Abstract
High-fidelity surface functional group (e.g., N-hydroxysuccinimide (NHS) reactive ester) patterning is readily and reliably achieved on commercial poly(ethylene glycol) (PEG)-based polymer films already known to exhibit high performance non-fouling properties in full serum and in cell culture conditions. NHS coupling chemistry co-patterned with methoxy-capped PEG using photolithographic methods is directly spatially imaged using imaging time-of-flight secondary ion mass spectrometry (ToF-SIMS) and principal components statistical analysis. Patterned NHS surface reactive zones are clearly resolved at high sensitivity despite the complexity of the polymer matrix chemistry. ToF-SIMS imaging also reveals the presence of photo-resist residue remaining from typical photolithography processing methods. High cross-correlation between various ion-derived ToF-SIMS images is observed, providing sensitive chemical corroboration of pattern chemistry and biological reactivity in complex milieu. Surface-specific protein coupling is observed first by site-selective reaction of streptavidin with NHS patterns, followed by identical patterns of biotinylated Alexa-labeled albumin coupling. This suggests that streptavidin immobilized on the patterns remains bioactive. Fluorescently labeled full serum is shown to react selectively with NHS-reactive regions, with minimal signal from methoxy-capped regions. Insufficient serum is adsorbed under any conditions to these surfaces to support cell attachment in serum-containing media. This reflects the high intrinsic non-adsorptive nature of this chemistry. Fibroblasts attach and proliferate in serum culture only when a cell adhesion peptide (RGD) is first grafted to NHS regions on the PEG-based surfaces. Longer-term serum-based cell culture retains high cell-pattern fidelity that correlates with chemical imaging of both the NHS and RGD patterns and also lack of cell adhesion to methoxy-capped regions. Cell staining shows orientation of adherent cells within the narrow patterned areas. Cell patterns are consistently retained beyond 15 days in serum media.
Keywords: TOF-SIMS imaging, biomaterials, bio-immobilization, PEG, cell pattern
1. Introduction
Thin hydrophilic films, coatings and hydrogels are commonly exploited to provide biologically “non-fouling” surface chemistries for biomedical and biotechnology applications. Among the many chemistries reported, poly(ethylene glycol) (PEG) polymers and PEG-like materials are likely of greatest interest: this PEG biomaterials design strategy, with an extensive history, has been frequently reviewed.[1-4] Numerous surface treatment methods to produce PEG interfaces have been reported, including PEG grafting,[5-16] adsorptive chemistries,[17-22] self-assembled monolayers (SAMs),[23, 24] and plasma (e.g., “glyme”) surface treatments.[25] These methods seek the notable surface resistance to protein and cellular non-specific binding known for these systems. Importantly, despite the intense amount of work in this area, few commercial PEG surface products are available that retain the benefits known for PEG surface architectures. These surface chemistries, when scaled beyond research production, often do not exhibit the non-fouling performance required for biomedical applications in sensors, assays, and medical devices. Despite many claims, examples of robust, specifically reactive PEG-based thin films suitable and proven for commercial biomedical applications are rare.
Recently, a multi-component PEG-based cross-linked polymer surface chemistry was reported as a commercial coating formulation developed specifically for in vitro bioassay applications.[26] The thin film coating chemistry, applied with conventional, industrially accepted processing techniques, combines covalent substrate attachment, reversible functional group reactivity and cross-linking chemistries within the PEG coating matrix. Expected PEG-based bio-fouling performance was reported for over 400 industrial lots of this chemistry, including significant inhibition of protein adsorption, and microbial and mammalian cell attachment.[26, 27] Significantly, selective and reversible reactive group (e.g., N-hydroxysuccinimide (NHS), vinylsulfone, biotin, etc.) functionalization allows specific attachment of DNA, antibodies, and cell adhesion peptides (e.g., RGD). This is possible from solution-phase immobilization as well as with standard microarray printing methods that provide spatial control required for highly reproducible microarray assays and patterned immobilization.[28-31] In these cases, high specific surface attachment of various biochemistries is obtained with very low background signals characteristic of non-specific bio-fouling in biological media (e.g., serum).
Surface patterning is routinely used to immobilize bioactive molecules such as proteins, oligonucleotides and small ligands,[32-34] to localize surface reactions for bioassays and to provide desired cell and bacterial adhesion. Such patterning is exploited for biochips,[32, 35, 36] co-cultures,[37-39] tissue engineering,[40] cell-based biosensors,[41] and studies of extracellular effects on cell behavior.[42-44] Biomolecule and cell surface patterning is often achieved by microcontact printing,[29, 45] microfluidics methods,[46] or use of photoactivation or deactivation of functional groups.[47, 48] Conventional photolithographic methods have also been applied to create patterned cell substrates.[29, 49-51] Analytical methods to determine immobilization densities, spatial fidelity, compare different spatial chemical patterns or assert control or consistency of immobilization are tedious, requiring comparison of qualitative signals for immobilized chemistry (e.g., fluorescent labels or intrinsic surface spectroscopic signals) with tedious, expensive radiolabel quantitation.[52, 53]
This current study extends previous surface immobilization results reported for a new PEG-based commercial non-fouling polymer surface for applications in diagnostics and bio-immobilization. [26] We demonstrate surface patterning of this chemistry to produce large-area co-planar surfaces with selective NHS-reactivity co-existing with highly biologically inert (non-fouling) regions. As a model for controlling regional and spatial biological reactivity on a commercially scaled surface chemistry, this photolithographed patterned surface of spatially defined reactive and non-reactive polymer is immobilized with peptide cell ligands for serum-based cell cultures. Significantly, highly specific surface analytical data demonstrate the fidelity of these spatially distinct chemical surface patterning features and their correlation with biological reactivity to cultured cells on a non-adherent background. Specifically, state-of-the-art time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging data[36, 54] accurately detail the different surface chemistries spatially correlated to desired surface reactions. Since the polymer surface is highly endowed with tethered PEG, non-specific reactions of proteins, organisms and cells are well controlled.[26] Photoresist-based patterning allows spatial control of NHS chemistry in the PEG matrix to permit spatial control of immobilization and cell adhesion in serum-containing media. Previous surface analytical results show that X-ray photoelectron spectroscopy (XPS) does not have sufficient molecular specificity to differentiate between covalently attached NHS groups and physically adsorbed but hydrolyzed NHS.[54] However, ToF-SIMS, with its enhanced molecular specificity, surface selectivity, and higher spatial resolution over XPS,[36, 55] was very useful in providing semi-quantitative analysis of NHS-grated PEG surfaces.[54] ToF-SIMS imaging was therefore used here in tandem with XPS, fluorescence microscopy, and biological assays to characterize patterned commercial Optichem® PEG surfaces.
