH and UCH-L3 (but not UCH-L1) released small amounts of Lys11-linked polymers from UBE2S (Fig. 6c).
Validation of Cezanne as a Lys11-specific deubiquitinase
The catalytic OTU domain of Cezanne is closely related to those of TRABID and A20 (Fig. 7a), which preferentially cleave Lys63- and Lys48-linked polyubiquitin, respectively 33. These three proteins share an extended catalytic domain of 320-350 residues compared to other OTU domains (~150 Aa), and also the domain structure of the full-length enzymes is similar (Fig. 7b). We analyzed the linkage preference of the isolated OTU domains in more detail using ubiquitin tetramers of defined linkages. Cezanne cleaved Lys11 linkages with considerably higher activity compared to linear, Lys48-, and Lys63-linked chains (Fig. 7c). We reduced the enzyme:substrate ratio from 1:7 to 1:45 (3 μM tetramers were hydrolyzed with 60 nM Cezanne) and even at this lower concentration Cezanne hydrolyzed Lys11 tetramers within 10 minutes (Fig 7c, Supp. Fig. 8d). In contrast, the A20 and TRABID OTU domains did not cleave Lys11-linked tetramers efficiently, but remained Lys48 and Lys63 selective, respectively (Fig. 7d, e). Finally, we exploited the distinct electrophoretic mobility of different chain linkages. Ubiquitin tetramers were mixed and incubated with Cezanne, and only the band for Lys11-linked tetraubiquitin disappeared, while Lys48-, Lys63-linked and linear tetraubiquitin levels remained unaffected (Fig. 7f). Furthermore, no cleavage of Lys6- or Lys29-linked diubiquitin is observed (S. Virdee, J. Chin and DK, unpublished). This identifies Cezanne as the first described DUB with Lys11-preference. It also highlights that OTU domains are furnished with highly selective catalytic cores.
Figure 7.
Cezanne cleaves Lys11-linkages preferentially
(a) Phylogenetic tree of the OTU family DUBs 41, showing the close relationship of A20, TRABID and Cezanne. (b) Schematic presentation of domain structures of the OTU family DUBs Cezanne, A20 and TRABID. (c-e) Specificity analysis of Cezanne (c), A20 (d) and TRABID (e) against Lys11-, Lys48-, Lys63- and linear ubiquitin tetramers, resolved on 4-12% SDS-PAGE gels and visualized by silver staining. In this comparison, Cezanne shows specificity for Lys11-linked chains, while A20 and TRABID remain Lys48- and Lys63-specific, respectively. (f) Cezanne prefers Lys11-linkages in a mixture of differently linked ubiquitin tetramers. The different electrophoretic mobility of the individual tetraubiquitin molecules allows identification of the linkage type.
A20 and TRABID are ‘signaling’ deubiquitinases, with roles in NF-κB and Wnt-signaling, respectively 29,34. Also Cezanne has been linked to NF-κB signaling, since it is upregulated by tumor necrosis factor (TNF)α35. Activation of the NF-κB transcription factor by cytokines such as TNFα relies on ubiquitination of receptor interacting proteins including RIP1, leading to activation of downstream kinase cascades36. Overexpression of Cezanne leads to deubiquitination of RIP135, however, this inhibitory effect of Cezanne has been attributed to its non-specific activity against Lys48- and Lys63-linkages35. We also find that high concentrations of Cezanne cleave these chain types (Fig. 6b, Supp. Fig. 8c and data not shown). However, our data show that the preferred substrate of Cezanne is Lys11-linked polyubiquitin, and this chain type has not been implicated in NF-κB signaling so far. Lys11-linkage preference suggests that Cezanne may stabilize negative regulators of NF-κB signaling since Lys11-linkages can act as proteasomal targeting signals. It is however also possible that Lys11-linkages may have non-degradative roles. Identification of new Cezanne substrates will provide deeper insights into the roles of Lys11-linkages in vivo. It will also be important to understand the molecular basis of Cezanne specificity, and to analyze further OTU DUBs as well as enzymes from other DUB families for specificity against Lys11-linked chains.
