Abstract
The reproducibility of serial measurements using a volumetric proton MR Spectroscopic Imaging (MRSI) acquisition implemented at 3 Tesla and with lipid suppression by inversion-recovery has been evaluated. Data were acquired from two subjects at five time points, and processed using fully-automated procedures that included rigid registration between studies. This data were analyzed to determine coefficients of variance (COV) for each metabolite and for metabolite ratio images based on an individual voxel analysis, as well as for average and grey-matter and white-matter values from atlas-defined brain regions. The volumetric MRSI acquisition was found to obtain data of sufficient quality for analysis over 70±6% of the total brain volume, and spatial distributions of the resultant COV values were found to reflect the known distributions of susceptibility-induced magnetic field inhomogeneity. Median values of the resultant voxel-based COVs were 6.2%, 7.2%, and 9.7% for N-acetylaspartate, creatine, and choline respectively. The corresponding mean values obtained following averaging over lobar-scale brain regions within the cerebrum were 3.5%, 3.7%, and 5.2%. These results indicate that longitudinal volumetric MRSI studies with post-acquisition registration can provide an intra-subject reproducibility for voxel-based analyses that is comparable to previously-reported single-voxel MRS measurements, while additionally enabling increased sensitivity by averaging over larger tissue volumes.
Keywords: Brain, Proton MR Spectroscopic Imaging, Intra-subject reproducibility
INTRODUCTION
Detection of metabolic alterations by Magnetic Resonance (MR) Spectroscopy provides a valuable adjunct to diagnostic studies using structural MRI methods. When implemented using volumetric proton MR Spectroscopic Imaging (MRSI) of the brain, these methods enable metabolic mapping of multiple brain regions with relatively high spatial resolution. One application of these methods is for non-invasive monitoring of temporal changes of metabolism with disease or treatment, and examples of clinical studies using longitudinal MRSI measurements in the brain include monitoring of brain tumor treatment (1), progression of disease or injury (2,3), and evaluation of new pharmaceutical treatments (4-6). While many of these previous longitudinal studies made use of manual identification of regions of interest in each of the acquired MRSI data, it is also possible to align all MRI and MRSI series between studies taken at different times, thereby making automated MRSI analysis approaches possible for detection of metabolic changes over time. Methods for achieving the inter-study alignment have included post-acquisition registration (1), use of a stereotactic head frame for repositioning and immobilization (7), and a priori image registration (8).
Of primary importance in longitudinal imaging studies is the reproducibility of the measurement, and several studies have reported variances of metabolite measures obtained from repeated MRSI acquisitions in the same subject (7-13). These previous reports have described a range of acquisition parameters and signal calibration methods, with the majority using PRESS-volume selected MRSI implemented at 1.5 T. A summary of the findings from these previous reports clearly identifies issues of concern that are common to similar studies using single-voxel measurements, namely signal-to-noise ratio (SNR) and quality and consistency in the data acquisition afforded by automated set-up approaches as being of importance (14,15). Additional considerations include instrumental variability, ability of data analysis methods to tolerate signal artifacts, and accuracy of methods for correction of partial volume signal loss (16). Issues that are more specific to MRSI include that the measurement variances will also be spatially variant (17) and that data may be integrated over larger regions and separately reported for grey-matter (GM) and white-matter (WM), requiring that a reporting of results include maximum variances as well as mean values over larger brain regions.
This study reports on the intra-subject reproducibility of a whole-brain MRSI approach, similar to that reported by Gu et al. (13). By using inversion-recovery based lipid suppression the presented MRSI method avoids the requirement for volume-preselection, and provides maximum spatial coverage. This is implemented at 3T and with phased-array detection, which represents improved sensitivity over previous reports done at 1.5T while still being valid for instrumentation that is widely available for diagnostic studies. An additional feature of the methodology used was a relatively high degree of automation applied both at the time of data acquisition, which used standard setup procedures, and data processing, which was fully automated using the MIDAS (Metabolic Imaging and Data Analysis System) package (17,18). Results are presented from both a voxel-based measurement as well as averaged over lobar-scale tissue volumes.