2. Results and Discussion
2-1. XPS and ToF-SIMS Analysis of PEG-based hydrogel (Optichem®) surfaces
2-1-1. XPS analysis of unpatterned Optichem® polymer surfaces
Unpatterned Optichem® surfaces were analyzed (1) as received (“fresh”) and (2) after deliberate NHS hydrolysis (immersed in purified water overnight). The XPS-measured nitrogen concentrations for the fresh and hydrolyzed polymer surfaces were 1.3 ± 0.1 and 1.0 ± 0.2 atomic percent, respectively. The detected XPS nitrogen signals are attributed to the combination of several known nitrogen species within the polymer chemistry.[26, 36] The relatively small decrease in XPS-measured nitrogen upon NHS hydrolysis indicates that most nitrogen species reside within the polymer matrix, not the NHS terminal groups.
2-1-2. ToF-SIMS analysis of unpatterned OptiChem® surfaces
Two commercial Optichem® polymer coatings on glass slides were analyzed: a non-fouling, non-reactive methoxy-PEG (MeO-capped) Optichem® where NHS groups were quenched by exposure to 2-methoxyethylamine, and an amine-reactive NHS-capped Optichem® coating. Prior to ToF-SIMS analysis of patterned Optichem® surfaces, unpatterned NHS-activated, hydrolyzed and MeO-capped surfaces were examined to determine the characteristic molecular fragments for each surface species. The NHS-activated and hydrolyzed surfaces were easily separated by principal component analysis (PCA) of the negative secondary ion spectra (manuscript in preparation). The characteristic positive and negative mass fragments from NHS are consistent with the species previously identified from NHS-containing self-assembled PEG monolayers.[11, 54] In particular, in the negative secondary ion spectra, key NHS fragments are observed at m/z 98 and 114, and key organic acid fragments resulting from hydrolysis are observed at m/z 43 and 58 (see Table S1 in Supplementary Information). We note that m/z peaks at 43 and 58 are also detected from the NHS-capped surfaces, which are produced from the ester linkage fragments that attach the NHS to the PEG chains. However, the m/z 98 and 114 fragment peak intensities relative to the 43 and 58 fragments are significantly higher on the NHS-activated surfaces. Thus, as proposed previously,[54] a multivariate peak ratio of NHS-related peaks to the carboxylic acid/ester species fragments can be used to monitor the relative surface concentration of NHS groups.
2-1-3. ToF-SIMS imaging of patterned OptiChem® surfaces
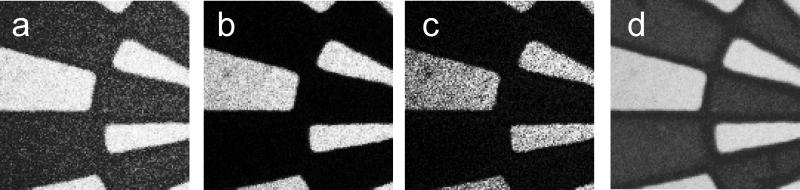
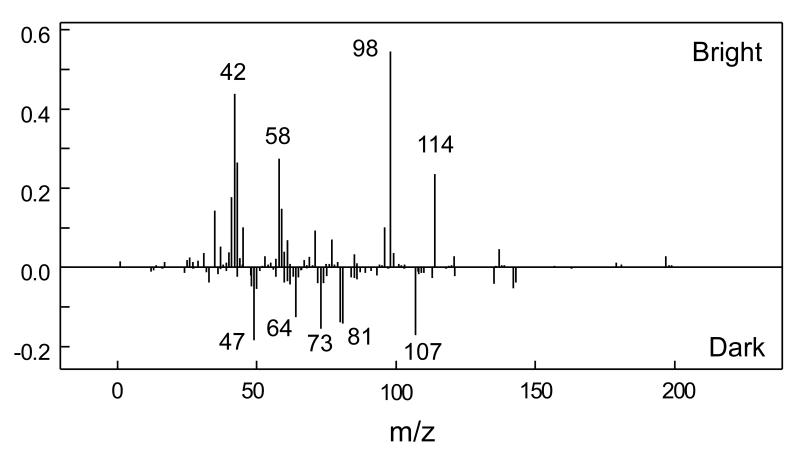
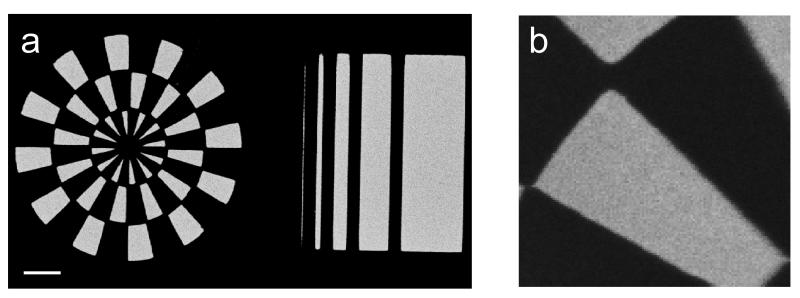
Figure S1 (see Supplementary Information) shows the intended surface chemistry pattern prepared as a photomask on a transparent polymer sheet (printed at 5080 DPI using a commercial process). The diameter of the patterned wheel mask is 3 mm. The narrowest bar feature width is 6.35 μm, increasing by a factor of 2 moving left to right across the striped pattern in Figure S1. NHS-patterned polymer Optichem®-coated substrates were prepared by conventional mask photolithography using this pattern (see Scheme S1 in Supplementary Information). Imaging of selected 500 μm × 500 μm regions of patterned surfaces using the ToF-SIMS Bi3+ ion source and their fragment pattern analysis by PCA methods generated surface images. (All ToF-SIMS images shown in this study are 500 μm × 500 μm.) Figures 1a, b and c depict the negative ion images of m/z 42, 98 and 114, characteristic of the NHS molecular fragment. This supports successful NHS patterning at least at μm resolution on the OptiChem® surface.[54] The PC-1 scores map in Figure 1d derived from the negative secondary ion images between m/z 1 and 200 in combination with the PC-1 loading plot showing raw ion data (Fig. 2), also confirmed successful NHS patterning; it also provides additional information about the presence of other species in the bright and dark regions of the image. Bright regions in Figure 1d correspond to the NHS patterns, as the characteristic NHS fragments at m/z 98 and 114 have high positive loadings in Figure 2. However, negatively loaded peaks in Figure 2 (i.e., m/z 107) map to the dark regions in Figure 1d and do not correlate with ToF-SIMS peaks observed from MeO-capped unpatterned control. This can be explained further from an additional PC-3 surface image (see Figure S2 in Supplementary Information) that exhibits 3 actual regions of distinct chemistry on the patterned surface, namely; i) NHS-modified regions; ii) MeO-capped regions; and iii) a narrow interfacial region between these regions (i) and (ii). The PCA loadings plot for PC-3 (data not shown) indicates that the major negative ion mass fragment contributing to the dark region at the pattern edge (barely observable in Fig. 1d but shown in Supplementary Information Figure S2) is m/z 107. Separate analysis of the photoresist coating as a control showed that this peak is attributed to a narrow zone of photoresist chemistry (Microposit (Shipley) SPR-220). Full details of this photoresist ToF-SIMS surface analysis will be presented elsewhere (manuscript in preparation).