Discussion
We have generated defined Lys11-linked ubiquitin chains that enabled detailed structural and functional analyses of this abundant ubiquitin chain type. Our structural analysis indicates that Lys11-linked polymers are dynamic and adopt compact conformations (Fig. 4,5) that are distinct from known polyubiquitin structures1. The crystal structure may have captured one (or potentially two) compact conformation(s) of Lys11-linked diubiquitin. Solution studies and crystallographic analysis indicate that Lys29 and Lys33 reside at an interface while Ile44 of the ubiquitin hydrophobic patch is not involved.
Additional studies will be required to determine preferred domain orientations in Lys11-linked diubiquitin in solution and at the single molecule level. Furthermore, the structure of longer Lys11-linked ubiquitin polymers is not clear, although the crystal packing suggests models for higher order oligomers. Recent attempts using computational modeling to predict diubiquitin conformations also found that Lys11-linked chains are likely compact, yet the proposed model is different, as it relied on the assumption that the hydrophobic patch including Ile44 is an integral part of a symmetric interface37.
It is important to analyze how the structural features of Lys11-linkages can be recognized, and to identify and understand UBDs that bind this chain type specifically. With a partially exposed hydrophobic patch, interaction of Lys11-linked chains with UBDs is likely to result in new interaction modes. In particular, proteins with tandem UBDs may be well suited to interact with adjacent exposed Ile44 patches in Lys11-linked polyubiquitin. Alternatively, novel classes of UBDs may selectively recognize Lys11-linked polyubiquitin. Structures of Lys11-linked diubiquitin bound to UBDs will give further insights into the conformational variability of this linkage type.
UBE2S and Cezanne – specific regulators of Lys11-linkages
The intrinsic specificity of UBE2S to assemble Lys11-linked ubiquitin chains had been noted 10,12,14, and we confirm and extend these findings. The conserved C-terminal Lys-rich tail is a unique feature of UBE2S not found in other E2 enzymes. We identify this Lys-rich tail to be a target of cis autoubiquitination, and removal of this sequence does not alter Lys11-specificity of UBE2S. To achieve its specificity, UBE2S must position the acceptor ubiquitin towards the catalytic center in a defined orientation that favors Lys11-linkage assembly. However, when the local concentration of ubiquitin is increased near the active site, e.g. in the UBE2S UBD fusion protein, UBE2S becomes less specific and also assembles Lys63-linkages. This indicates that subtle differences in affinity regulate the Lys11-preference of UBE2S. Furthermore, UBE2S lacks key residues predicted to interact with RING E3 ligases, and UBE2S does not interact with RING domain proteins in recent yeast-2-hybrid analyses38,39. Currently, the only known E3 machinery to utilize UBE2S is the APC/C, in which UBE2S acts as a Lys11 chain elongating enzyme to extend short ubiquitin polymers assembled by UBE2C or UBE2D/UbcH5 on APC/C substrates12-14. UBE2S is overexpressed in many cancers, which may be consistent with its role in cell cycle regulation (http://www.proteinatlas.org/).
The marked preference of the Cezanne OTU domain for Lys11-linkages is equally striking. Which features of a Lys11-linked chain does Cezanne recognize? Like other DUBs, OTU domains interact extensively with a distal ubiquitin26 and these interactions would presumably disrupt a Lys11-interface. It is hence more likely that Cezanne recognizes the sequence context around Lys11, or local structural features in a proximal ubiquitin in a specific manner26. To fully understand Cezanne linkage-selectivity, complex structures with Lys11-linked diubiquitin need to be obtained. Specific DUBs have recently been used to distinguish linkage types of ubiquitinated proteins40, and therefore, Cezanne may expand the toolkit currently available for such studies.
Independent regulation of proteasomal degradation
It is now accepted that ubiquitin chains can mediate degradative but also important non-degradative functions 2, and this has partly been explained by structural differences between Lys48- and linear/Lys63-linked ubiquitin chains, resulting in differential recognition of these chain types by UBDs and DUBs 33.