METHODS
MRSI and MRI studies were carried out using healthy subjects under procedures approved by the institutional human subjects research review committee and with informed consent obtained from all participants. Two subjects, 1 male 26 year old and 1 female 28 year old, were studied at five separate occasions at approximately weekly intervals.
MRI and proton MRSI studies of the brain were carried out at 3 Tesla (Siemens Trio), with an eight-channel phased-array head coil. MRSI data were acquired using a volumetric spinecho acquisition that used two-dimensional phase encoding and echo-planar readout, frequencyselective water suppression, and lipid inversion nulling. The acquisition used a 73° excitation, TE = 70 ms, TR = 1710 ms, TI = 198 ms, and a total acquisition time of 26 minutes. After correction for oversampling in the readout spatial and spectral dimensions the resultant image was equivalent to 50×50 voxels in-plane and 18 slices, over a field-of-view (FOV) of 280×280×180 mm, with selection of a slab of 135 mm covering the cerebrum. These spatial sampling parameters, and associated data processing, were optimized to acquire data at a higher spatial resolution than warranted from sensitivity considerations in order to maximize spatial coverage via minimization of intra-voxel magnetic-susceptibility induced spectral distortions (19). Following correction of B0 inhomogeneity frequency shifts and spatial smoothing the effective voxel volume was approximately 1 mL. The MRSI acquisition included a second dataset that was obtained in an interleaved manner without water suppression and using 20° excitation and gradient-echo observation. This data provided a water reference signal with identical spatial parameters as the metabolite MRSI, and which was used for several processing functions to enhance the metabolite image reconstruction and analysis. Each study included a T1-weighted MRI (MPRAGE, Magnetization Prepared Rapid Gradient Echo) at 1 mm isotropic resolution (TR/TE/TI=2150/4.4/1100 ms, flip angle 8°). The total time required for the overall imaging protocol was approximately 40 min.
Reconstructions of the metabolite and water-reference images were carried out in a fully-automated manner using the MIDAS package (17,18). Parametric spectral analysis (20) was used to determine volumetric maps of N-acetylaspartate (NAA), total creatine (Cre), and total choline (Cho), and all parameter maps were interpolated to 64×64×32 points. The processing included k-space extrapolation (21) to reduce ringing artifacts from subcutaneous lipids and metabolite signal intensity normalization using tissue water as an internal reference, which was derived from the water-reference MRSI signal. Rigid registration was applied to all image series, from all studies in a single subject, to align them with the T1-weighted MRI obtained in the first study. The processing also included calculation of the relative tissue volume contribution to each MRSI voxel. This was determined by applying a tissue segmentation procedure (22,23) to the T1-weighted MRI to map grey-matter white-matter, and CSF, followed by a resampling and convolution by the MRSI spatial response function to coincide with the MRSI voxel volume and location (18).
Using all studies from each subject, a voxel-based analysis of the metabolite and metabolite ratio images was applied to determine the mean and standard deviation at each location. For the individual metabolite images, this evaluation was applied twice, without and with correction for CSF volume contribution, which was applied as Met’=Met/(1-fcsf) for 0 ≤ fcsf ≤ 0.3, where Met is the uncorrected metabolite value, fcsf is the fraction of CSF in the MRSI voxel. Voxels with a linewidth greater than 13 Hz, or a voxel tissue fraction (total of grey-matter and white-matter) of less than 70% were excluded from the calculation. A second analysis with a linewidth threshold of 15 Hz was also used to evaluate the relative contribution to the resultant variability. Results were computed only if four or more studies had a valid result at that same voxel location. Maps of the coefficient of variation (COV), in percent, at each voxel were then generated, and these results from all subjects were then combined to determine the distributions of COVs. This analysis was also applied to the tissue segmentation images and the water image created from the water-reference MRSI data.