Figure 1.
a-c: Raw ToF-SIMS negative ion images (each 500 μm × 500 μm) for NHS- and MeO-capped patterned surfaces at m/z 42, 98 and 114, respectively, where bright regions correspond to each fragment map. Image d is the PC-1 scores map of negative ion ToF-SIMS spectra derived from principal component analysis of raw ion images between m/z 1 and 200 (see Figure 2) where bright regions correspond to NHS-correlated ion fragments and dark regions correspond to ions correlated with MeO-capped chemistry.
Figure 2.
PCA-derived loadings plot for PC-1 from ToF-SIMS image data on patterned NHS surfaces. Peaks with positive loadings correspond to bright NHS-modified regions in the Figure 1d image. Peaks with negative loadings correspond to dark MeO-capped surface regions in Figure 1d.
2-2. Chemical imaging of protein patterns from bulk surface immobilization
2-2-1. Streptavidin surface immobilization and coupling with biotinylated bovine serum albumin (BSA)
This polymer surface is known to effectively resist non-specific protein adsorption after reactive tethering of protein to its surface.[26] The photolithographic process utilized in this work protects NHS-active regions using photoresist while allowing specific methoxylation (chemical inactivation) of unprotected regions. This ultimately results in a spatially resolved pattern clearly separating MeO-capped (non-fouling) and NHS amine-reactive regions. Figure 3a shows fluorescence images of a patterned surface treated with solution-phase streptavidin and then exposed to Alexa555-labeled biotinylated BSA; images clearly demonstrate that streptavidin was bound specifically to the NHS regions. However, some pattern deviations are evident: the “aperture artifact” at the pattern’s center and absence of 3 wheel ‘spokes’ suggest over-development during the photomasked patterning process. The photomask shows a smaller aperture central to the wheel pattern; longer photo-development enlarges this aperture with further loss of the adjacent narrowest striped bars. Images also show clear interfaces of distinct chemistry between NHS and MeO patterned surfaces as evidenced in Figures 1 and 2. Photolithography creates clear separation of MeO-capped and NHS regions protected by photoresist during the methoxylation process. ToF-SIMS imaging was used to analyze immobilized proteins in surface-templated patterns. Figure 3b shows the positive PC-1 scores image from the ToF-SIMS data for the streptavidin-immobilized surface. The corresponding mass fragments from amino acids in the protein (m/z 110, 120, 130, 136, 159 and 170) listed in Table S2 (Supplementary Information) observed in the PC-1 loadings plots are consistent with previous SIMS studies of proteins (see Figure S3 in Supplementary Information).[56-59] The fact that the amino acid fragments were only detected in the NHS surface regions indicates that streptavidin is immobilized selectively in those regions by bulk reaction and that biotinylated BSA interacts specifically with patterned streptavidin.
Figure 3.
Chemical imaging of protein-patterned surface chemistry. (a) Fluorescence image of a patterned surface (wheel feature is 3mm diameter) treated with solution phase streptavidin and then exposed to biotinylated BSA labeled with Alexa555. Scale bar: 500 μm. (b) Correlated PC-1 scores map of ToF-SIMS positive image from PCA treatment of the streptavidin-immobilized surface image data (image: 500 μm × 500 μm, streptavidin fragments mapped to lighter regions).
Figure 4 compares specific and non-specific protein uptake onto patterned and unpatterned surface chemistries compared to bare glass adsorptive controls. Figure 4a shows surface normalized fluorescence intensities for Alexa dye-labeled biotin-BSA binding streptavidin immobilized by bulk exposure from solution to unpatterned NHS and MeO-capped (control) surfaces, and patterned NHS and MeO-capped regions. Untreated bare glass slides adsorbed with streptavidin and then reacted with dye-labeled biotinylated BSA served as negative controls. The influence of the photolithographic treatment was assessed in Figure 4 by comparing OptiChem® surfaces processed using fully transparent and fully opaque photomask controls to create 100% MeO-capped and 100% NHS-reactive surfaces, essentially analogous to fresh NHS- and MeO-capped surfaces but treated identically as all other lithographed cohorts. Typical fluorescence background signal from the low fluorescence uncoated glass slides before protein exposure was about 100 fluorescence units (RFU). Figure 4a data for specific protein-immobilization shows that NHS-patterned regions exhibit substantial protein fluorescence intensity from covalent coupling, while that on MeO-capped regions on the same patterned surfaces was negligible. The ratio of each signal represents a very efficient simultaneous reduction in non-specific protein adsorption and selective NHS-mediated covalent immobilization. Trends for these relative intensities agreed with identical protein assays on the uniform NHS and MeO-capped surfaces from both photo-exposed and non-exposed treatments.
Figure 4.
Comparison of normalized surface fluorescence intensities for proteins both specifically (a), and non-specifically (b), reacted with various treated OptiChem® surfaces: (a) Alexa555- labeled biotin-BSA reacted with bulk-phase immobilized streptavidin on photo-patterned (n=15, ±S.D.), and unpatterned (n=3, ±S.D.) control NHS- and MeO-capped surfaces: both fresh, i.e., untreated by photolithography (i.e., see NHS or MeO data), and also exposed to lithographic processing using control 100% opaque or 100% transparent photomasks, i.e., see NHS or MeO patterning process data), and bare glass (controls). (b) Non-specific protein-surface reactivity assay using complete serum: Alexa555-labeled neat goat serum adsorbed both to bare glass (control) and to the various OptiChem® surfaces.
2-2-2. Interaction of dye-labeled serum proteins with patterned surfaces
Figure 4b summarizes surface fluorescence intensity data across the same patterned and unpatterned surfaces compared to glass for adsorbed Alexa555-labeled complete goat serum. NHS-reactive surfaces exhibited an adsorbed or immobilized protein fluorescence intensity nearly 20 times lower than the serum signal on bare glass. This reflects a relatively lower density of protein NHS covalent immobilization and intrinsically low adsorption compared to glass. MeO-capped OptiChem® surfaces exposed to goat serum consistently showed variable and spatially heterogeneous fluorescence intensities resulting in large standard deviations, although absolute RFU intensities were not substantial.
Data in Figure 4 are consistent with Figure 1 and previous reports of specific and non-specific adsorption.[26] Serum contains thousands of different globular proteins with substantially higher intrinsic surface adsorption activity than streptavidin; non-specific protein adsorption occurs readily on most surfaces. NHS-capped surface regions showed significant fluorescence intensity while MeO-capped coating showed low fluorescence signal. Thus, suppressed non-specific protein adsorption was maintained in full serum, a condition that few surfaces can control, even after the patterning process involving UV light exposure and subsequent sonication in solvents required to remove the photoresist. The thin hydrogel coating, applicable to a variety of materials, appears sufficiently chemically stable to retain both non-fouling and specific immobilization properties in co-planar regions under rigorous processing conditions.