It is however not clear, whether the different degradative chain types, such as Lys48- and Lys11-linked polymers, are redundant, or independently regulated. Our data argue against redundancy in proteasomal degradation. We show that Lys11-linked chains are structurally distinct from Lys48-linked chains, and hence represent non-identical entities within cells. Furthermore, we identify enzymes to selectively assemble and disassemble Lys11-linkages. Hence, it appears that (at least) two structurally distinct proteasomal degradation signals exist in cells, which can be independently regulated. Key players in such independent regulation are linkage-specific DUBs. Our data show that several DUBs would be unable to remove ubiquitin chains from Lys11-modified proteasomal substrates, e.g. those that are degraded during the cell cycle, while enzymes like Cezanne may specifically target Lys11-modified substrates. Alternatively, proteasomal shuttling factors, the ubiquitin receptors of the proteasome itself, and proteasome bound DUBs may have intrinsic specificity for particular degradative chain types, actively regulating residence times of substrates at the proteasome. This could create a hierarchy of proteasomal degradation with the outcome that some degradation signals are more efficient than others.
What are the advantages for the cell to maintain independent proteasomal degradation signals? A hierarchy in proteasomal degradation would be attractive in order to respond to stimuli. For example, in times of stress or during the cell cycle, degradation of signaling proteins may need to be prioritized, which could be mediated by dedicated, more efficient linkage types.
Often, E3 ubiquitin ligases are found in complex with deubiquitinases, and in many cases cross-regulation of these opposing activities are required to balance these systems 26. However, in order to degrade such machineries, different types of ubiquitin linkages could be utilized that are not recognized by the ligase-associated DUBs, and hence would result in non-reversible degradation signals. Using different degradative chain types may represent an attractive mechanism to degrade degradation machineries.
Our data highlight once again that differently linked ubiquitin chains must be regarded as independent posttranslational modifications, as they are specifically generated, recognized, and reversed by defined enzymes and mechanisms.
Supplementary Material
Acknowledgements
We would like to thank Elaine Stephens, Sew Peak-Chew and Farida Begum for mass-spectroscopic analysis, Mark Allen (MRC CPE) for help with isotope labeling, Tom Nicholson (ENZO LifeSciences) for providing ubiquitin mutants, Philip Cohen (Dundee), Sylvie Urbe, Michael Clague (Liverpool), Keith D. Wilkinson (Emory), Sokol Todi (Ann Arbor) and Benedikt Kessler (Oxford) for constructs, Takeshi Tenno (Kobe) and Masahiro Shirakawa (Kyoto) for kindly providing NMR data for Lys48 and Lys63-linked diubiquitin and members of the Komander lab, especially Yogesh Kulathu, Yu Ye, Masato Akutsu, Satpal Virdee and Arsalan Eslambolchi for providing reagents and many discussions. Anja Bremm is a MRC Career Development Fellow.
Appendix
Experimental Procedures
Autoubiquitination assays
Analytical assays were carried out in 30 μl reactions at 37 °C containing 250 nM ubiquitin-activating enzyme (E1), 2.8 μM (UBE2S) or 3.4 μM (UBE2C) ubiquitin conjugating enzyme (E2), 19.5 μM ubiquitin, 10 mM ATP, 40 mM Tris (pH 7.5), 10 mM MgCl2 and 0.6 mM DTT. After 1 h the reaction was stopped by addition of 10 μl 4 × LDS sample buffer (Invitrogen), resolved by SDS-PAGE on 4-12 % gradient gels and subjected to Western blot analysis using rabbit polyclonal anti-ubiquitin antibody (Upstate).