Data processing and analysis functions within the MIDAS package include multi-voxel analysis based on lobar level anatomic regions. For this analysis, all signal-normalized metabolite images, metabolite ratio images, spectral fitting parameters, and MRSI-resolution tissue fraction maps, were spatially transformed to a common spatial reference frame, for which the BrainWeb simulated MRI (24) was used as the target. This in turn was associated with a brain atlas that mapped each of the left and right frontal, temporal, occipital, and parietal lobes, and the cerebellum. For this registration procedure, the T1-weighted MRI from the first study was used to determine the non-linear spatial transformation parameters to the reference MRI (25,26), and this spatial transformation applied to all MRSI-resolution images, with interpolation to 2 mm isotropic voxels. Following spatial transformation, all voxels within each of the atlas-defined lobar scale regions were selected and a regression analysis carried out of the CSF-corrected metabolite values against the relative white-matter voxel fraction, to obtain the metabolite measures corresponding to 100% GM and 100% WM.
MRI and spectroscopic results were reviewed using display modules within the MIDAS package as well as the MRIcro program (www.mricro.com).
RESULTS
In Figure 1 are shown example images for NAA, with selected axial slices from a single study; the mean-value image generated by summation over five studies; and the resultant COV image, which has been displayed with an image scale between 0% and 10%. Note that the mean-value and COV images cover a smaller spatial extent due to the action of the voxel exclusion tests. This and the other metabolite COV images showed lower values in central white-matter, which is indicative of the relatively good magnetic field inhomogeneity seen in these regions. In Figure 2 are shown examples of spectra obtained from two voxel locations with each repeated measurement. Spectra were selected from a central WM region, corresponding to an 8-Hz linewidth, and in the region of the caudate nucleus, corresponding to a linewidth of 13 Hz. This result indicates the overall quality of this data, with variability increasing in areas known to be more challenging for MRS measurements, and including both changes of linewidth and spectral baseline.
Figure 1.
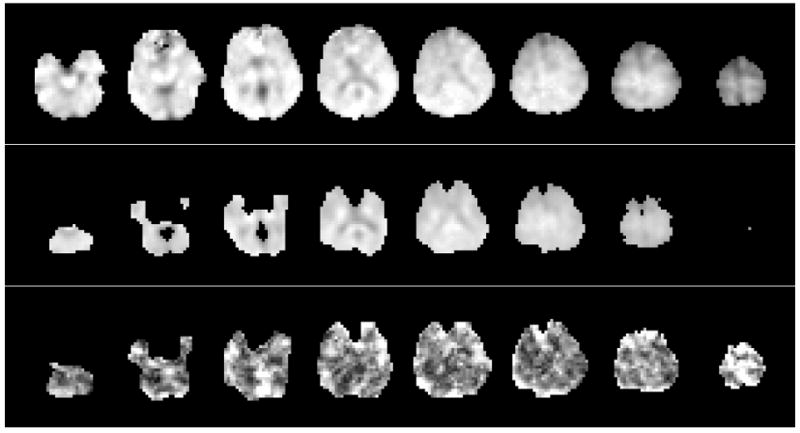
Example images for NAA, showing (a) the metabolite image for a single subject, with every 2nd axial slice shown at 11 mm spacing; (b) the mean-value NAA image calculated from five repeated studies with voxel exclusion criteria as described in the text; and (c) the COV image, shown with a range from 0% (black) to 10% (white).
Figure 2.
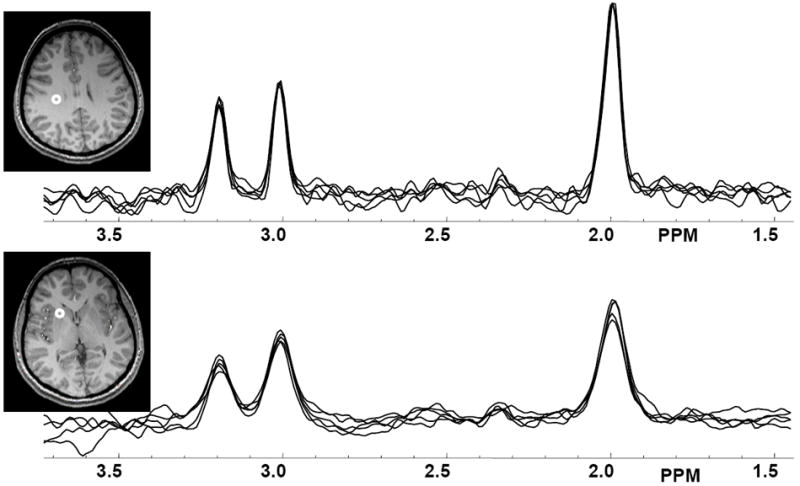
Example spectra from five repeated MRSI measurements, selected from central white matter (top) and in the region of the caudate nucleus (bottom), with voxel locations indicated on the MRI.