2-3. RGD peptide tethering on patterned polymer surfaces, cell adhesion and proliferation, and pattern fidelity in serum-based cell culture
2-3-1. Cell patterning on NHS-patterned OptiChem® surfaces with and without RGD cell adhesion peptides
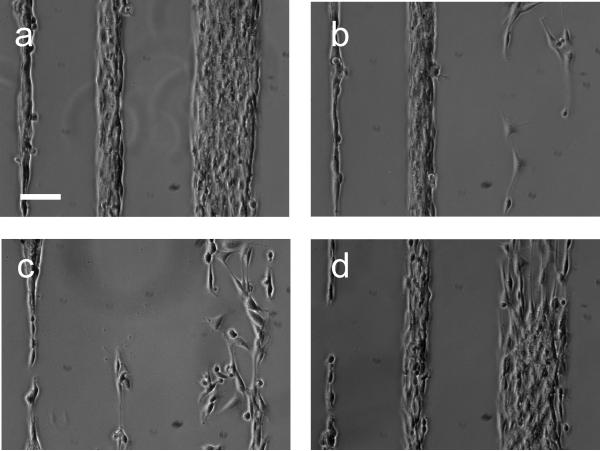
Surface modification with RGD peptide allows cell adhesion on a non-adhesive PEG layer.[26, 30, 60-62] Microscopic surface images for fibroblast cell culture at 24 h in serum-containing media for non-patterned (control) surfaces are shown in Figure S4 (Supplementary Information). Consistent with previous data,[26] no fibroblast cell adhesion was observed on either MeO-capped (Fig. S4a) or NHS-capped (Fig. S4b) surfaces in serum without RGD modification, supporting the precedent that PEG-based materials very effectively prevent cell adhesion, correlated with the lack of observed serum protein adsorption (vida infra). While no cells adhered on MeO-capped surfaces after RGD modification and serum exposure (Fig. S4c), RGD-modified NHS surfaces (Fig. S4d) facilitate cell adhesion from serum media, indicating sufficient RGD peptide density selectively on the NHS-reactive chemistry by bulk solution phase coupling.
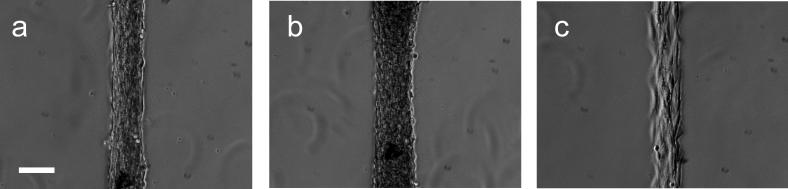
Figure 5a-e show microscopic images of fibroblast cell culture from serum media on the patterned polymer surfaces. Figure 5a shows no cell attachment and indicates that photo-patterning preserved the non-fouling surface properties unsupportive to cell culture without RGD, even in the presence of serum. After RGD modification (Fig. 5b-e), cells adhered in serum with high fidelity to the patterns on these surfaces corresponding clearly with NHS/RGD patterns (compare with Fig. 1d). After 48 h-incubation in serum-containing media, cells remained within the pattern (Fig. 5d, e). On the narrowest cell pattern (Fig. 5e), cells spread and oriented along the pattern axis and proliferated within the pattern. Figure 6 shows the corresponding ToF-SIMS image from mass fragments originating from the GRGDS peptide-immobilized pattern (sum of negative ion images from m/z 42, 45, 58 and 59; see also Figure S5 in Supplementary Information for fragment analysis) and demonstrates that RGD peptide solution exposure modifies NHS region specifically to facilitate reliable cell patterning in serum media. Direct correlation of the patterned chemistry with Figure 1d for NHS and Figure 5 cell adhesion patterns indicates that RGD coupled specifically to NHS regions.
Figure 5.
Phase contrast microscopy images of region-selective fibroblast adhesion and proliferation in serum-containing cultures on (a) patterned NHS-capped surface before, and (b-e) after RGD bulk phase surface modification. Images were taken at 24 h (a) and 48h (b-e) after cell seeding in DMEM/FBS. Scale bars: (a-d): 200 μm; (e) 50 μm.
Figure 6.

The ToF-SIMS image (500 μm × 500 μm) obtained by summing the images from negatives ion at m/z 42, 45, 58 and 59 showing the selective immobilization of RGD peptide (bright regions) from bulk solution to a patterned NHS-surface (MeO-capped darker regions).
2-3-2. Proliferation, orientation and pattern fidelity of adherent cells in longer-term serum cultures
Fibroblast cells adhering to patterns in serum-containing media (10% fetal bovine serum (FBS)) retain pattern fidelity for long term culture. Previous studies have shown adverse effects on cell adhesive patterns from non-specifically adsorbed serum proteins on substrates that permit invasion of adherent cells over time into non-adhesive regions (loss of pattern fidelity).[40, 63] In addition, endogenous cell extracellular matrix production, proliferation and migration contribute ultimately to cell pattern failure in cultures over time, typically within a few days.[29, 64, 65] Figure 7 shows cell-surface pattern fidelity in serum-containing media over time at 3 locations indicated by squares on the surface pattern legend (top, Fig. 7a). Seeded fibroblasts adhere rapidly (hours) and become confluent within the patterns 2-4 days after seeding, depending on the seeding density and pattern width. Figure 8 shows fluorescence images of these patterned adherent cells on culture Day 4 stained with rhodamine-phalloidin to visualize actin stress fibers. Adherent cells on narrow line patterns orient (as shown by stained actin fibers in Fig. 8a) after 4 days: these cells migrate within the RGD pattern and reorganize their spread footprint to align, then proliferate only within the narrow patterned area. Compared with such orientation within narrow lines, cells within larger patterned areas proliferate randomly but remain within the RGD-patterned adhesive patterned area (Fig. 8b).
Figure 7.
Cell pattern images from fibroblast-seeded serum cultures over time. (a) image locator with three squares with capital letters reflecting places on the pattern where cell images were acquired in (b).
(b) Phase contrast microscopy images of patterned fibroblast cells from 3 pattern locations (locations A, B, C) 2-4 days after cell seeding and culture in DMEM/FBS media. Scale bars: 100 μm.
Figure 8.
Fluorescence images of stained adherent fibroblast cells on narrow line patterns (see Figure 7a, location C, approx. line width 100 μm) (a), and wide block patterns (see Figure 7a, position B, approx. line width 3200 μm) (b). Actin stress fibers were stained with rhodamine-phalloidin (bright). Scale bars: 50 μm.