Lys11-linked diubiquitin synthesis
Large-scale ubiquitin chain assembly was carried out in 1 ml reactions. Ubiquitin dimers were synthesized by incubating 250 nM E1 enzyme, 4.8 μM UBE2SΔC, 1.5 mM ubiquitin, 10 mM ATP, 40 mM Tris (pH 7.5), 10 mM MgCl2 and 0.6 mM DTT at 37 °C overnight. Subsequently, 50 mM DTT was added to the reaction before further dilution with 14 ml of 50 mM ammonium acetate (pH 4.5) to precipitate enzymes. The solution was filtered through a 0.2 μm syringe filter and Lys11-linked diubiquitin was purified by cation exchange using a MonoS column (GE Healthcare) and concentrated to 5 mg ml−1.
Generation of UBE2S – UBP fusion
The engineered UBE2S-UBD fusion protein made use of a naturally occurring NcoI restriction site in the human UBE2S sequence just before the Lys-rich tail (residue 196). The IsoT ZnF UBP domain (residues 173-289) was amplified from cDNA with primers UBP-FW 5′- CCAAGGTTCCATGGTACGGCAGGTGTCTAA-GCATGCC-3′ and UBP-RV 5′-GCCTAGCGGCCGCTTATGTCTTCTGCATCTTCAGCATGTCGATG-3′). The amplified fragment was ligated into the NcoI/NotI restriction sites present in the pGEX6P1-UBE2S expression plasmid.
Lys11-linked tetraubiquitin synthesis
Ubiquitin tetramers were synthesized by incubating 250 nM E1 enzyme, 4.8 μM UBE2S-UBD, 2.9 mM ubiquitin, 400 nM AMSH, 10 mM ATP, 40 mM Tris (pH 7.5), 10 mM MgCl2 and 0.6 mM DTT in a 1 ml reaction at 37 °C. After 1.5 h 400 nM AMSH was added again to counteract the formation of Lys63-linked ubiquitin chains. After 3 h, 50 mM DTT was added to the reaction before further dilution with 14 ml of 50 mM ammonium acetate (pH 4.5). Lys11-linked di-, tri- and tetraubiquitin were purified by cation exchange using a MonoS column (GE Healthcare) (Fig. 3). It was also possible to use Lys11-linked diubiquitin as input material to obtain tetraubiquitin. We encountered problems with loss-of-specificity in UBE2S enzymes, and careful analysis by MS/MS of the produced chains, and specificity validation using ubiquitin Lys-only mutants was required for each individual batch of UBE2S.
NMR analysis
13C,15N-labeled ubiquitin K63R or K11R mutant was expressed from a pET17b plasmid in Rosetta2 (DE3) pLacI cells. A 100 ml overnight culture grown in LB medium was pelleted and resuspended in modified K-MOPS minimal media42, lacking nitrogen and carbon sources. This was used to inoculate 3 L modified K-MOPS media supplemented with 13C glucose and 15N ammonium chloride. Protein expression was induced after 16 h growth at 30 °C with 0.4 mM IPTG, and cells were harvested after a further 4 h. Mutant ubiquitin was purified according to43. To obtain only distally labeled Lys11-linked diubiquitin, wild-type ubiquitin was mixed with 13C,15N-labeled ubiquitin K11R mutant in a 1:2 ration in a chain assembly reaction. Labeled samples were dialyzed at 100 μM concentration against phosphate buffered saline (150 mM NaCl, 18 mM Na2HPO4, 7 mM NaH2PO4 × 2H2O, pH 7.2) or against 150 mM NaCl, 50 mM NH4Ac (pH 3.5) in 3 kDa cut-off Slide-A-Lyzer dialysis cassettes (Thermo Scientific). NMR experiments were acquired on Bruker DRX600MHz and AV2+ 700MHz spectrometers equipped with cryogenic triple resonance TCI probes at 298K. Data were processed in Topspin 2.1 (Bruker, Karlsruhe) and analyzed in Sparky (Goddard & Kneller, UCSF). Weighted chemical shift perturbations were measured in 15N fast HSQC experiments44 and defined as ((Δ1H)2)0.5 + ((Δ15N/5)2)0.5 [ppm]45. Standard triple resonance experiments (HNCACB, CBCA(CO)NH and HNCA) were used to assign all mono- and di-ubiquitin K63R or K11R species and confirm the identity of shifted and doubled resonances (see Supplementary data).