Relative to the total brain volume, as determined by the sum of the GM and WM maps derived from the MRI-based tissue segmentation, the fraction of the brain volume that returned fitted spectral results with ≤13 Hz linewidth and averaged over all studies was 70±6%. Following application of the voxel exclusion based on the requirement for a minimum of four studies having a valid result at a given voxel in order to compute the COV, the average brain volume fraction was reduced slightly to 67%.
In Figure 3 are shown box-and-whisker plots that summarize the COV distributions for each of the metabolite images with a 13 Hz maximum spectral linewidth threshold. Also shown are the results for the MRSI-resolution tissue segmentations, which represent the variation over the repeated studies for the tissue fractional volume calculated from the MRI-derived tissue segmentation, for which all voxels within the brain were selected. To the right of Figure 3 are shown COV distributions for the spectral linewidth (LW) and the integrated water-reference MRSI signal, for which all voxels that were passed to the spectral fitting routine were included with this selection being based on a <16 Hz threshold applied to the water-reference MRSI data.
Figure 3.
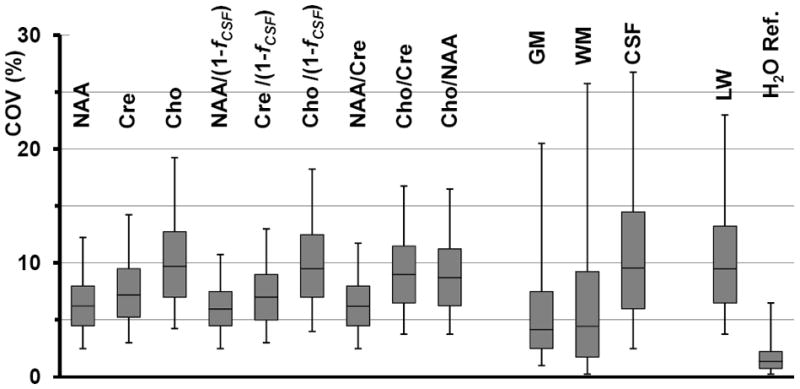
COV values for the voxel-based analysis of: each of the individual metabolites normalized to tissue water; the CSF-corrected values; the metabolite ratio images; the results of tissue segmentation at the MRSI resolution; the linewidth (LW); and the signal from the waterreference MRSI (H2ORef.). The box-and-whisker plots show from bottom to top the 5th percentile; 25th percentile; median; 75th percentile; and 95th percentile of the number of voxels analyzed.
These results indicate that the majority of voxels within the NAA, Cre, and NAA/Cre images are obtained with COVs of under 10% (median value of 6.2%, 7.2%, and 6.2% respectively), while variances are higher for the Cho image (median 9.7%) and metabolite ratios that include Cho. It can also be seen that application of the CSF correction makes little difference to the individual-metabolite COVs, which is a consequence of the fact that the correction was only applied at voxels with a relatively small CSF contribution (≤ 30%), and that the variability of the tissue segmentation results for voxels with a large tissue fraction was relatively small. Results obtained using a 15 Hz threshold for the voxel selection (not shown) resulted in a small increase in the brain volume used for the COV evaluation to 75%, a minimal change of the median values, and a 2% increase of the 95 percentile range of the COVs for NAA, Cre, and Cho. The median value for the COV of the water image was 1.4%, lending support to the use of this tissue water measurement for providing a reference signal for longitudinal studies; however, it is noted that this result may no longer apply in cases of pathology.