After 5 days of culture in serum-containing media, cells exceeded 100% confluence on all RGD-functinalized patterns. In larger cell patterns at high cell density (Fig. S6a in Supplementary Information), confluent cell sheets peeled spontaneously from patterned surfaces (Day 5), leaving exposed surface within the patterns. As shown in Figure S6b, cell patterns are readily re-established 24 h after a second cell seeding in serum-containing media, adhering to previously peeled surfaces (Day 6). It is possible that RGD modification or cell-deposited matrix proteins remain on the NHS-reacted regions and available to re-engage newly seeded cells after initially seeded cells peel. In addition, no newly seeded cells adhered on non-adhesive MeO-capped regions even at Day 5 in serum culture. Significantly, fragments of peeled non-adherent cell sheets harvested from the pattern cultures using pipette transfer and seeded onto fresh tissue culture plastic in serum rapidly adhered and proliferated normally (data not shown). Figure 9 shows fibroblast cell patterns at location A (see Fig. 7a) after 5-day serum culture. At Day 6 and 7, confluent cell sheets peeled spontaneously from the surface (Fig. 9b,c). In this case, no new cells were seeded, and the remaining adherent cells proliferated selectively within the RGD-modified region and re-formed cell patterns spontaneously and repeatedly (Fig. 9d). The MeO-capped surfaces remained non-adhesive, and cell patterns re-established without invasion into inert surface areas for a total of 15 days before the experiments were terminated. This supports the idea that cell sheets at confluence peeled under excessive cell density and surface occupancy constraints, and not because of either cell necrosis or hydrolysis of RGD-conjugated NHS groups that might also cause cell release
Figure 9.
Microscopic images of patterned adherent fibroblast cells at location A (see Figure 7a) from Day 5 to Day 9 in continuous culture in DMEM/FBS, showing consistent pattern fidelity even after endogenous cell peeling at confluence (image c) and spontaneous self re-seeding to restore the original patterns (image d) (a: day 5, b: day 6, c: day 7, d: day 9). Scale bar: 100 μm.
While cell detachment was observed on larger area patterns, other cell patterns maintained pattern fidelity without either invasion into inert areas and cell detachment for up to 11 days as shown in Figure 10. The cell pattern at location C was confluent at Day 4 and remained at Day 10 (Fig. 10a). As shown in Figure 10b, the cell pattern widened slightly at Day 11 and then detached at Day 12. While cell pattern width changed slightly, no cells spread into the MeO-capped regions. In fact, when the remaining adherent cells formed the pattern again (at Day 15), cells remained strictly within the pattern at 100% re-confluence and no cells spread into adjacent MeO-capped regions (Fig. 10c). All other patterns also showed no cell invasion to inert surface areas for least 15 days after cell seeding in serum-containing media before experiments were terminated. All residual adherent cells continuously re-formed patterns repeatedly and maintained patterns consistent with the surface patterns expected for specific cell attachment.
Figure 10.
Microscopic images of patterned cells at location C (see Figure 7a) from Day 10 to Day 15 in continuous culture in DMEM/FBS, showing consistent longer term pattern fidelity in DMEM/FBS (a: day 10, b: day 11, c: day 15). Scale bar: 100 μm.
Other surface chemistries have been reported to produce large-scale cell sheets in serum cultures, harvested from surfaces at confluence using non-enzymatic induction to prompt cell-surface peeling without destroying cell-cell contacts (e.g., temperature or electrochemical stimuli).[66-68] Applications for such viable cell constructs include regenerative medicine and in vitro tissue surrogates. Cell sheet peeling from large-scale areas here, while unexpected without any type of stimulus is similar to one other report where short RGD peptide spacers were attributed to induce cell tension resulting in observed cell peeling in culture.[69] Additionally, this seems to be the first cell patterning study to demonstrate immediate spontaneous re-seeding and repeated cell culture confluence within previously peptide- and cell- patterned areas in complex biological media.
3. Conclusions
This study has shown that imaging ToF-SIMS methods provide high-resolution chemical mapping capabilities for complex surface chemistries. PCA processing provides highly sensitive supporting data complementary to raw ion images for visualizing surface patterns through complex sequences of chemistry and surface bio-immobilization. Cross-correlation of the chemical images links the intended photolithographed patterns to the existence of clearly patterned surface-reactive chemistry, site-specific chemical coupling of bioactive species, low biological fouling, and site selection of patterns by cultured cells. Pattern fidelity is retained in long-term, serum-containing cultures for these commercial PEG-based surfaces. Given PEG’s popularity as a chemistry of choice in biotechnology and biomaterials applications, and the current scarcity of such coating chemistry in widely disseminated manufactured formats, this study is unique with practical impacts for demonstrating the utility of a PEG commercial surface for highly specific surface patterning for long-term selective biological immobilization in physiological media, and in the application of new ToF-SIMS imaging tools to track and validate pattern fabrication and performance.
4. Experimental
Materials
All the reagents were used as received. Telechelic NHS-terminated PEG (NHS-PEG-NHS (molecular weight: 3400 Da), α-{6-[(2,5-dioxo-1pyrrolidinyl)oxy]-6oxohexyl}-ω-{6-[(2,5-dioxo-1-pyrrolidinyl)oxy]-6-oxohexyloxy}-polyoxyethylene was purchased from NOF Corporation (Tokyo, Japan). Silane reagents (3-trimethoxysilylpropyl)diethylenetriamine (aminosilane) and 6-azidosulfonylhexyltriethoxy silane (azidosilane) were purchased from Gelest (Morrisville, PA). Polyoxyethylene sorbitan tetraoleate, anhydrous dimethylsulfoxide (DMSO), anhydrous dimethylacetamide, ACS-grade isopropanol, 2-methoxyethylamine, and neat goat serum were from Sigma (St. Louis, MO). Photoresist Microposit (formerly Shipley) SPR™-220-3.0 and Developer Microposit MF™-319 were purchased from Microchem (Newton, MA). Streptavidin was purchased from Prozyme (San Leandro, CA). Biotinylated BSA was obtained from Pierce (Rockford, IL). AlexaFluor555 carboxylic acid succinimidyl ester (Alexa555-NHS), rhodamine-phalloidin were obtained from Invitrogen (Carlsbad, CA). RGD peptide (Gly-Arg-Gly-Asp-Ser, GRGDS) was purchased from American Peptide Company (Sunnyvale, CA). Substrates consisted of low-fluorescence borosilicate glass microscope slides (Schott Glass, D263, 75.6 × 25.0 × 1.0 mm). The commercial high-performance, non-fouling PEG-based crosslinked transparent, amine-reactive polymer coating (Optichem®, Accerl8 Technology, Denver, CO) was reported previously in detail.[26] Amine-reactivity is imparted by use of NHS-terminated PEG within the thin film matrix, providing reliable, high-density amine-reactivity to externally applied chemistry (e.g., peptides, proteins). NHS group content can be controlled, hydrolyzed away, capped with non-reactive groups using conventional carbodiimide chemistry as previously described.[70]
Reaction-specific photolithographic patterning of Optichem® coatings
Schematics of the process used to obtain the desired NHS reactive group surface pattern as produced by conventional photolithography is shown in Scheme S1 (see Supplementary Information). Photoresist resin (Microposit (Shipley) SPR-220) mixed with isopropanol (ratio of 9:11 v/v) was pipet-dispensed onto OptiChem® polymer-coated low fluorescence 1×3 inch glass slides and each slide spun at 3500rpm for 75 sec. Instead of soft-baking on a hot plate, slides were cured at 100°C in vacuum (ca. 0.1mmHg) for 5 min. After cooling, a commercially printed photomask generated from computer graphics and printed at 5080 DPI (with 5 μm resolution) onto a polymer transparent film was placed on top of the coating and exposed to UV irradiation for 30 sec. After 75 sec of immersion in developer solution, slides were rinsed with water and dried under N2 gas. The exposed (developed) areas were selectively reacted with 2-methoxyethylamine by slide immersion into a 50mM solution in 50 mM borate buffer at pH 9 for 1 hour to create MeO-reacted regions. After rinsing with water and drying, the slide was lightly sonicated in DMSO, acetone and then isopropanol for 20 sec each to remove the photoresist, washed with water and dried by centrifugation. This exposed the NHS-capped regions. The influence of the photolithographic treatment on the chemistry was assessed by comparing OptiChem® surfaces processed using fully transparent and fully opaque photomask controls to create 100% MeO-capped and 100% NHS-capped surfaces, essentially analogous to fresh unpatterned NHS- and MeO-capped surfaces but treated identically as all other lithographed cohorts. Pattern line resolution produced on the polymer surface differed slightly from the 5 μm mask resolution. While both pattern precision and line resolution can be improved by improving the photomask quality and further optimization of the patterning procedure, this issue does not affect any results or conclusions from this current study.