Crystallization and structure determination of Lys11-linked diubiquitin
Crystals of Lys11-linked diubiquitin formed from 3 M NaCl and 0.1 M citric acid (pH 3.5) after 1 day. Before freezing in liquid nitrogen, crystals were soaked in mother liquor containing 15 % ethylene glycol. Diffraction data were collected to 2.2 Å on ESRF beamline ID14-EH2 (Grenoble). Pointless46 identified a orthorhombic space group (P2221), and molecular replacement in MolRep47 with monoubiquitin (pdb-id 1ubq, 48) as a search model identified 12 ubiquitin molecules. Subsequent rounds of model building in COOT49, and refinement in PHENIX50 using NCS, simulated annealing (initially) and TLS B-factor refinement at later stages, were performed. Restraints for the isopeptide linkage were generated using phenix.elbow. Data collection and refinement statistics can be found in Table 1.
In vitro deubiquitination assays
Polyubiquitinated UBE2S was obtained by incubating 250 nM E1 enzyme, 4.2 μM UBE2S, 1.5 mM ubiquitin, 10 mM ATP, 40 mM Tris (pH 7.5), 10 mM MgCl2 and 0.6 mM DTT in a 1 ml reaction at 37 °C overnight. Polyubiquitinated TRAF6 was assembles by incubating 250 nM E1 enzyme, 3 μM GST-TRAF61-285, 160 μM UBE2N/Ubc13, 160 μM UBE2V1/Uev1a, 10 mM ATP, 40 mM Tris (pH 7.5), 10 mM MgCl2 and 0.6 mM DTT in a 1 ml reaction at 37 °C for 3 h. UBE2V1 was added to the reaction after 1 h. Separation of free ubiquitin polymers from polyubiquitinated UBE2S or TRAF6 was achieved by gel filtration chromatography using a Superdex75 prep grade column (GE Healthcare).
Deubiquitination assays were performed according to33. Essentially, DUBs were diluted to 0.2 mg ml−1 in 150 mM NaCl, 25 mM Tris (pH 7.5) and 10 mM DTT and activated at 23 °C for 10 min. Subsequently, 10 μl of diluted enzyme were mixed with 5 μg diubiquitin or polyubiquitinated UBE2S/TRAF6 and 3 μl of 10 × DUB buffer (500 mM NaCl, 500 mM Tris (pH 7.5) and 50 mM DTT) in a 30 μl reaction. Ubiquitin cleavage was detected either by silver staining or by Western analysis using polyclonal anti-ubiquitin antibody (Upstate).
Further details on molecular biology, protein purification and mass-spectroscopy can be found in Supplementary Experimental Procedures.
Experimental Procedures References
- 42.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–47. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 44.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–8. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 45.Hajduk PJ, et al. NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J Med Chem. 1997;40:3144–50. doi: 10.1021/jm9703404. [DOI] [PubMed] [Google Scholar]
- 46.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 47.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–4. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 48.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Footnotes
Accession codes
Coordinate and structure factors have been deposited with the protein data bank accession number 2xew.
References
- 1.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–53. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 6.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–50. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 7.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–53. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–42. [PubMed] [Google Scholar]
- 10.Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271:2823–31. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- 11.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–65. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williamson A, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnett MJ, et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–9. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–39. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–16. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 18.Reyes-Turcu FE, et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 19.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–92. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varadan R, et al. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–63. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 21.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–47. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 22.Tenno T, et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–75. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang N, et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–90. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–98. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–14. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 26.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 27.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009 doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komander D, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 29.Tran H, Hamada F, Schwarz-Romond T, Bienz M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22:528–42. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- 31.Edelmann MJ, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–90. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 32.Winborn BJ, et al. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–43. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–73. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–41. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 35.Enesa K, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283:7036–45. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 36.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 37.Fushman D, Walker O. Exploring the Linkage Dependence of Polyubiquitin Conformations Using Molecular Modeling. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Wijk SJ, et al. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markson G, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009 doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.