Abstract
Epigenomes are comprised, in part, of all genome-wide chromatin modifications including DNA methylation and histone modifications. Unlike the genome, epigenomes are dynamic during development and differentiation in order to establish and maintain cell type-specific gene expression states that underlie cellular identity and function. Chromatin modifications are particularly labile, providing a mechanism for organisms to respond and adapt to environmental cues. Results from studies in animal models clearly demonstrate that epigenomic variability leads to phenotypic variability including susceptibility to disease that is not recognized at the DNA sequence level. Thus, capturing epigenomic information is invaluable for comprehensively understanding development, differentiation, and disease. Herein, we provide a brief overview of epigenetic processes, how they are relevant to human health, and review studies utilizing technologies that enable epigenome mapping. We conclude by describing feasible applications of epigenome mapping, focusing on epigenome-wide association studies (eGWAS), which have the potential to revolutionize current studies of human diseases and will likely promote the discovery of novel diagnostic, preventative, and treatment strategies.
Keywords: Epigenetics, genetics, gene expression, gene regulation
In contrast to the genome, which remains largely unchanged in most cells of an organism, epigenomes are the product of a gradual commitment of cell lineages to more constrained patterns of gene expression throughout development that is, in part, shaped by the environment. An epigenome can be defined as the combination of all genome-wide chromatin modifications in any given cell type that directs its unique gene expression pattern. Epigenomes are labile during development1, are responsive to extrinsic factors2, are altered in disease3, and even differ among individuals with identical genetic composition4.
The principal chromatin modifications include DNA methylation and post-translational modifications of histones that package the DNA in nucleosomal units. Although both DNA methylation and histone modifications appear to be meiotically and/or mitotically heritable, only the former epigenetic process is backed with strong mechanistic support for heritability5. As we discuss in this review, accumulating evidence from mammalian studies indicate that variability of epigenetic modifications of chromatin during development and in response to distinct environmental factors directly contribute to adult phenotypic variability and disease susceptibility that could not previously be accounted for by DNA sequence alone. Thus, characterizing genome-wide chromatin modification patterns (i.e. epigenome mapping) may aid in the discovery of disease-causing genes in humans for which non-genetic factors are clearly involved and confound DNA sequence-based association studies of disease6, 7. Epigenome mapping across different cell types and developmental periods from normal individuals, in a manner analogous to the efforts of sequencing the human genome, is an essential prerequisite.
Technological advances allowed characterization of the first human epigenome in CD4+ T cells, which describes 38 histone modifications including both histone methylation and acetylation8–10. Similar technologies have now enabled truly genome-wide analyses of DNA methylation11, 12. Taking advantage of these and other technologies, the recently launched International Human Epigenome Consortium (IHEC) aims to map 1000 reference epigenomes within a decade13. Herein, we provide a brief overview of epigenetic processes and how they are relevant to human health and review initial studies utilizing epigenome mapping. In particular, we discuss the evidence supporting that epigenetic processes offer a mechanistic link between genetic determinants and environment during development, and address how results of epigenome mapping might be applied to measure previously unobservable potential inter-individual variability that could account for differences to disease susceptibility.
Epigenetic Processes
In general, epigenetic processes including DNA methylation and histone modifications are thought to modulate the accessibility of cis-DNA elements to trans-acting factors via regulating chromatin structure14. In plants, RNA interference contributes to epigenetic regulation, however whether there is a conserved mammalian equivalent remains unclear15 and will not be further discussed here. Similarly, there is considerable debate of whether chromatin remodeling1 should be considered an epigenetic mechanism as it is unknown that this process can transmit memory of cell fate from one generation to the next.
Epigenetic regulation of chromatin structure consequently influences gene expression. This type of control is exemplified by the phenomenon of genomic imprinting, the mono-allelic expression of genes in a parent-of-origin-dependent manner. Critical cis-elements that regulate expression of imprinted genes called imprinting control regions (ICRs) exhibit parental-specific chromatin modifications including DNA methylation and histone modifications that govern their activity16. Similarly, as a form of dosage compensation between female and male eutherians, X-chromosome inactivation (XCI), mediated primarily by Xist and Tsix genes, is accompanied by chromosome-specific histone modifications associated with heterochromatin and gene silencing at the two-cell stage of early embryonic development17. Once established, XCI is maintained by DNA methylation-mediated gene silencing in cells of the embryo proper. As genomic imprinting and XCI have been reviewed extensively elsewhere16, 17, we focus on the global distribution and functional importance of DNA methylation and histone modifications as they relate to cellular phenotype.
DNA methylation
Perhaps the best understood epigenetic mechanism is DNA methylation, which in mammals occurs almost exclusively within 5’-cytosine-guanine-3’ dinucleotides (CpGs), although CpNpG methylation has also been detected18. In general, DNA methylation is typically associated with gene silencing by affecting the binding of methylation-sensitive DNA binding proteins and/or by interacting with various modifications of histone proteins that alter DNA accessibility to promoters19. Once established by the de novo methyltransferases DNMT3a and DNMT3b20, DNA methylation is maintained through mitosis primarily by the DNMT1 enzyme which associates with PCNA and the replication foci and has a significant preference for action on hemi-methylated DNA following DNA replication21. This mechanism allows for perpetuation of the DNA methylation state in newly formed cells, a well-established mode of epigenetic inheritance.
Several studies suggest multiple functional roles for DNA methylation including silencing transposable elements22, mediating developmental gene regulation23, and reducing transcriptional noise24. Indeed, DNA methylation in mammals is essential for embryonic development25, differentiation26, and cell cycle control27. Additionally, DNA methylation plays critical roles in maintaining transcriptional silencing of genes on the inactive X-chromosome and imprinted genes28. Human diseases have been associated with abnormal DNA methylation patterns including cancer29, ICF (immunodeficiency, centromeric instability, facial anomalies) syndrome, ATRX (alpha-thalassemia, mental retardation) syndrome, and fragile X syndrome30. Even mutations in a gene that encodes a protein that ‘reads’ DNA methylation signals and thought to act as a transcriptional silencer, MeCP2, are associated with the autism spectrum disorder, Rett syndrome31. In plants, DNA methylation deficiency results in spurious transcription initiation from cryptic sites, demonstrating a role for methylation in reducing transcriptional noise24. A central theme of these findings is that DNA methylation functions to maintain a repressed chromatin state, thereby stably silencing promoter activity32. It is important to note, however, that DNA methylation is not always associated with gene silencing. Some studies have demonstrated that DNA methylation can augment expression of an imprinted gene by blocking the binding of repressor proteins to silencer elements within the gene33. Furthermore, additional novel roles of DNA methylation have been suggested by recent epigenome mapping studies (discussed below).
Histone modifications
In addition to the covalent modification of DNA, histone proteins that package nuclear DNA are known to be subjected to a wide array of post-translational modifications on specific residues along their NH2-terminal ‘tails’ that project outside of the nucleosome core. Such modifications include acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and potentially others that remain to be discovered34, 35. Since these modifications contribute to chromatin conformation, the potential information stored within different combinatorial patterns of these modifications led to the hypothesis of a ‘histone code’, which proposes that specific combinations of these modifications dictate locus-specific transcriptional competence36. Although the specific functional consequences of each of these modifications are insufficiently understood, many of the enzymes responsible for adding and removing them have been identified35. In addition, alterations of the genes encoding histone-modifying enzymes and perturbations to the modification patterns they produce are associated with disease37, underscoring the importance of histone modifications in normal development. For example, the Polycomb-group protein EZH2, a histone H3 lysine 27 (H3K27) methyltransferase, was overexpressed in prostate cancer38, while global loss of H4K16 acetylation and H4K20 trimethylation (H4K20me3) was observed in lymphoma and colon cancer39.