XPS Analysis of Polymer Surfaces
XPS measurements were performed on a Kratos Axis Ultra DLD X-ray photoelectron spectrometer employing a hemispherical analyzer for spectroscopy and a spherical mirror analyzer for imaging as described previously.[36]
ToF-SIMS Analysis of Polymer Surfaces
ToF-SIMS data for patterned and unpatterned surfaces were acquired on an ION-TOF 5-100 instrument (ION-TOF GmbH, Münster, Germany) using a Bi3+ primary ion source. Positive and negative ion images and spectra were acquired with a pulsed 25 keV, 1.3 pA primary ion beam in high current bunched mode from 500 μm × 500 μm areas on sample surfaces. All images obtained contained 128 × 128 pixels within this area. These analysis conditions resulted in spatial resolution of approximately 4 μm. Data were collected using an ion dose below the static SIMS limit of 1×1012 ions/cm2. A low-energy electron beam was used for charge compensation on the polymer surface samples. The mass resolutions (m/Δm) for the negative secondary ion spectra were typically between 6000 and 7500 for the (m/z) 25 peak. The mass resolutions (m/Δm) for the positive secondary ion spectra were typically between 7000 and 8500 for the (m/z) 27 peak. PCA was performed on ToF-SIMS data as described previously[71, 72] using a series of scripts written by NESAC/BIO for MATLAB (MathWorks, Inc., Natick, MA). PCA data were then used to build PCA scores image maps of the surfaces.[36, 54] ToF-SIMS image line resolution was calculated to be 6.0 ± 0.4 μm as derived from multiple line scans, approximately the same resolution as the photomask.
Streptavidin immobilization
Streptavidin (5 mg/ml in phosphate buffered saline (PBS)) was 10-fold diluted in 55.6 mM phosphate buffer (pH 8.5). The solution was degassed and three 40μL drops were pipetted onto an OptiChem® coated surface. Another coated glass slide was then placed on top with the coated surfaces facing and the streptavidin solution between. The slide sandwiches were left at a room temperature (ca 22°C) at 100% humidity for 1 hour. Slides were then washed with PBS containing 0.01% Tween 20, rinsed with water, dried by centrifugation and immediately treated with a solution of biotinylated BSA.
Conjugation of Alexa555 to biotinylated BSA
To 100 μL of 10 mg/mL biotinylated BSA in PBS, 10 μL of 1 M Na2CO3 and then 20 μL of 10 mg/mL Alexa555-NHS in DMSO were added and gently shaken at room temperature for 1 h. Dye-conjugated biotin-BSA was separated by gel permeation chromatography using a Sephadex™ G-50 desalting column. BCA determination of final protein content was ~0.5 mg/mL and that of Alexa555 was 3 × 10−2 mM.
Reaction of biotinylated BSA with streptavidin-treated OptiChem® slides
In an Evergreen tube, Alexa555-biotinylated BSA was diluted in ~30 mL of 25 mM HEPES containing 20% glycerol, 50 mM KCl, 0.1% Triton X-100 and 0.1% BSA (pH 7.6) to make approximately 1μg/mL of biotinylated BSA. Each streptavidin slide was inserted into the tube and gently shaken at room temperature for 1 h. The slide was rinsed with PBS containing 0.01% Tween20 and water, and dried by centrifugation. Fluorescence images of dye-labeled BSA bound to surfaces were produced using a TECAN LS400 scanner (excitation 546 nm, emission 590 nm).
Interaction of neat Alexa555-goat serum with patterned OptiChem®
Neat goat serum was labeled with Alexa555 in the same manner as biotinylated BSA described above. OptiChem®-coated slides were immersed and exposed to approximately 1 mg/mL of Alexa555-labeled undiluted goat serum in PBS at 22°C for 1 h. After rinsing with PBS with 0.01% Tween 20 and then water, slides were dried by centrifugation. Fluorescence images and intensity maps of the serum-bound surfaces were obtained using a TECAN LS400 scanner (excitation 546nm, emission 590nm). Scanner PMT was adjusted so that Alexa555 fluorescence signal after adsorption to bare glass controls was ~20,000 RFU.
Preparation of RGD-grafted polymer surfaces
MeO-capped and NHS-capped unpatterned and patterned surfaces were reacted with RGD peptides by bulk aqueous immobilization. GRGDS (100μM) was dissolved in 50mM sodium borate buffer, and 50μL of the solution was dropped at 3 locations on each surface. The solution spots were sandwiched by placing another cover glass slide on top of the droplets, forcing the drops to form a thin liquid film between the coating and the top glass plate, and incubated for 20.5 h.[26] The slides were finally washed with sterile PBS to remove excess unreacted GRGDS.
Adhesion and proliferation of fibroblast cells on patterned surfaces
NIH-3T3 fibroblasts (ATCC, Manassas, VA) were expanded in cultures on tissue culture plastic in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS and 1% antibiotic/antimycotic at 5% CO2, 37 °C. In all cell culture procedures, serum-containing media described above (DMEM/FBS) was used. Cells were trypsinized and seeded onto the various Optichem® polymer substrates at a density of 3×104 cells/cm2 in DMEM/FBS and incubated for 2 h in DMEM/FBS. Substrates were then rinsed with PBS to remove unattached cells, and incubated further in DMEM/FBS. Microscopic observations were carried out with a Nikon TE2000-U inverted microscope equipped with a Coolsnap ES CCD camera and controlled with Metamorph software. Media was changed every 5 days. At 5 days after initial seeding and incubation, new cells were seeded onto the cell-patterned surfaces at a concentration of 3×104 cells/cm2 in DMEM/FBS and incubated for 2 h, and rinsed with PBS.