In Table 1 are shown the results for the lobar-scale region analyses. These show the COVs for the GM and WM metabolite values derived from the tissue regression analysis, and the average value obtained by integration over all voxels within each region. The COVs were calculated separately for the left and right sides of each brain region and the average value reported. In addition, the average value over the whole cerebrum is shown. Results indicate that the lowest COVs, of under 4%, were obtained for average values of NAA, Cre, and NAA/Cre. These values are slightly increased for the WM result of the regression analysis, and worst for the GM result, which reflects the increased uncertainties introduced by the segmentation of CSF and GM fractions; the increased spectral linewidth in these regions, as indicated by the relative LW values for GM and WM in Table 1; and the smaller number of voxels having large GM volume fractions.
TABLE 1.
Average COVs (%) derived from the lobar brain region analyses.
| Frontal | Temporal | Parietal | Occipital | Cerebellum | Cerebrum | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GM | WM | Ave | GM | WM | Ave | GM | WM | Ave | GM | WM | Ave | GM | WM | Ave | GM | WM | Ave | |
| NAA | 3.6 | 2.8 | 3.0 | 5.9 | 2.8 | 3.6 | 5.5 | 3.1 | 3.5 | 6.3 | 4.0 | 4.0 | 4.3 | 5.6 | 3.9 | 5.3 | 3.2 | 3.5 |
| Cre | 3.7 | 3.3 | 3.0 | 5.0 | 4.7 | 3.7 | 6.2 | 3.6 | 3.7 | 4.8 | 5.8 | 4.5 | 3.4 | 17.2 | 3.0 | 4.9 | 4.3 | 3.7 |
| Cho | 4.9 | 4.7 | 4.1 | 9.0 | 5.8 | 4.9 | 12.8 | 3.8 | 5.1 | 11.8 | 5.5 | 6.5 | 4.5 | 28.3 | 3.9 | 9.6 | 5.0 | 5.2 |
| NAA/Cre | 2.4 | 1.8 | 1.3 | 3.3 | 3.7 | 1.9 | 4.2 | 2.7 | 1.9 | 5.9 | 3.2 | 2.0 | 4.2 | 11.0 | 4.0 | 3.9 | 2.8 | 1.8 |
| Cho/Cre | 4.2 | 2.9 | 2.9 | 7.3 | 6.1 | 2.9 | 8.2 | 3.0 | 2.7 | 9.5 | 6.5 | 3.7 | 3.1 | 10.2 | 3.3 | 7.3 | 4.6 | 3.0 |
| Cho/NAA | 3.2 | 3.3 | 2.9 | 6.6 | 5.3 | 2.5 | 6.0 | 3.1 | 2.7 | 12.1 | 6.6 | 3.3 | 5.8 | 31.2 | 5.4 | 7.0 | 4.6 | 2.9 |
| LW | 6.3 | 3.0 | 4.5 | 5.9 | 3.1 | 2.8 | 11.8 | 4.1 | 5.8 | 7.6 | 6.1 | 5.5 | 3.9 | 5.3 | 3.0 | 7.9 | 4.1 | 4.7 |
| H2ORef | 0.4 | 0.3 | 0.3 | 0.5 | 0.4 | 0.7 | 0.5 | 0.2 | 0.7 | 1.1 | 0.7 | 1.4 | 0.5 | 1.8 | 1.0 | 0.6 | 0.4 | 0.8 |
LW = Spectral linewidth, and H2ORef = the water reference MRSI signal.
All COVs for the region-based measurements of WM values and the average metabolite results were significantly less than the corresponding median values obtained from the voxelbased analysis, illustrating the increased potential for detection of subtle metabolic alterations following integration over larger brain regions. Also indicated in Table 1 is the excellent reproducibility of the water signal, again supporting the use of this data for metabolite signal normalization.