One of the best understood histone modification is the reversible acetylation of lysine residues on histones H3 and H440. Acetylation is accomplished by specific histone acetyltransferases (HATs) that neutralizes the net positive charge of the histone tail and facilitates access of transcription factors to the underlying DNA41. While histone acetylation is clearly associated with transcriptional activation42, the reverse reaction catalyzed by histone deacetylases (HDACs) increases the tail’s positive charge, lowers the transcription potential of the underlying DNA, and is associated with a transcriptionally repressed state43. Distinct from acetylation, methylation of histones can be associated with transcriptional activation or repression depending on the specific residue targeted and degree of methylation35, 36. For example, H3K4me3 is frequently enriched at gene promoters, many of which are active44, 45. Localization of H3K4me3 at inactive promoters, however, facilitates the transient binding of HATs and HDACs to maintain the promoters in a repressed but poised state for future activation46. How histone methylation accomplishes gene repression is not fully understood, however it is known that methylation of certain lysine residues, such as H3K9, acts as a docking site for heterochromatin protein 1 (HP1) which in turn recruits histone methyltransferases47. Other modifications are not restricted to promoters, including the ‘activating’ modification H3K36 which is distributed over the gene bodies of actively expressed genes8, 48. Although histone phosphorylation, ubiquitylation, and sumoylation of specific serine, threonine, or lysine residues are involved in transcriptional regulation, modifications on other residues are associated with mitosis, DNA repair, and apoptosis34. The precise functions of many of these modifications remain under investigation.
New evidence suggests the possibility that histone modifications could be mitotically heritable, though this is debated. In yeast for example, the heterochromatic states mediated by the interactions of hypoacetylated histones and silent-information-regulator proteins or H3K9 methylation and the Swi6 chromodomain are maintained through cell division49. In flies, H3K27 and H3K4 methylation, catalyzed by Polycomb-group and trithorax-group protein complexes respectively, mediate mitotic inheritance of lineage-specific gene expression patterns50. This in part accomplished by EED, a constituent of the Polycomb repressive complex 2 (PRC2), which binds specifically to histone tails, activates the methyltransferase activity of PRC2, and facilitates propagation of the H3K27me3 mark upon mitosis51, 52. The extent to which other histone modifications are heritable remains unclear. However, histone modifications along with DNA methylation undergo considerable changes during development53 when the epigenome is particularly vulnerable to environmental influences, as later discussed.
Epigenome Mapping
Recent technological advances in DNA sequencing, in part, has enabled epigenome mapping. Data derived from these technologies provide unprecedented insight into the distribution, interplay, and potential novel functions of chromatin modifications and associated proteins. While other reviews explain these technologies in greater detail54–58, we focus on the currently available methods that utilize next-generation sequencing to map epigenomes and discuss unexpected findings.
Detecting DNA methylation genome-wide
Several strategies that allow detection of DNA methylation include digestion of DNA by methylation-sensitive or –insensitive restriction endonucleases59, the chemical modification of DNA by sodium bisulfite60, immunoprecipitation of 5-methylcytosine to separate unmethylated and methylated fractions of the genome61, and enrichment of methylated DNA using DNA binding proteins62. These strategies have been coupled to high-throughput technologies. Although the genomic coverage and resolution varies among the different strategies, each offers unique advantages. Thus, these approaches are complementary and would potentially generate mutually reinforcing and comprehensive genome-wide DNA methylation data.
Of these strategies, one that provides the best resolution of cytosine methylation incorporates sodium bisulfite treatment of DNA. Sodium bisulfite chemically converts all unmethylated cytosines to uracil by hydrolytic deamination of the 5,6-dihydrocytosine-6-sulphonate product at the C4 position of the pyrimidine ring. However, a methyl group at the C5 position inhibits this reaction, protecting methylated cytosines from conversion63, 64. Upon PCR amplification using primers corresponding to the bisulfite-converted sequences of interest, the products are traditionally cloned and sequenced to evaluate CpG methylation throughout the amplified region65. Several applications exploiting this ‘gold-standard’ bisulfite-conversion method have been adapted to evaluate methylation globally using next-generation sequencing technology including a shotgun-sequencing approach66, a reduced representation shotgun-sequencing approach67, and a targeted approach using bisufite padlock probes68. Bisulfite treatment of genomic DNA combined with ultra-high-throughput sequencing using Illumina technology allows sensitive measurement of cytosine methylation on a genome-wide scale within specific sequence contexts at single nucleotide resolution66. This technology has been successfully applied to the Arabidopsis thaliana genome66, 69, to the mouse genome66, and more recently to the human genome11, 12, impressively accounting for >90% of all cytosines genome-wide. However, in order to yield comprehensive and accurate methylation data, a major prohibitive disadvantage of this approach is the high cost since a very large amount of sequencing is required. To overcome this limitation, a semi-random approach termed ‘reduced representation bisulfite sequencing’ (RRBS) has been devised67. In RRBS, genomic DNA is first digested with specific restriction enzymes whose recognition sequences are disproportionately represented in GC-rich DNA, such as BglII or MspI, fragments of a certain size range are then selected and subjected to adaptor ligation, treated with sodium bisulfite, PCR amplified and sequenced. Although this approach provides useful nucleotide-level quantitation of cytosine methylation, the selection of fragments in a given size range explicitly excludes entire genome coverage and introduces a bias towards CpG-rich regions of the genome that typically include unmethylated CpG islands in normal cells. However, RRBS has proven useful for cancer studies that require the rapid, high-throughput analysis of aberrant DNA methylation patterns in multiple samples70. An alternative approach that eliminates the potential bias of methods targeting CpG islands yet offers nucleotide-level resolution involves the use of bisulfite padlock probes (BSPPs) to capture selected locations of the genome for methylation profiling68. However, the genome-wide coverage of DNA methylation is limited to the number of BSPPs used per reaction, thus precluding a complete analysis of DNA methylation throughout the genome.
Other strategies avoid the use of sodium bisulfite altogether and instead enriches for methylated DNA. One approach takes advantage of an antibody targeted to single methylated cytosines in order to directly capture methylated sequences within genomic DNA. Termed methylated DNA immunoprecipitation (MeDIP), this approach allows for relative quantitation of DNA methylation genome-wide when coupled to comparative genomic hybridization microarrays as was applied to human61 and plant71 samples. More recently, MeDIP has been adapted to high-throughput sequencing using Illumina technology (MeDIP-Seq)72, providing a more complete genome-wide coverage of DNA methylation than microarrays can achieve. Another way to enrich for methylated DNA is using a methyl-binding domain (MBD) fused to human IgG73 or MBD proteins bound to a sepharose matrix62 before hybridizing onto an array. Irrespective of the platforms used to measure output DNA methylation signals, limitations of these affinity approaches include the bias that methylated CpG-rich sequences give higher enrichments than equally methylated yet CpG-poor sequences, the inability to measure DNA methylation of individual repetitive elements, and the lower resolution of methylation detection relative to bisulfite-sequencing. To overcome these limitations, a novel approach called Methyl-MAPS (methylation mapping analysis by paired-end sequencing) has recently been developed that provides single nucleotide resolution of DNA methylation, covers >80% of CpG sites within mammalian genomes, and enables quantitation of methylation at repetitive elements in addition to single-copy loci by combining enzymatic fractionation and deep sequencing74.