Fluorescent staining of patterned cells on surfaces
After patterning cells on the RGD-modified surface, cytoskeletal organization was investigated by staining for actin stress fibers.[73, 74] Cells cultured on the patterned surfaces were rinsed with PBS and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. After washing with PBS, cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min. After blocking with 2% BSA in PBS for 30 min, cells were stained with rhodamine-phalloidin for 1 h at room temperature. Fluorescence images were acquired using a Nikon TE2000-U inverted microscope equipped with a Coolsnap ES CCD camera.
Supplementary Material
Acknowledgments
Dr. G. Harbers is gratefully acknowledged for technical support. This research was supported by the National ESCA and Surface Analysis Center for Biomedical Problems (NIH grant EB-002027) and NIH grant EB-001473.
References
- [1].Harris JM. Poly(ethylene glycol) chemistry : biotechnical and biomedical applications. Plenum Press; 1992. [Google Scholar]
- [2].Hoffman AS. J Biomat Sci-Polym E. 1999;10:1011. doi: 10.1163/156856299x00658. [DOI] [PubMed] [Google Scholar]
- [3].Leckband D, Sheth S, Halperin A. J Biomat Sci-Polym E. 1999;10:1125. doi: 10.1163/156856299x00720. [DOI] [PubMed] [Google Scholar]
- [4].Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. Langmuir. 2001;17:5605. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- [5].Gombotz WR, Wang GH, Horbett TA, Hoffman AS. J Biomed Mater Res. 1991;25:1547. doi: 10.1002/jbm.820251211. [DOI] [PubMed] [Google Scholar]
- [6].Mao G, Castner DG, Grainger DW. Chem. Mater. 1997;9:1741. [Google Scholar]
- [7].Sofia SJ, Premnath V, Merrill EW. Macromolecules. 1998;31:5059. doi: 10.1021/ma971016l. [DOI] [PubMed] [Google Scholar]
- [8].Emoto K, Van Alstine JM, Harris JM. Langmuir. 1998;14:2722. [Google Scholar]
- [9].Lee SW, Laibinis PE. Biomaterials. 1998;19:1669. doi: 10.1016/s0142-9612(98)00044-1. [DOI] [PubMed] [Google Scholar]
- [10].Jo S, Park K. Biomaterials. 2000;21:605. doi: 10.1016/s0142-9612(99)00224-0. [DOI] [PubMed] [Google Scholar]
- [11].Xia N, Hu YH, Grainger DW, Castner DG. Langmuir. 2002;18:3255. [Google Scholar]
- [12].Schlapak R, Pammer P, Armitage D, Zhu R, Hinterdorfer P, Vaupel M, Fruhwirth T, Howorka S. Langmuir. 2006;22:277. doi: 10.1021/la0521793. [DOI] [PubMed] [Google Scholar]
- [13].Bearinger JP, Castner DG, Golledge SL, Rezania A, Hubchak S, Healy KE. Langmuir. 1997;13:5175. [Google Scholar]
- [14].Dalsin JL, Lin LJ, Tosatti S, Voros J, Textor M, Messersmith PB. Langmuir. 2005;21:640. doi: 10.1021/la048626g. [DOI] [PubMed] [Google Scholar]
- [15].Wazawa T, Ishizuka-Katsura Y, Nishikawa S, Iwane AH, Aoyama S. Anal. Chem. 2006;78:2549. doi: 10.1021/ac052102j. [DOI] [PubMed] [Google Scholar]
- [16].Kingshott P, Thissen H, Griesser HJ. Biomaterials. 2002;23:2043. doi: 10.1016/s0142-9612(01)00334-9. [DOI] [PubMed] [Google Scholar]
- [17].Lee JH, Kopecek J, Andrade JD. J Biomed Mater Res. 1989;23:351. doi: 10.1002/jbm.820230306. [DOI] [PubMed] [Google Scholar]
- [18].Malmsten M, Lassen B, Holmberg K, Thomas VV, Quash G. J Colloid Interface Sci. 1996;177:70. doi: 10.1006/jcis.1996.0007. [DOI] [PubMed] [Google Scholar]
- [19].Li JT, Carlsson J, Lin JN, Caldwell KD. Bioconjugate Chem. 1996;7:592. doi: 10.1021/bc960048v. [DOI] [PubMed] [Google Scholar]
- [20].Kenausis GL, Voros J, Elbert DL, Huang NP, Hofer R, Ruiz-Taylor L, Textor M, Hubbell JA, Spencer ND. J Phys Chem B. 2000;104:3298. [Google Scholar]
- [21].Ruiz-Taylor LA, Martin TL, Zaugg FG, Witte K, Indermuhle P, Nock S, Wagner P. Proc. Natl. Acad. Sci. U.S.A. 2001;98:852. doi: 10.1073/pnas.98.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang NP, Michel R, Voros J, Textor M, Hofer R, Rossi A, Elbert DL, Hubbell JA, Spencer ND. Langmuir. 2001;17:489. [Google Scholar]
- [23].Mrksich M, Whitesides GM. Poly(Ethylene Glycol) 1997;680:361. [Google Scholar]
- [24].Ostuni E, Chapman RG, Liang MN, Meluleni G, Pier G, Ingber DE, Whitesides GM. Langmuir. 2001;17:6336. [Google Scholar]
- [25].Johnston EE, Bryers JD, Ratner BD. Langmuir. 2005;21:870. doi: 10.1021/la036274s. [DOI] [PubMed] [Google Scholar]
- [26].Harbers GM, Emoto K, Greef C, Metzger SW, Woodward HN, Mascali JJ, Grainger DW, Lochhead MJ. Chem. Mater. 2007;19:4405. doi: 10.1021/cm070509u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Saldarriaga Fernandez IC, van der Mei HC, Lochhead MJ, Grainger DW, Busscher HJ. Biomaterials. 2007;28:4105. doi: 10.1016/j.biomaterials.2007.05.023. [DOI] [PubMed] [Google Scholar]
- [28].Mrksich M. Chem. Soc. Rev. 2000;29:267. [Google Scholar]
- [29].Rozkiewicz DI, Kraan Y, Werten MWT, de Wolf FA, Subramaniam V, Ravoo BJ, Reinhoudt DN. Chem. Eur. J. 2006;12:6290. doi: 10.1002/chem.200501554. [DOI] [PubMed] [Google Scholar]
- [30].Hern DL, Hubbell JA. J. Biomed. Mater. Res. 1998;39:266. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- [31].Plummer ST, Wang Q, Bohn PW, Stockton R, Schwartz MA. Langmuir. 2003;19:7528. [Google Scholar]
- [32].Xu G, Goldberg M. Method of Photolithographic Production of Polymer Array. 20040023367 US Patent. 2004
- [33].Blawas AS, Reichert WM. Biomaterials. 1998;19:595. doi: 10.1016/s0142-9612(97)00218-4. [DOI] [PubMed] [Google Scholar]