DISCUSSION
Based on median COV values, this study indicated better performance than previous studies of intra-subject MRSI reproducibility at either 1.5T or 3T (7-13), supporting the use of the post-acquisition image registration approach in comparison to stereotactic repositioning (7) or a priori registration at the time of data acquisition (8). Probably the most important factor that impacts reproducibility is the SNR and its impact on spectral fitting. In this regard, this study has benefited from the increased sensitivity provided by measurement at 3T, while also using a shorter TE than those used for the two previous studies carried out at 3T (12,13). However, it should also be noted that this study did not consider the relative performance of short-TE MRS where additional gains of SNR are achievable, although at the expense of increased difficulties for spectral analysis that can result in a counterproductive degradation of performance (7). Although the MRSI acquisition parameters were chosen to obtain data with higher spatial resolution than optimally required from sensitivity considerations (19), it is believed that the smaller voxel size, together with the water-referenced postprocessing procedures used, ultimately benefits spectral analysis through the resultant improvement in spectral linewidth.
For an image-based analysis using the 3T acquisition methods described in this report it is evident that the magnitude of metabolic changes that may be considered significant will be spatially variant, as well as depending on the volume over which the change occurs and the appropriate spatial extent used for the analysis (27). However, to summarize the general findings, these studies indicate that when using individual voxel-based analyses, changes of NAA, Cre, or NAA/Cre on the order of 7% to 15% should be detectable between two studies in the same subject, while changes in the order of 15% to 20% may be required for Cho and ratios including Cho. By integrating results over larger volumes greater sensitivity can be obtained, with changes on the order of 5% being considered detectable for all metabolites in white-matter.
The voxel-based COV values determined in this study, which reflect an effective voxel volume of approximately 1 mL, are comparable to those determined for 8 mL single-voxel measurements at 1.5T (28,29), and although higher than values reported by Simmons et al. (14) that used a relatively high SNR measurement, the volume-integrated results from this study are comparable to the values reported by Simmons et al. The COVs from this study are also smaller than a short-TE (30 ms) single-voxel measurement at 3T (30), which likely reflects the increased difficulties of spectral analysis of short-TE 1H MRS. In consideration of the choice of technique to employ for longitudinal MRS measurements, the MRSI approach described in this report provides information over a greater spatial extent and with improved spatial resolution than single voxel measurements, although at the expense of increased acquisition time, which can detrimentally impact reproducibility due to increased susceptibility to subject movement.
Several improvements to the MRSI methodologies used in this study are possible that may further improve the reproducibility. Firstly, improved automatic shimming and optimized water suppression may potentially improve the reliability for Cho observation and for all metabolites in neocortical GM regions. Second, improved tissue segmentation methods may improve the tissue-regression analysis result for GM, for example by making use of multiple MRI image contrasts and incorporation of a priori information into the segmentation algorithm, which was not used in this study. Third, there remain applications where volume selection methods can be used instead of the lipid inversion-nulling approach used in this study, which would provide SNR improvements on the order of 38% for NAA and 49% for Cho (based on T1 values of 1.3 and 1.1 seconds respectively (31)); this approach would, however, still require the whole-brain water MRSI data to be obtained in order to apply the MRI to MRSI spatial registration procedure. Fourth, the processing and analysis methods could be tailored to the spatial scale of the target measurement. For example, if tissue regression analysis methods are to be applied to determine GM and WM metabolite values over larger brain volumes, there may be an advantage to applying heavier spatial smoothing (after correction of B0-induced spectral shifts) prior to the spectral analysis (32). Statistical methods can also be developed that would increase sensitivity for detection of smaller metabolic changes over larger brain regions (27).
Limitations of this study include the small number of subjects used; that the results only apply to 67% of the total brain volume that corresponds to regions from which spectral linewidths of ≤13 Hz were obtained; the higher likelihood of compliance from healthy volunteers to remain stationary during the MR study; and the reliance on tissue water as an internal reference for signal normalization, which may be invalid in the presence of pathology due to both changes of relaxation rates and water density. This study has also primarily considered measurements over a large volume of the brain and not made measurements in specific brain regions such as the hippocampus, since acquisition methods can be specifically optimized to improve the spectral quality in such regions. However, the reproducibility of such targeted MRSI studies, given all other experimental conditions remaining unchanged, would likely be comparable to the median values determined in this study.
CONCLUSION
This study has demonstrated that longitudinal volumetric MRSI studies implemented at 3T can achieve COV’s for a voxel-based image analysis in central white-matter of under 10%, and that by taking advantage of the multi-voxel nature of the MRSI measurement by averaging over larger regions, COVs on between 3% for NAA and 6.5% for Cho can be achieved. While this study considered normal subjects, it provides information on precision to support extension of these MRSI methods for longitudinal studies in patients.
