Abstract
A tetrahydroquinoline oxocarbazate (PubChem CID 23631927) was tested as an inhibitor of human cathepsin L (EC 3.4.22.15) and as an entry blocker of severe acute respiratory syndrome (SARS) coronavirus and Ebola pseudotype virus. In the cathepsin L inhibition assay, the oxocarbazate caused a time-dependent 17-fold drop in IC50 from 6.9 nM (no preincubation) to 0.4 nM (4-h preincubation). Slowly reversible inhibition was demonstrated in a dilution assay. A transient kinetic analysis using a single-step competitive inhibition model provided rate constants of kon = 153,000 M−1s−1 and koff = 4.40 × 10−5 s−1 (Ki = 0.29 nM). The compound also displayed cathepsin L/B selectivity of >700-fold and was nontoxic to human aortic endothelial cells at 100 μM. The oxocarbazate and a related thiocarbazate (PubChem CID 16725315) were tested in a SARS coronavirus (CoV) and Ebola virus-pseudotype infection assay with the oxocarbazate but not the thiocarbazate, demonstrating activity in blocking both SARS-CoV (IC50 = 273 ± 49 nM) and Ebola virus (IC50 = 193 ± 39 nM) entry into human embryonic kidney 293T cells. To trace the intracellular action of the inhibitors with intracellular cathepsin L, the activity-based probe biotin-Lys-C5 alkyl linker-Tyr-Leu-epoxide (DCG-04) was used to label the active site of cysteine proteases in 293T lysates. The reduction in active cathepsin L in inhibitor-treated cells correlated well with the observed potency of inhibitors observed in the virus pseudotype infection assay. Overall, the oxocarbazate CID 23631927 was a subnanomolar, slow-binding, reversible inhibitor of human cathepsin L that blocked SARS-CoV and Ebola pseudotype virus entry in human cells.
Cathepsin L is one of the 11 members of human lysosomal cysteine proteases (i.e., B, C, F, H, K, L, O, S, V, W, and X) that fall in the C1 family (papain family) of the CA clan (Rossi et al., 2004). These enzymes were traditionally linked to nonspecific proteolytic activity within lysosomes. More recently, cathepsin L has been implicated in regulatory events relating to cancer, diabetes, immunological responses, degradation of the articular cartilage matrix, and other pathological processes (Chapman et al., 1997; Turk and Guncar, 2003; Maehr et al., 2005; Vasiljeva et al., 2007), including osteoporosis, rheumatoid arthritis, and tumor metastasis (McGrath, 1999; Turk et al., 2001; Potts et al., 2004; Schedel et al., 2004; Palermo and Joyce, 2008). Inhibitors of cathepsin L block viral entry of severe acute respiratory syndrome coronavirus (SARS-CoV) and Ebola virus and impair conversion of Hendra virus glycoprotein into the mature, active form (Chandran et al., 2005; Pager and Dutch, 2005; Simmons et al., 2005). With respect to the development of antiviral agents, inhibitors of human cathepsin L are not subject to resistance because of rapid mutations of the viral genome. Cathepsin L is therefore an attractive target for drug development.
In 2002, SARS-CoV infection resulted in 8096 cases of infection and 774 deaths (Enserink, 2003). After virus uptake following angiotensin-converting enzyme 2 receptor binding, cathepsin L-mediated proteolysis induces conformational changes in the SARS-CoV S glycoprotein to trigger the endosomal membrane fusion process (Simmons et al., 2005). Likewise, cleavage of the Ebola glycoprotein by cathepsin L reveals the receptor interaction domain within GP, and cathepsin L activity is required for entry into the host cell (Chandran et al., 2005; Schornberg et al., 2006; Lee et al., 2008). The Ebola virus causes fatal hemorrhagic fever. The first outbreaks were reported in Sudan and Zaire in 1976 (Feldmann et al., 2003). Inhibition of cathepsin L thus holds promise for therapeutic intervention for both SARS-CoV and Ebola virus infection.