These epigenome mapping techniques afforded an unprecedented opportunity to observe the distribution of DNA methylation over the majority of the genome leading to some surprising discoveries. The majority of gene promoters were depleted of DNA methylation, whereas there was markedly increased cytosine methylation within gene bodies. The large scope of DNA methylation within gene bodies, as observed for a third of all genes in the Arabidopsis genome66, 69 and for a substantial number of genes in humans11, 12, 68, 75, suggests a potentially novel role for DNA methylation that seemingly contradicts the dominant model for DNA methylation-mediated gene silencing. Specifically, higher levels of DNA methylation were detected within gene bodies of actively expressed genes compared to silent genes. Suppression of spurious initiation of transcription within highly active genes76, modulation of transcriptional elongation77, regulation of pre-mRNA splicing12, and tissue-specific alternate promoter usage (Maunakea, A.K. et al, Nature, in press) are potential novel roles of gene body methylation. That substantial gene body methylation is an evolutionarily conserved feature of eukaryotic genomes78–80 further supports a biological role and warrants continued investigation.
Another unexpected common finding of these genome-wide DNA methylation studies was that, in contrast to the long-held view that cytosines within CpG islands remain free of DNA methylation, a subset of CpG island-containing promoters were in fact methylated in normal cells81–84. Consistent with previous gene-centric studies in cancer, this normal promoter methylation was, in general, negatively correlated with the expression state of the underlying gene. In studying normal embryonic stem cell differentiation, gain or loss of promoter methylation was observed concomitant with loss or gain of expression, respectively, of the associated gene11, 12, many of which have previously defined roles in embryonic stem cell function. Interestingly, it was also recognized that unlike somatic cells, embryonic stem cells harbored substantial levels of non-CpG cytosine methylation11, 12. Genome-wide, non-CpG methylation was diminished in human ES cell-differentiated fibroblasts12 and significantly gained in induced pluripotent embryonic like-stem cells generated from primary human fibroblasts11, which together indicates a novel role of non-CpG methylation in the origin and maintenance of the pluripotent stem cell state.
Finally, a landmark study using a more limited genome-wide DNA methylation fingerprinting technique called AIMS, observed remarkable differences in the DNA methylation profiles of monozygotic twins, particularly those that were older, had different lifestyles, and had spent less of their lives together4. These results implicate a probabilistic accumulation of epigenetic variability likely due to environmental differences experienced by these genetically identical individuals during their lifetime and may be involved in the etiology of monozygotic twin discordance for common diseases and traits85. Further support of this notion comes from a recent DNA methylation profiling study that uncovered epigenetic differences that might contribute to the discordance for the autoimmune disease systemic lupus erythematosus (SLE) among monozygotic twins86.
Additional epigenome mapping of DNA methylation profiles across a larger number of cell types and individuals, coupled with global gene expression data, should reveal a more comprehensive understanding of the prevalence of normal promoter methylation, non-CpG methylation, and inter-individual variability to elucidate the epigenetic determinants of normal differentiation and disease susceptibility. Notably, the studies of DNA methylation described above focus exclusively on 5-methylcytosine. However, 5-hydroxymethylcytosine has recently been detected in mammalian DNA87, 88, yet its in vivo function remains unclear in part because current technologies, including the gold-standard bisulfite-sequencing approach, fail to distinguish between the two types of DNA modifications89. MeDIP approaches using recently introduced anti-5-hydroxymethylcytosine antibodies may overcome this critical limitation.
Detecting histone modifications genome-wide
Most of the existing methods for studying histone modifications on a genomic scale combine the use of chromatin immunoprecipitation (ChIP) with high-throughput technologies including DNA microarrays (ChIP-chip) or massively parallel sequencing (ChIP-Seq). ChIP technique relies on the isolation of individual chromatin fragments using an antibody specific to a particular feature of the chromatin fragments, including DNA-binding proteins, histone modifications, and nucleosomes. Since these techniques have been reviewed elsewhere56, 90, 91, we will only mention that applications of these techniques across multiple species including yeast92–94, fly95, mouse96, 97, and human44, 98–100 have revealed that distinct genomic regions such as enhancers, promoters, and gene bodies have distinct histone modification patterns8, 9, 101–104. Thus, mapping these modifications globally may provide a more precise functional annotation of genomes. In addition, detecting lesser-known histone modifications including phosphorylation and sumoylation using chemical approaches such as peptide synthesis, mutagenesis, and in vitro nucleosomal arrays could complement in vivo ChIP results in elucidating their function(s)105.
Our epigenome mapping data of 38 histone modifications including methylation and acetylation using ChIP-Seq in normal primary human CD4+ T cells have revealed remarkably consistent categorical patterns across gene regions depending on the transcriptional activity of the underlying gene8, 9. For example, histone modifications across actively expressed genes can be separated into at least four categories based on their general patterns and may correspond to their distinct functions in transcription (Fig. 1). ‘Active’ histone modification marks highly enriched within gene promoters, for instance, may be involved in transcription initiation (e.g. H3K4me3), whereas those that are intragenic may be involved in elongation, termination, or pre-mRNA splicing (e.g. H3K36me3)8, 9, 46, 106, 107. Similarly, other histone modifications across silent genes adopt distinct patterns possibly impairing or preventing transcription initiation when enriched at promoters (e.g. H3K27me3) or disrupting elongation when enriched throughout gene bodies. ‘Silent’ histone marks such as H3K27me3 seem to function dominantly over ‘active’ histone marks such as H3K4me3, as exemplified by the co-localization of both marks (i.e. bivalent domains) over inactive promoters8, 44, 97, 103. These results suggest that histone modifications delineate functional features of genes, including their structure and intrinsic regulatory elements. Indeed, an integrated analysis of these epigenome profiles with that of non-promoter DNase hypersensitive sites, putatively marking cis-regulatory elements, showed enrichment of specific histone modifications within these sites including H3K9me1 and H3K4me1/me2/me38. More generally, based on their association with gene expression status, all histone acetylations and most histone methylations we examined were considered as ‘active’ marks, while only five histone methylations (i.e. H3K9me2, H3K9me3, H3K27me2, H3K27me3 and H4K20me3) were considered as ‘repressive’ marks9. Additionally, previous observations indicated that certain histone acetylations and methylations are enriched at and potentially mark enhancer elements8, 100, 102; however there is some debate over the precision of these marks. For example, one study found that enhancers are associated with H3K4me1 but not H3K4me3 marks101, whereas we and others have shown that H3K4me2/3 and H3K9me1 marks are present at many potential distal regulatory regions including well-characterized enhancers for IFNγ, Th2 cytokine genes, and FOXA18, 9, 108. These studies suggest that enhancers may be associated with a variety of epigenetic patterns in likely a context-dependent manner. It should be noted, however, that although these studies show consistent association of specific histone marks with transcriptional states, the precise functional role of these modifications with transcription and whether or not they are a cause or consequence of gene expression states are still unclear.