- [34].Revzin A, Tompkins RG, Toner M. Langmuir. 2003;19:9855. [Google Scholar]
- [35].Houseman BT, Gawalt ES, Mrksich M. Langmuir. 2003;19:1522. [Google Scholar]
- [36].Lee CY, Harbers GM, Grainger DW, Gamble LJ, Castner DG. J. Am. Chem. Soc. 2007;129:9429. doi: 10.1021/ja071879m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bhatia SN, Balis UJ, Yarmush ML, Toner M. Faseb J. 1999;13:1883. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- [38].Khademhosseini A, Suh KY, Yang JM, Eng G, Yeh J, Levenberg S, Langer R. Biomaterials. 2004;25:3583. doi: 10.1016/j.biomaterials.2003.10.033. [DOI] [PubMed] [Google Scholar]
- [39].Hui EE, Bhatia SN. Langmuir. 2007;23:4103. doi: 10.1021/la0630559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hatakeyama H, Kikuchi A, Yamato M, Okano T. Biomaterials. 2007;28:3632. doi: 10.1016/j.biomaterials.2007.04.019. [DOI] [PubMed] [Google Scholar]
- [41].Falconnet D, Csucs G, Grandin HM, Textor M. Biomaterials. 2006;27:3044. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- [42].Ito Y. Biomaterials. 1999;20:2333. doi: 10.1016/s0142-9612(99)00162-3. [DOI] [PubMed] [Google Scholar]
- [43].Doh J, Irvine DJ. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5700. doi: 10.1073/pnas.0509404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu WF, Chen CS. Adv. Drug Deliv. Rev. 2007;59:1319. doi: 10.1016/j.addr.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yang ZP, Chilkoti A. Adv. Mater. 2000;12:413. [Google Scholar]
- [46].Tan W, Desai TA. Tissue Eng. 2003;9:255. doi: 10.1089/107632703764664729. [DOI] [PubMed] [Google Scholar]
- [47].Mooney JF, Hunt AJ, McIntosh JR, Liberko CA, Walba DM, Rogers CT. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12287. doi: 10.1073/pnas.93.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nakanishi J, Kikuchi Y, Takarada T, Nakayama H, Yamaguchi K, Maeda M. J. Am. Chem. Soc. 2004;126:16314. doi: 10.1021/ja044684c. [DOI] [PubMed] [Google Scholar]
- [49].Doh J, Irvine DJ. J. Am. Chem. Soc. 2004;126:9170. doi: 10.1021/ja048261m. [DOI] [PubMed] [Google Scholar]
- [50].Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- [51].Itoga K, Kobayashi J, Yamato M, Kikuchi A, Okano T. Biomaterials. 2006;27:3005. doi: 10.1016/j.biomaterials.2005.12.023. [DOI] [PubMed] [Google Scholar]
- [52].Gong P, Harbers GM, Grainger DW. Anal. Chem. 2006;78:2342. doi: 10.1021/ac051812m. [DOI] [PubMed] [Google Scholar]
- [53].Mann BK, West JL. J. Biomed. Mater. Res. 2002;60:86. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- [54].Cheng F, Gamble LJ, Grainger DW, Castner DG. Anal. Chem. 2007;79:8781. doi: 10.1021/ac0715423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Belu AM, Graham DJ, Castner DG. Biomaterials. 2003;24:3635. doi: 10.1016/s0142-9612(03)00159-5. [DOI] [PubMed] [Google Scholar]
- [56].Mantus DS, Ratner BD, Carlson BA, Moulder JF. Anal.Chem. 1993;65:1431. doi: 10.1021/ac00058a021. [DOI] [PubMed] [Google Scholar]
- [57].Wagner MS, Castner DG. Langmuir. 2001;17:4649. [Google Scholar]
- [58].Lhoest JB, Wagner MS, Tidwell CD, Castner DG. J. Biomed. Mater. Res. 2001;57:432. doi: 10.1002/1097-4636(20011205)57:3<432::aid-jbm1186>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- [59].Kim YP, Hong MY, Shon HK, Moon DW, Kim HS, Lee TG. Appl. Surf. Sci. 2006;252:6801. [Google Scholar]
- [60].Houseman BT, Mrksich M. Biomaterials. 2001;22:943. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- [61].Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. J. Am. Chem. Soc. 1998;120:6548. [Google Scholar]
- [62].Saneinejad S, Shoichet MS. J. Biomed. Mater. Res. 1998;42:13. doi: 10.1002/(sici)1097-4636(199810)42:1<13::aid-jbm3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- [63].Nelson CM, Raghavan S, Tan JL, Chen CS. Langmuir. 2003;19:1493. [Google Scholar]
- [64].Sharma S, Tan W, Desai TA. Bioconjugate Chem. 2005;16:18. doi: 10.1021/bc049885k. [DOI] [PubMed] [Google Scholar]
- [65].Luk YY, Kato M, Mrksich M. Langmuir. 2000;16:9604. [Google Scholar]
- [66].Yang J, Yamato M, Shimizu T, Sekine H, Ohashi K, Kanzaki M, Ohki T, Nishida K, Okano T. Biomaterials. 2007;28:5033. doi: 10.1016/j.biomaterials.2007.07.052. [DOI] [PubMed] [Google Scholar]
- [67].Guillaume-Gentil O, Akiyama Y, Schuler M, Tang C, Textor M, Yamato M, Okano T, Vörös J. Adv. Mater. 2008;20:560. [Google Scholar]
- [68].Canavan HE, Cheng XH, Graham DJ, Ratner BD, Castner DG. Langmuir. 2005;21:1949. doi: 10.1021/la048546c. [DOI] [PubMed] [Google Scholar]
- [69].Harbers GM, Healy KE. J. Biomed. Mater. Res. 2005;75:855. doi: 10.1002/jbm.a.30482. [DOI] [PubMed] [Google Scholar]
- [70].Gong P, Grainger DW. Surf. Sci. 2004;570:67. [Google Scholar]
- [71].Wickes BT, Kim Y, Castner DG. Surf. Interface Anal. 2003;35:640. [Google Scholar]
- [72].Wagner MS, Graharn DJ, Castner DG. Appl. Surf. Sci. 2006;252:6575. [Google Scholar]
- [73].Berg MC, Yang SY, Hammond PT, Rubner MF. Langmuir. 2004;20:1362. doi: 10.1021/la0355489. [DOI] [PubMed] [Google Scholar]
- [74].Veiseh M, Veiseh O, Martin MC, Asphahani F, Zhang MQ. Langmuir. 2007;23:4472. doi: 10.1021/la062849k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.