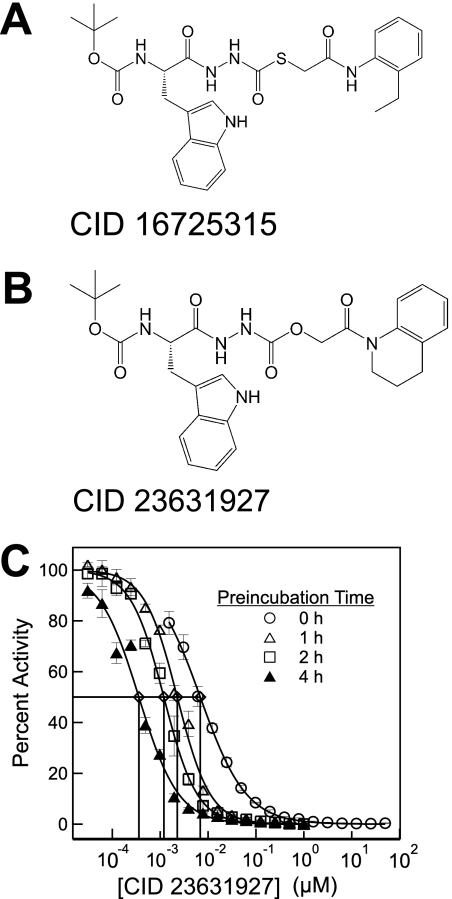
High throughput screening for cathepsin L inhibitors first identified [(S)-2-{N′-[(2-ethyl-phenylcarbamoyl)- methylsulfanylcarbonyl]-hydrazino}-1-(1H-indol-3-ylmethyl)-2-oxo-ethyl]-carbamic acid tert-butyl ester (PubMed CID 16725315; Fig. 1A) as a novel thiocarbazate compound exhibiting potent inhibition against cathepsin L (Myers et al., 2008a; Shah et al., 2008). Molecular docking simulation of the thiocarbazate against papain structure revealed key contacts for this ligand-protein interaction. (Beavers et al., 2008) Addition medicinal chemistry efforts resulted in the synthesis of an even more potent tetrahydroquinoline oxocarbazate compound, N′-[(S)-2-tert- butoxycarbonylamino-3-(1H-indol-3-yl)-propionyl]-hydrazinecarboxylic acid 2-(3,4-dihydro-2H-quinolin-1-yl)-2-oxo-ethyl ester (PubMed CID 23631927; Fig. 1B) (Beavers et al., 2008; Myers et al., 2008a,b).
Fig. 1.
A, thiocarbazate CID 16725315. B, oxocarbazate CID 23631927. C, activity of CID 23631927 against human cathepsin L after preincubation with the enzyme target for 0 (○), 1 (△), 2 (□), and 4 h (▲).
In this article, a full kinetic characterization of the oxocarbazate inhibitor will be reported, along with a measure of the potency in blocking entry of SARS-CoV and Ebola virus pseudotypes. This compound displayed an IC50 of ∼200 and 300 nM in preventing 293T cell infection by the Ebola and SARS-CoV pseudotypes, respectively. Labeling of intracellular active cathepsin L using an activity based probe, DCG-04, permitted quantitative estimation of the interaction of cathepsin L inhibitors with the target enzyme in HEK 293T cells.
Materials and Methods
Cathepsin L Assay.
As described in prior work (Shah et al., 2008), the cathepsin L assay was carried out using 1 μM Z-Phe-Arg-7-amido-4-methylcoumarin (Z-Phe-Arg-AMC; Sigma, St. Louis, MO) and 8.7 ng/ml human liver Cathepsin L (Calbiochem, San Diego, CA). Assay buffer consisted of 20 mM sodium acetate, 1 mM EDTA, and 5 mM cysteine, pH 5.5. The enzyme was incubated in the buffer for 30 min to ensure that the catalytic cysteine was in the reduced form. The enzymatic reaction (25°C) was read on an Envision florescent microplate reader (PerkinElmer Life and Analytical Sciences, Waltham, MA), and the fluorescence signal was measured at the excitation and emission wavelengths of 355 and 460 nm, respectively. CID 23631927 was stored as 10 mM dimethyl sulfoxide (DMSO) aliquots (−80°C) and used fresh in each experiment. Samples of CID 23631927 can be obtained from the National Institutes of Health Molecular Libraries Small Molecule Repository (BioFocus, South San Francisco, CA).