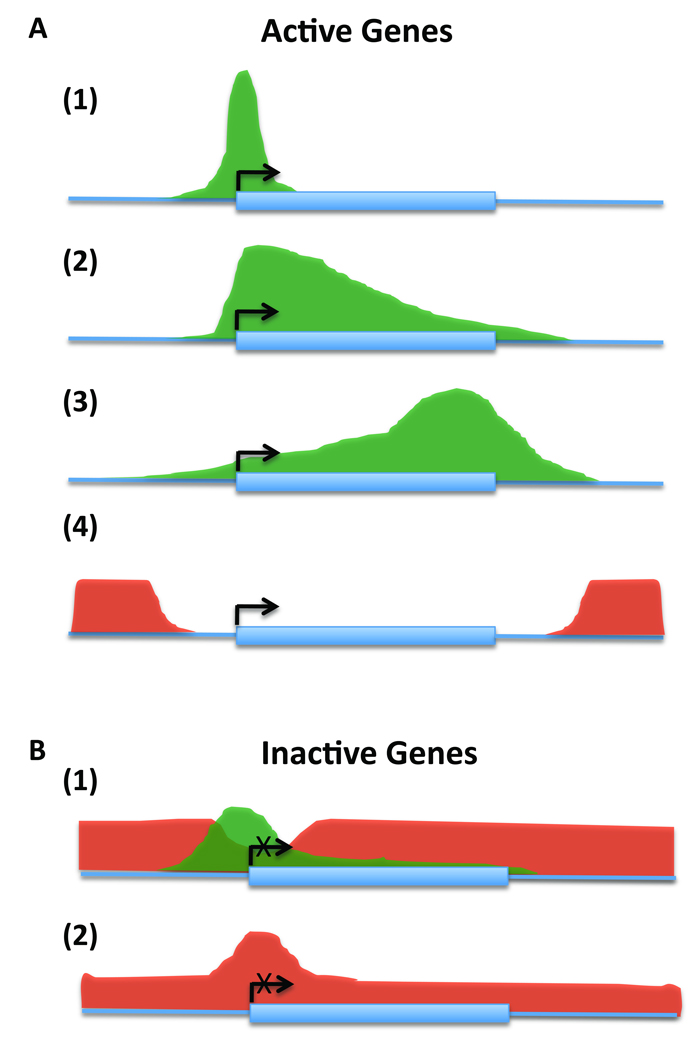
Figure 1. Distinct histone modification patterns delineate gene structure and associates with gene expression states.
Histone modifications associated with actively expressed genes can be separated into four major categories based on their distribution relative to gene structure as in (A): (1) highly enriched at gene promoters but depleted in gene bodies (e.g. H3K4me3, H3K4me2), (2) relatively depleted at gene promoters but highly enriched proximal to the 5’-end with a gradual 5’-3’ decrease in signal (e.g. H4K20me1, H2BK5me1), (3) 5’-3’ gradual increase in signal throughout the gene body, peaking towards the 3’-end (e.g. H3K36me3), and (4) depletion throughout the gene region with relative enrichment at the 5’ and 3’ boundaries of genes (e.g. H3K27me3). In addition, histone modifications across silent genes adopt two distinct patterns (B): (1) enrichment of a repressive mark (e.g. H3K27me1) over the gene with an active mark (e.g. H3K4me3) surrounding the promoter and (2) modestly enriched at gene promoters with declining signal throughout the gene region (e.g. H3K27me2, H3K27me3). Arrows depict active expression whereas crossed-out arrows depict silencing. Green histone patterns denote ‘active’ marks, whereas red denotes ‘silent’ marks. These generalized patterns were based on 38 different histone modifications we examined in T cells in relationship to genome-wide gene expression8.
In addition to these static observations, comparative ChIP-Seq analyses in stem/progenitor cells and differentiated cells revealed the remarkably dynamic processes of histone modifications genome-wide10, 109. The co-localization of the transcriptionally permissive H3K4me3 and repressive H3K27me3 histone modification marks were detected at promoters in a subset of genes in embryonic stem cells97, 110 and somatic cells8, 44. This bivalent configuration is presumed to maintain the underlying gene in a transcriptionally silent but poised state8, 97. In ES cells, bivalent domains target and potentially regulate the expression of key developmental genes as resolution of bivalent marks to a monovalent state appear to be associated with the establishment of lineage-specific gene expression patterns during differentiation97, 111, 112, a potentially universal paradigm applicable to multipotent cells. One recent example of such a resolution occurred exclusively on the transcriptionally competent tissue-specific imprinted allele of Grb10 whose promoter is bivalently marked in multipotent embryonic neuronal precursor cells and appears only to resolve to a monovalent state (i.e. H3K4me3) upon neuronal commitment permitting Grb10 expression in differentiated neurons, whereas the allele remained bivalent and transcriptionally silent in all other somatic cells113. While some bivalent domains resolve upon differentiation, others arise de novo as observed in ES cell-differentiated neuronal progenitor cells45, 114. For example, in the neuronal progenitor cell stage bivalent domains were established at the promoters of glial-specific genes, including those the encode for the myelin basic protein and glial fibrillary acidic protein, which resolved to a repressive chromatin state (i.e. H3K27me3) when these cells differentiated into neurons45. In addition, genes that maintain stem cells in a pluripotent state, including Oct3/4 and Nanog, become silenced by H3K9 methylation upon differentiation into neuronal progenitors and non-bivalent promoters of lineage-specific genes exhibited changes in histone modification patterns according to their transcriptional status during differentiation45. Similar dynamics were recapitulated in hematopoietic stem cells and their more lineage-restricted erythrocyte progenitors115 (depicted in Fig. 2). Additionally, our recent epigenomic data on various T helper cell subsets demonstrated that signature cytokine genes adopted a histone modification pattern consistent with their expression status within their respective lineage upon differentiation, whereas promoters of key transcription factor genes were marked by both H3K4me3 and H3K27me3 even when they were not expressed in the cell116. These bivalently modified transcription factor genes can be induced under appropriate growth conditions and underlie the plasticity of T cells. Altogether, these studies support a recurring theme for histone modifications in restricting multipotency and defining the developmental potential of progenitor cells. These modifications also interact with DNA methylation to reinforce lineage commitment as was observed in neuronal progenitors114.
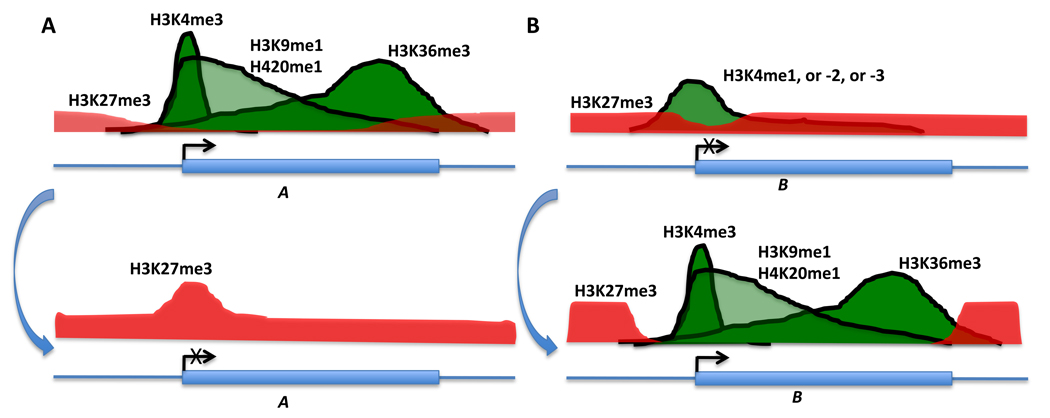
Figure 2. Dynamics of histone modification patterns in association with changes in gene expression during differentiation.
During the differentiation of the multipotent CD34+/CD133+ hematopoietic stem cells (HSCs) into more lineage-restricted CD36+/CD34− erythrocyte precursor cells, dramatic changes of gene expression are accompanied by changes of histone modifications. In general, upon differentiation, histone modifications associated with gene silencing (e.g. H3K27me3) and active gene expression (e.g. H3K4me3, H3K36me3, H3K9me1, and H4K20me1) are gained and lost, respectively, in down-regulated genes (A, e.g. CD34), whereas the opposite is true for up-regulated genes even when the gene promoter region harbors activating histone marks (e.g. H3K4me1) and silencing marks such as H3K27me1 (B, e.g. CD36). Differentiation of CD34+ to CD36+ cells is depicted from top to bottom (blue arrow) for both A and B. Other loci experienced similar changes of these and other histone marks including genes in the HOX cluster, transcription factors Gata1 and Gata3, and other lineage-specific genes115.