Acknowledgments
The spatial registration component of the MIDAS package was provided by Dr. Colin Studholme.
Sponsors: This study was supported by NIH grant R01EB0822.
References
- 1.Nelson SJ, Vigneron DB, Dillon WP. Serial evaluation of patients with brain tumors using volume MRI and 3D 1H MRSI. NMR Biomed. 1999;12:123–138. doi: 10.1002/(sici)1099-1492(199905)12:3<123::aid-nbm541>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 2.Suhy J, Miller RG, Rule R, Schuff N, Licht J, Dronsky V, Gelinas D, Maudsley AA, Weiner MW. Early detection and longitudinal changes in amyotrophic lateral sclerosis by (1)H MRSI. Neurology. 2002;58:773–779. doi: 10.1212/wnl.58.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz Maniega S, Cvoro V, Chappell FM, Armitage PA, Marshall I, Bastin ME, Wardlaw JM. Changes in NAA and lactate following ischemic stroke: a serial MR spectroscopic imaging study. Neurology. 2008;71:1993–1999. doi: 10.1212/01.wnl.0000336970.85817.4a. [DOI] [PubMed] [Google Scholar]
- 4.Sajja BR, Narayana PA, Wolinsky JS, Ahn CW. Longitudinal magnetic resonance spectroscopic imaging of primary progressive multiple sclerosis patients treated with glatiramer acetate: multicenter study. Mult Scler. 2008;14:73–80. doi: 10.1177/1352458507079907. [DOI] [PubMed] [Google Scholar]
- 5.Kalra S, Cashman NR, Genge A, Arnold DL. Recovery of N-acetylaspartate in corticomotor neurons of patients with ALS after riluzole therapy. Neuroreport. 1998;9:1757–1761. doi: 10.1097/00001756-199806010-00016. [DOI] [PubMed] [Google Scholar]
- 6.Jang JH, Kwon JS, Jang DP, Moon WJ, Lee JM, Ha TH, Chung EC, Kim IY, Kim SI. A proton MRSI study of brain N-acetylaspartate level after 12 weeks of citalopram treatment in drug-naive patients with obsessive-compulsive disorder. Am J Psychiatry. 2006;163:1202–1207. doi: 10.1176/ajp.2006.163.7.1202. [DOI] [PubMed] [Google Scholar]
- 7.Langer DL, Rakaric P, Kirilova A, Jaffray DA, Damyanovich AZ. Assessment of metabolite quantitation reproducibility in serial 3D-(1)H-MR spectroscopic imaging of human brain using stereotactic repositioning. Magn Reson Med. 2007;58:666–673. doi: 10.1002/mrm.21351. [DOI] [PubMed] [Google Scholar]
- 8.Ratai EM, Hancu I, Blezek DJ, Turk KW, Halpern E, Gonzalez RG. Automatic repositioning of MRSI voxels in longitudinal studies: impact on reproducibility of metabolite concentration measurements. J Magn Reson Imaging. 2008;27:1188–1193. doi: 10.1002/jmri.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chard DT, McLean MA, Parker GJ, MacManus DG, Miller DH. Reproducibility of in vivo metabolite quantification with proton magnetic resonance spectroscopic imaging. J Magn Reson Imaging. 2002;15:219–225. doi: 10.1002/jmri.10043. [DOI] [PubMed] [Google Scholar]
- 10.Li BS, Babb JS, Soher BJ, Maudsley AA, Gonen O. Reproducibility of 3D proton spectroscopy in the human brain. Magn Reson Med. 2002;47:439–446. doi: 10.1002/mrm.10081. [DOI] [PubMed] [Google Scholar]
- 11.Tedeschi G, Bertolino A, Campbell G, Barnett AS, Duyn JH, Jacob PK, Moonen CT, Alger JR, Di Chiro G. Reproducibility of proton MR spectroscopic imaging findings. AJNR Am J Neuroradiol. 1996;17:1871–1879. [PMC free article] [PubMed] [Google Scholar]
- 12.DeVito TJ, Nicolson Y, Bureau Y, Williamson PC, Drost DJ. Reproducibility of multislice 2D proton MRSI at 3.0T. International Society for Magnetic Resonance in Medicine, Kyoto. 2004:2459. [Google Scholar]