Epigenome, Environment, and Disease
During and following the programmed changes to the mammalian epigenome in development, epigenetic processes respond to extrinsic factors including environment, diet, and even behavior. This plasticity is thought to allow the organism to respond and adapt quickly to external stimuli, yet also confer the organism, and even in some cases its offspring, with the ability to ‘memorize’ contacts with such stimuli into adulthood117, 118. Recent studies in mammalian systems provide explicit examples of this metastability that contrasts with the neo-Darwinian concept of inheritance119. In particular, evidence that fetal environment influences epigenetic processes that subsequently alter susceptibility to chronic disorders are mounting. These results have important implications for human health and provide further impetus to map human epigenomes.
Perhaps the most compelling initial data demonstrating that epigenetic modifications are sensitive to environmental stimuli comes from studies of so-called metastable epialleles identified in mice. As the name suggests, metastable epialleles are alleles that are variably expressed due to epigenetic modifications that are established in early development, giving rise to heritable phenotypic mosaicism between cells and between individuals in the absence of genetic heterogeneity120. So far, such alleles include murine Avy, AxinFu, and CabpIAP, all of which are associated with contraoriented IAP insertions whose activities are regulated by DNA methylation and influence gene expression121. DNA methylation levels at these IAP elements vary stochastically and depend upon maternal nutrition and environmental exposures during early development, contributing directly to variable expressivity and phenotypic variegation122, 123. For instance, agouti dams fed a diet high in folic acid and betaine, which are metabolized into methyl donors required for the catalytic processes of DNA and histone methylation, shifts the coat color distribution of offspring from yellow toward brown and decreases their risk for obesity by increasing DNA methylation near the viable yellow agouti (Avy) locus124. Maternal dietary supplementation with genistein found in soy produced similar results125. Surprisingly, opposite results were observed in offspring of dams fed a diet comprising an endocrine active compound bisphenol A (BPA)126, linking the impact of environmental exposure to xenobiotics to potentially deleterious consequences on the epigenome and health of mammals. Indeed, fetal exposure to xenobiotics alters epigenetic states in the offspring that produces deleterious affects on the health of the offspring and on the health of their descendants through transgenerational transmission of the epigenetic (i.e. DNA methylation) perturbation in the germline127, 128. Intriguingly, BPA-induced hypomethylation in the fetal epigenome could be abolished by maternal dietary supplementation with methyl donors, demonstrating that changes in diet can offset the potentially deleterious effects of environmental toxicants on the developing fetus. How this works at the molecular level is still not quite understood, but other examples of the plasticity of the fetal epigenome abound and are not necessarily restricted to metastable epialleles.
Animal models mimicking human pathology by alterations in maternal nutrition during pregnancy and weaning have consistently demonstrated significant responses on epigenetic modifications to chromatin and subsequent effects of offspring phenotype. Of particular interest are models recapitulating hypertension, which is a major risk factor for cardiovascular and cerebrovascular disease129 and to which genetic and environmental factors clearly contribute130. In rodents, subjecting mothers to relative undernutrition during pregnancy may predispose the developing offspring to cardiovascular disease later in life131. For example, uteroplacental insufficiency reduced the activity of DNMT1 in the kidneys of offspring, causing hypomethylation of the p53 promoter and increased p53 expression, presumably contributing to elevated p53-mediated apoptosis thereby reducing glomeruli number and causing hypertension132. In primates, a maternal high-fat diet contributes to increased histone H3 acetylation and decreased histone deacetylase activity consistent with increased expression of genes relevant to impaired lipid metabolism in the fetus133. These and other similar studies unequivocally show that maternal nutrition environment can significantly impact the fetal epigenome contributing directly to the health of the offspring.
Although maternal nutrition contributes to epigenetic modifications of the developing embryo in utero, maternal behavior can alter epigenetic patterns of gene expression in young neonates that, once established, also persists into adulthood. So far, examples of this phenomenon are limited to studies of rodents. Rat offspring of mothers with low pup licking and grooming (LG) behavior showed high levels of DNA methylation of the hippocampal glucocorticoid receptor (GR) gene promoter, concomitant with low expression levels of the associated gene, and exhibited poor response to stress later in life, whereas the opposite was observed in offspring whose mothers displayed more frequent LG behavior134, 135. Additionally, cross-fostering experiments, where pups of low LG mothers were swapped with pups of high LG mothers and vice versa, convincingly revealed that the offspring GR methylation pattern was mediated by maternal behavior, rather than genetics, in the early postnatal environment during a critical window of postnatal development. That is, the DNA methylation pattern of the GR promoter was exclusively labile during the first week of birth, but once established remained unchanged into adulthood134, 135. Whether maternal phenotype contributes to epigenetic and phenotypic variability of offspring in humans, however, has yet to be directly determined.
Taken together, results from these studies lead to the conclusion that epigenetic modifications caused by the environment of the organism at particularly vulnerable or epigenetically labile periods of development are involved in the etiology of adult disease, which may be easily prevented with dietary supplementation at these critical developmental windows. Importantly, in specific cases, the induced metastable epigenetic alteration is transgenerationally heritable. A priori knowledge as to which epigenetically labile loci are associated with disease outcome in adulthood is essential for preventative or therapeutic intervention. This can be achieved by epigenome mapping studies in development.
Relevance to Human Disease
The above results from animal models are relevant to humans and are entirely compatible with the developmental origins of disease hypothesis postulating that nutrition and other environmental factors during human prenatal and early postnatal development influence ‘developmental plasticity136 and alter susceptibility to disease including adult cardiovascular disease, type 2 diabetes, and obesity137, 138. The role of epigenetic variability in contributing to disease susceptibility in humans has largely been unexplored. Yet results of epidemiological studies implicate epigenetic mechanisms in the etiology of disease.
Analogous to rodent studies of maternal behavior and offspring’s response to stress134, 135, the severity of symptoms arising from post-traumatic stress disorder (PTSD) that influences maternal cortisol production during pregnancy can predict the levels of cortisol excretion in babies, while women who experience severe stress during pregnancy give birth to offspring that experience altered activity of the hypothalamic-pituitary-adrenal (HPA) axis, which is regulated by the GR gene, in childhood139, 140. That low offspring cortisol levels are significantly associated with maternal PTSD implicate the involvement of epigenetic mechanisms in mediating this response. Also, consistent with the described animal studies supporting a major role of the intrauterine environment contributing to the health of progeny via epigenetic alterations, lower birth weight in African American populations, in particular, predicts higher blood pressure, elevated cortisol reactivity, and early signs of diabetes as children141, 142 and other related cardiovascular conditions as adults143. Additionally, studies of multigenerational trends of birth outcomes suggest a potentially non-genetic mode of inheritance since maternal fetal growth rate predicts that of their offspring144, 145. Together these and other epidemiological data provide preliminary support that environmental influences impacting on one generation, a likely source of non-genetic variability, can have lasting effects on the phenotype of subsequent generations thereby contributing to the inheritance of disease risk.
In cancer, substantial alterations of DNA methylation and histone modifications have been described. Global hypomethylation and site-specific gene hypermethylation of DNA are so widespread that they are now considered hallmarks of cancer146 and environmental exposure to carcinogens have been linked to these alterations147, 148. Importantly, changes in the epigenome that alter the expression of MLH1 and MSH2, tumor-suppressor mismatch repair genes, have been shown to be inherited through the germline149, 150. Also, loss of imprinting, associated with aberrant DNA methylation of critical cis-regulatory elements, of IGF2 is associated with increased cancer risk in children with Beckwith-Wiedemann syndrome151 and is associated with increased cancer incidence and family history of cancer in adults152, 153. These studies demonstrate that altered epigenetic processes might be causally linked to tumorigenesis. Indeed, animal models with deficient DNMT1 are highly susceptible to cancer development154, 155. The molecular mechanism(s) underlying this susceptibility requires further elucidation, however there is evidence linking the bioavailability of methyl donors to cancer risk either through diet and/or variants in the activity of enzymes involved in one-carbon metabolism156.
The hypothesis that epigenetic processes are key mediators that integrate and interpret environmental factors and regulate the expression of target genes involved in human disease is gaining momentum and has also been reviewed elsewhere7, 117, 157, 158. Persistent epigenetic variability that occurs early in development in response to maternal nutrition and the environment are associated with disease susceptibility, at least in animal models. Incorporating epigenetic variation into genetic studies of common diseases would be a powerful approach that could effectively take into account confounding factors such as environment that sequence-based studies of disease do not7. Such studies will require epigenome mapping.
Epigenome-wide association studies of disease
Genomic studies attempting to identify susceptibility genes for diseases, particularly those involving complex systems and traits, are technically demanding and most often yield poor results primarily due to the confounding influences of variables, such as environment, that are not directly accounted for159. Epigenome mapping provides a measurable readout of most extrinsic and intrinsic factors impacting gene expression variability related to phenotype7. Thus, integrating DNA sequence, transcriptome, and copy number variation information with epigenome mapping data will provide a powerful approach, called epigenome-wide association studies (eGWAS), towards comprehensively understanding disease. Indeed, accounting for non-genetic factors improves the statistical power of studies measuring gene expression variability in populations160. Furthermore, transcriptional variability can be inherited, presumably by epigenetics161. To complement proposed theoretical frameworks7, 162 and ongoing eGWAS efforts163, 164 for integrating genetic and epigenetic information on a genome-wide scale, we briefly highlight three feasible additional applications of eGWAS given current technology.
Genome-wide association studies (GWAS) using epigenetic modifications can increase statistical power to detect disease-causing genomic loci. In conventional GWAS, a large number of single nucleotide polymorphisms (SNPs), approximately one million, are genotyped in thousands of cases and controls, and are statistically tested one-by-one for an association with the disease status165. Due to a large number of statistical tests performed, the resulting p-values must be adjusted for multiple testing. Typically, only a few SNPs are found to be significantly associated with a disease, and the cumulative effect size of these SNPs constitutes only a very small fraction of the total expected effect size due to genetic factors166. The missing fraction of the genetic factors that contribute to the disease risk is believed to come from epistatic interactions of multiple genetic loci. In principle, one can attempt an epistatic association analysis with different combinations of SNPs. However, this becomes exponentially infeasible both computationally and statistically. As described in the previous sections, extrinsic signals are transmitted through intracellular transduction networks, which in turn are fed into gene regulatory cascades that impinge on the chromatin state via various epigenetic modifications (Fig. 3). Thus, the epigenomic variations caused by epistatic interactions of multiple genetic loci may be identified through epigenome mapping, which could pinpoint the affected regions and thus increase the statistical power of capturing the complex disease-causing epistatic interactions associated with genetic and environmental factors.
Epigenetic modifications could serve as proxies for unknown environmental factors associated with disease. Since there can be multiple unknown environmental factors that predispose to disease, it can be difficult to profile all possible factors for association with the disease status. The environment influences epigenetic states and thus these states are inherently quantifiable variables that can be used as proxies for diverse unknown disease predisposing environmental factors. Mapping these observable states would enable conventional DNA sequence-based studies to take into account the many unknown environmental variables that otherwise confound such studies.
Epigenetic modifications are observable marks of functional regulatory elements in the genome. It is widely believed that the majority of disease susceptibility loci for common diseases are in non-coding functional regulatory elements of the genome. Indeed, population studies have focused on identifying cis-acting regulatory variation that might associate with gene expression variability and disease susceptibility167. Therefore, the task of finding such regulatory elements is paramount. Computational methods for regulatory element identification based on DNA sequence alone are plagued by high false positive rates168. One of the reasons for the failure of DNA sequence-based methods for detecting regulatory elements is that regulatory information is non-locally encoded in linear DNA sequence. An epigenome integrates non-local information into local states. Indeed, it was observed that there are some regions in the genome, including known enhancer elements, where histone modification patterns are conserved between humans and mice while the underlying local DNA sequence is not96, 102. Complex interactions of regulatory elements in the genome with the local chromatin environment and the gene regulatory network create distinct chromatin states in the vicinity of these elements. As we have discussed, histone modification patterns have been used for predictions of functional elements101, 102, 169, thus epigenome mapping studies will further aid in the identification of functional regulatory sequences throughout the genome which may provide insight into disease.
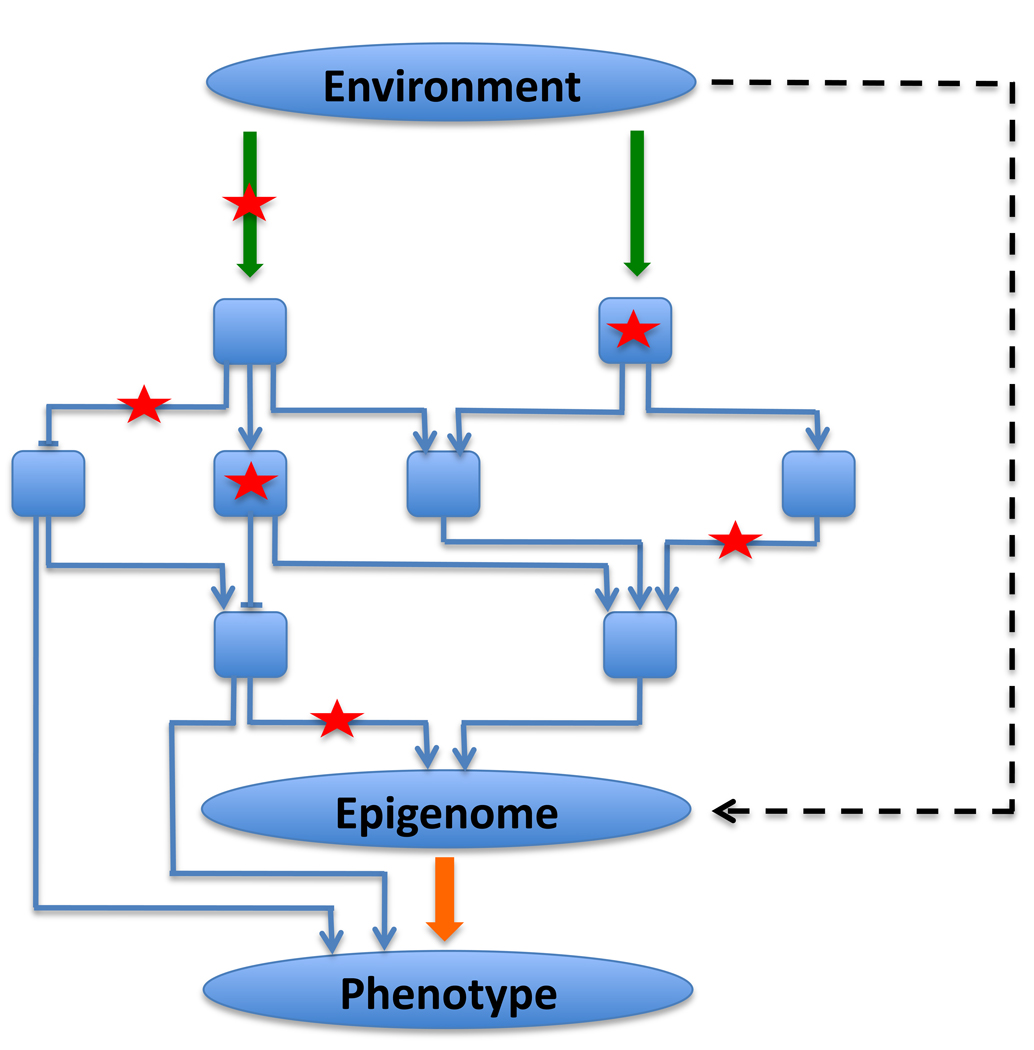
Figure 3. Integrating epigenome information in studies of disease.