- 13.Gu M, Kim DH, Mayer D, Sullivan EV, Pfefferbaum A, Spielman DM. Reproducibility study of whole-brain 1H spectroscopic imaging with automated quantification. Magn Reson Med. 2008;60:542–547. doi: 10.1002/mrm.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons A, Smail M, Moore E, Williams SC. Serial precision of metabolite peak area ratios and water referenced metabolite peak areas in proton MR spectroscopy of the human brain. Magn Reson Imaging. 1998;16:319–330. doi: 10.1016/s0730-725x(97)00280-4. [DOI] [PubMed] [Google Scholar]
- 15.Wardlaw JM, Signorini DF, Marshall I. Re: Serial precision of metabolite peak area ratios and water referenced metabolite peak areas in proton MR spectroscopy of the human brain Simmons et al 1996, vol 16, pages 319-330 1998. Magn Reson Imaging. 1998;17:483–487. doi: 10.1016/s0730-725x(98)00174-x. [DOI] [PubMed] [Google Scholar]
- 16.Kreis R. Issues of spectral quality in clinical 1H-magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004;17:361–381. doi: 10.1002/nbm.891. [DOI] [PubMed] [Google Scholar]
- 17.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR Spectroscopic Imaging (MRSI) Magn Reson Med. 2009;61:548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn Reson Imaging. 2003;21:113–120. doi: 10.1016/s0730-725x(02)00645-8. [DOI] [PubMed] [Google Scholar]
- 20.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: Application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 21.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med. 1996;35:678–687. doi: 10.1002/mrm.1910350509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 24.Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imag. 1998;17:463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 25.Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med Phys. 1997;24:25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau F, Maudsley AA, Ebel A, Darkazanli A, Weber P, Sivasankaran K, Yu Y, Studholme C. Evaluation of sub-voxel registration accuracy between MRI and 3D MR spectroscopy of the brain. Proc Soc Photo Opt Instrum Eng San Diego. 2005:1213–1221. [Google Scholar]
- 27.Poline JB, Worsley KJ, Evans AC, Friston KJ. Combining spatial extent and peak intensity to test for activations in functional imaging. Neuroimage. 1997;5:83–96. doi: 10.1006/nimg.1996.0248. [DOI] [PubMed] [Google Scholar]
- 28.Marshall I, Wardlaw J, Cannon J, Slattery J, Sellar RJ. Reproducibility of metabolite peak areas in 1H MRS of brain. Magn Reson Imaging. 1996;14:281–292. doi: 10.1016/0730-725x(95)02084-7. see comments. [DOI] [PubMed] [Google Scholar]
- 29.Marshall I, Wild J. A systematic study of the lactate lineshape in PRESS-localized proton spectroscopy. Magn Reson Med. 1998;40:72–78. doi: 10.1002/mrm.1910400111. [DOI] [PubMed] [Google Scholar]
- 30.Wellard RM, Briellmann RS, Jennings C, Jackson GD. Physiologic variability of single-voxel proton MR spectroscopic measurements at 3T. AJNR Am J Neuroradiol. 2005;26:585–590. [PMC free article] [PubMed] [Google Scholar]
- 31.Ethofer T, Mader I, Seeger U, Helms G, Erb M, Grodd W, Ludolph A, Klose U. Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magn Reson Med. 2003;50:1296–1301. doi: 10.1002/mrm.10640. [DOI] [PubMed] [Google Scholar]
- 32.Hanson LG, Adalsteinsson E, Pfefferbaum A, Spielman DM. Optimal voxel size for measuring global grey and white matter proton metabolite concentrations using chemical shift imaging. Magn Reson Med. 2000;44:10–18. doi: 10.1002/1522-2594(200007)44:1<10::aid-mrm3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]