An epigenome can be thought of as a sensor of perturbations in the environment, in signal transduction, and in the gene regulatory networks within cells. DNA sequence variations in various components of these systems are shown as red stars. Sequence variations in genes coding for proteins and enzymes of signal transduction pathways (green arrows) and in genes coding for transcription factors (blue boxes) can modulate signaling output from these systems. Sequence variants in cis-regulatory elements (blue lines) can also modulate the signaling output from gene regulatory networks. Although individually subtle and weak, these sequence variants may have a large cumulative combinatorial effect on the epigenome. Thus, an epigenome may integrate epistatic interactions of many genetic loci. Perturbations in environment can affect epigenomes directly (black dotted arrow) or through signaling and gene regulatory cascades that impinge on the chromatin states. Phenotype/disease status of an organism is an outcome of complex interactions between genotype and environment. An epigenome is an intermediate phenotype that is causally ‘closer’ to environmental and genetic factors and thus can provide very useful information for studies attempting to identify disease susceptibility factors. In addition, environmental signals can regulate gene expression levels through post-translational modifications of proteins involved in cell signaling and gene regulation without impacting DNA sequence or epigenetic marks, bypassing the epigenome to directly influence gene expression and phenotype.
Future perspective
There is now precedence of inter-individual epigenetic variability4, 170, which may influence disease susceptibility7, 86. In fact, three classes of epigenetic variation and examples of each have been described119. As we have discussed, tools for detecting global epigenetic diversity are already in place, with many of them continually being improved upon. We expect that a large number of normal and disease epigenomes will be determined soon, as the IHEC is currently underway13. Such efforts will provide valuable resources for enabling epigenome-wide association studies. One of the challenges we will face is how to integrate this vast amount of epigenomic data with other datasets that include gene expression, DNA sequence, and even high-throughout genome-wide 3-dimentional chromatin structural information171, 172 in order to extract useful biological insights. This may be resolved as the genome-wide techniques continuously improve in sensitivity, throughput, and cost-efficiency in addition to the availability and standardization of relevant computational tools. Epigenome mapping in normal and disease states will revolutionize our understanding of normal development, provide key mechanistic insights into the processes of cellular differentiation and gene regulatory networks, and, importantly, enhance our knowledge of the epigenetic contribution to disease, thereby enabling the discovery of novel diagnostic, preventative, and treatment strategies.
Acknowledgments
Sources of Funding
This work was supported by the Division of Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Non-Standard Abbreviations and Acronyms
- eGWAS
epigenome-wide association studies
- IHEC
International Human Epigenome Consortium
- ICR
imprinting control region
- XCI
X-chromosome inactivation
- CpG
cytosine-guanine dinucleotide
- DNMTs
DNA methyltranferases
- ICF
immunodeficiency, centromeric instability, facial anomalies
- ATRX
alpha-thalassemia, mental retardation
- HAT
histone acetyltransferases
- HDAC
histone deacetylases
- PRC2
polycomb repressive complex 2
- RRBS
reduced representation bisulfite sequencing
- BSPP
bisulfite padlock probe
- MeDIP
methylated DNA immunoprecipitation
- MeDIP-Seq
methylated DNA immunoprecipitation with massively parallel DNA sequencing
- MBD
methyl-binding domain
- Methyl-MAPS
methylation mapping analysis by paired-end sequencing
- ES
embryonic stem (cells)
- AIMS
amplification of intermethylated sites
- ChIP
chromatin immunoprecipitation
- ChIP-Seq
chromatin immunoprecipitation with massively parallel DNA sequencing
- ChIP-chip
chromatin immunoprecipitation with microarray technology
- IAP
intercisternal A particle
- BPA
bisphenol A
- LG
licking and grooming
- PTSD
post-traumatic stress disorder
- HPA
hypothalamic-pituitary-adrenal axis
- GWAS
genome-wide association studies
- SNPs
single-nucleotide polymorphisms
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Subject Codes
[141] Functional genomics, [142] Gene expression, [143] Gene regulation
Disclosures
None.
References
- 1.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Jirtle RL, Sander M, Barrett JC. Genomic imprinting and environmental disease susceptibility. Environ Health Perspect. 2000;108:271–278. doi: 10.1289/ehp.00108271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol. doi: 10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turan N, Katari S, Coutifaris C, Sapienza C. Explaining inter-individual variability in phenotype: is epigenetics up to the challenge? Epigenetics. 5:16–19. doi: 10.4161/epi.5.1.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr Opin Genet Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbott A. Project set to map marks on genome. Nature. 463:596–597. [PubMed] [Google Scholar]
- 14.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 16.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prothero KE, Stahl JM, Carrel L. Dosage compensation and gene expression on the mammalian X chromosome: one plus one does not always equal two. Chromosome Res. 2009;17:637–648. doi: 10.1007/s10577-009-9063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark SJ, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 19.Lande-Diner L, Zhang J, Ben-Porath I, Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G, Cedar H. Role of DNA methylation in stable gene repression. J Biol Chem. 2007;282:12194–12200. doi: 10.1074/jbc.M607838200. [DOI] [PubMed] [Google Scholar]
- 20.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 21.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 22.Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 24.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 25.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 26.Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 27.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 28.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 30.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 31.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 32.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 33.Murrell A, Heeson S, Bowden L, Constancia M, Dean W, Kelsey G, Reik W. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2001;2:1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 37.Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer. 2005;41:2381–2402. doi: 10.1016/j.ejca.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 39.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 40.Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Curr Opin Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- 41.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 42.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. Embo J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 44.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golebiewska A, Atkinson SP, Lako M, Armstrong L. Epigenetic landscaping during hESC differentiation to neural cells. Stem Cells. 2009;27:1298–1308. doi: 10.1002/stem.59. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 48.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 49.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 50.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 51.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 52.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 54.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 55.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 56.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuddapah S, Barski A, Cui K, Schones DE, Wang Z, Wei G, Zhao K. Native chromatin preparation and Illumina/Solexa library construction. CSH Protoc. 2009;2009 doi: 10.1101/pdb.prot5237. pdb prot5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 60.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 62.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro R, Weisgras JM. Bisulfite-catalyzed transamination of cytosine and cytidine. Biochem Biophys Res Commun. 1970;40:839–843. doi: 10.1016/0006-291x(70)90979-4. [DOI] [PubMed] [Google Scholar]
- 64.Hayatsu H, Wataya Y, Kazushige K. The addition of sodium bisulfite to uracil and to cytosine. J Am Chem Soc. 1970;92:724–726. doi: 10.1021/ja00706a062. [DOI] [PubMed] [Google Scholar]
- 65.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R, Rehli M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 74.Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, Gingrich JA, Haghighi F, Nutter R, Bestor TH. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010 doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suzuki MM, Kerr AR, De Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007;17:625–631. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004 doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 79.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 81.Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, Albertson DG, Fridlyand J, Mao JH, Shchors K, Weiss WA, Costello JF. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- 83.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, Paterson AD, Popendikyte V. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29:169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- 86.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernstein BE, Humphrey EL, Liu CL, Schreiber SL. The use of chromatin immunoprecipitation assays in genome-wide analyses of histone modifications. Methods Enzymol. 2004;376:349–360. doi: 10.1016/S0076-6879(03)76023-6. [DOI] [PubMed] [Google Scholar]
- 91.Falk J. Using ChIP-based technologies to identify epigenetic modifications in disease-relevant cells. IDrugs. 13:169–174. [PubMed] [Google Scholar]
- 92.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 94.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 95.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 97.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 98.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 102.Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chatterjee C, Muir TW. Chemical approaches for studying histone modifications. J Biol Chem. 285:11045–11050. doi: 10.1074/jbc.R109.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mattout A, Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr Opin Cell Biol. doi: 10.1016/j.ceb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 110.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 111.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Atkinson SP, Koch CM, Clelland GK, Willcox S, Fowler JC, Stewart R, Lako M, Dunham I, Armstrong L. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells. 2008;26:1174–1185. doi: 10.1634/stemcells.2007-0497. [DOI] [PubMed] [Google Scholar]
- 113.Sanz LA, Chamberlain S, Sabourin JC, Henckel A, Magnuson T, Hugnot JP, Feil R, Arnaud P. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008;27:2523–2532. doi: 10.1038/emboj.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 115.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49:4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- 118.Morgan DK, Whitelaw E. The case for transgenerational epigenetic inheritance in humans. Mamm Genome. 2008;19:394–397. doi: 10.1007/s00335-008-9124-y. [DOI] [PubMed] [Google Scholar]
- 119.Richards EJ. Inherited epigenetic variation--revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 120.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 121.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 123.Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136:1706S–1710S. doi: 10.1093/jn/136.6.1706S. [DOI] [PubMed] [Google Scholar]
- 124.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl. 2006;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He FJ, MacGregor GA. Cost of poor blood pressure control in the UK: 62,000 unnecessary deaths per year. J Hum Hypertens. 2003;17:455–457. doi: 10.1038/sj.jhh.1001581. [DOI] [PubMed] [Google Scholar]
- 130.Staessen JA, Wang J, Bianchi G, Birkenhager WH. Essential hypertension. Lancet. 2003;361:1629–1641. doi: 10.1016/S0140-6736(03)13302-8. [DOI] [PubMed] [Google Scholar]
- 131.Bertram CE, Hanson MA. Animal models and programming of the metabolic syndrome. Br Med Bull. 2001;60:103–121. doi: 10.1093/bmb/60.1.103. [DOI] [PubMed] [Google Scholar]
- 132.Pham TD, MacLennan NK, Chiu CT, Laksana GS, Hsu JL, Lane RH. Uteroplacental insufficiency increases apoptosis and alters p53 gene methylation in the full-term IUGR rat kidney. Am J Physiol Regul Integr Comp Physiol. 2003;285:R962–R970. doi: 10.1152/ajpregu.00201.2003. [DOI] [PubMed] [Google Scholar]
- 133.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE. Developmental plasticity and human health. Nature. 2004;430:419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 137.Barker DJ. Intrauterine programming of coronary heart disease and stroke. Acta Paediatr Suppl. 1997;423:178–182. doi: 10.1111/j.1651-2227.1997.tb18408.x. discussion 183. [DOI] [PubMed] [Google Scholar]
- 138.Barker DJ. The developmental origins of chronic adult disease. Acta Paediatr Suppl. 2004;93:26–33. doi: 10.1111/j.1651-2227.2004.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 139.Yehuda R, Teicher MH, Seckl JR, Grossman RA, Morris A, Bierer LM. Parental posttraumatic stress disorder as a vulnerability factor for low cortisol trait in offspring of holocaust survivors. Arch Gen Psychiatry. 2007;64:1040–1048. doi: 10.1001/archpsyc.64.9.1040. [DOI] [PubMed] [Google Scholar]
- 140.Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res. 2008;167:121–135. doi: 10.1016/S0079-6123(07)67009-5. [DOI] [PubMed] [Google Scholar]
- 141.Covelli MM. The relationship of blood pressure and cortisol reactivity to family history of hypertension of African American adolescents. J Cardiovasc Nurs. 2006;21:347–353. doi: 10.1097/00005082-200609000-00004. [DOI] [PubMed] [Google Scholar]
- 142.Li C, Huang TK, Cruz ML, Goran MI. Birth weight, puberty, and systolic blood pressure in children and adolescents: a longitudinal analysis. J Hum Hypertens. 2006;20:444–450. doi: 10.1038/sj.jhh.1002021. [DOI] [PubMed] [Google Scholar]
- 143.Fan Z, Lipsitz S, Egan B, Lackland D. The impact of birth weight on the racial disparity of end-stage renal disease. Ann Epidemiol. 2000;10:459. doi: 10.1016/s1047-2797(00)00105-8. [DOI] [PubMed] [Google Scholar]
- 144.Lumey LH. Decreased birthweights in infants after maternal in utero exposure to the Dutch famine of 1944–1945. Paediatr Perinat Epidemiol. 1992;6:240–253. doi: 10.1111/j.1365-3016.1992.tb00764.x. [DOI] [PubMed] [Google Scholar]
- 145.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 146.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 147.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 148.Sutherland JE, Costa M. Epigenetics and the environment. Ann N Y Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 149.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 150.Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 151.DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am J Hum Genet. 2002;70:604–611. doi: 10.1086/338934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 153.Woodson K, Flood A, Green L, Tangrea JA, Hanson J, Cash B, Schatzkin A, Schoenfeld P. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J Natl Cancer Inst. 2004;96:407–410. doi: 10.1093/jnci/djh042. [DOI] [PubMed] [Google Scholar]
- 154.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 155.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 156.Giovannucci E. Alcohol, one-carbon metabolism, and colorectal cancer: recent insights from molecular studies. J Nutr. 2004;134:2475S–2481S. doi: 10.1093/jn/134.9.2475S. [DOI] [PubMed] [Google Scholar]
- 157.Gluckman PD, Hanson MA, Beedle AS. Non-genomic transgenerational inheritance of disease risk. Bioessays. 2007;29:145–154. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- 158.Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 159.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 160.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Simons JW. Epigenetic hereditary transcription profiles III, evidence for an epigenetic network resulting in gender, tissue and age-specific variation in overall transcription. Biol Direct. 2009;4:37. doi: 10.1186/1745-6150-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 163.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 86:411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 166.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pastinen T, Ge B, Hudson TJ. Influence of human genome polymorphism on gene expression. Hum Mol Genet. 2006;15(Spec No 1):R9–R16. doi: 10.1093/hmg/ddl044. [DOI] [PubMed] [Google Scholar]
- 168.Siggia ED. Computational methods for transcriptional regulation. Curr Opin Genet Dev. 2005;15:214–221. doi: 10.1016/j.gde.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 169.Won KJ, Chepelev I, Ren B, Wang W. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics. 2008;9:547. doi: 10.1186/1471-2105-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A. Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, Wei CL, Ruan Y, Sung WK. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]