Inhibitor Potency Determination.
A 16-point, 2-fold serial dilution dose-response assay was performed in triplicate. Each well of a 96-well assay plate (Corning Life Sciences, Acton, MA) contained 38 μl of water and 2 μl of inhibitor in DMSO. Positive and negative controls were present to serve as internal controls into which 2 μl of DMSO was transferred in place of the inhibitor. Ten microliters of 10 μM Z-Phe-Arg-AMC in 5× concentrated assay buffer and 50 μl of 17.4 ng/ml cathepsin L in assay buffer were added sequentially to initiate the protease reaction. A total of 50 μl of assay buffer was dispensed in place of enzyme into negative control wells. This resulted in a final dose response concentration range of 50 μM to 1.5 nM inhibitor (2% DMSO) in a 100-μl final reaction volume. A four-parameter logistic model (IDBS XLfit eq. 205; Guildford, Surrey, UK) was used to fit the measured data and find IC50 values.
Time-Dependent Inhibition and Reversibility Test.
Preincubation assays were performed to further establish the time dependence of inhibition. Enzyme and inhibitor were preincubated for various times in a 96-well microplate before the addition of substrate to initiate the enzymatic reaction. A total of 47.5 μl of cathepsin L (18.3 ng/ml) and 47.5 μl of CID 23631927 at various concentrations in assay buffer were incubated up to 4 h. A volume of 5 μl of Z-Phe-Arg-AMC substrate was then added, and the plate was monitored for AMC hydrolysis. A dilution assay protocol was deployed as described previously (Shah et al., 2008) to monitor reversibility. The concentrated mixture of the enzyme (870 ng/ml) and the inhibitor CID 23631927 (25 nM) were incubated for 1 h before being diluted 100-fold in a Corning 96-well plate with assay buffer containing 1 μM Z-Phe-Arg-AMC to a final volume of 200 μl.
Kinetic Data Fitting.
In the kinetic simulations, the concentrations of chemical species ([E], [S], [I], [P], [ES], [EI]) over time were calculated using a system of ordinary differential equations for each reaction step as adopted for the parameter fitting in prior work (Shah et al., 2008). Progress curves at each inhibitor concentration were fit into a five-parameter (k1, k−1, kon, koff, kcat) kinetic inhibition model using MATLAB 7.8.0 (The MathWorks, Inc., Natick, MA). The objective function used in the minimization search was computed as the sum of the square of the relative errors between the simulated values and experimental data.
Selectivity against Cathepsin B.
CID 23631927 was assayed for inhibition against cathepsin B. Human liver cathepsin B (65 ng/ml; Calbiochem, San Diego, CA) was used, and the corresponding substrate was 15 μM Z-Arg-Arg-AMC. The reactions were performed in 20 mM sodium acetate buffer containing 5 mM cysteine and 1 mM EDTA, pH 5.5.
Cytotoxicity.
CID 23631927 was tested for cytotoxicity against human aortic endothelial cells as described previously (Shah et al., 2008). Cells were seeded at 1000 cells in 25 μl per well in a 384-well plate. The plate was centrifuged and incubated at 37°C for 24 h. CID 23631927 and doxorubicin positive control were then serially diluted in EGM-2 endothelial cell media (Lonza Walkersville, Inc., Walkersville, MD). A total of 5 μl of these serial dilutions was added to the wells in triplicate, resulting in final concentrations of compound from 100 μM to 156 nM (0.17% DMSO). The plate was centrifuged and incubated at 37°C for 24 h. A total of 30 μl of CellTiter-Glo (Promega, Madison, WI) was added to each well and centrifuged. After 10 min, luminescence was measured using the Envision microplate reader.
Virus Pseudotype Preparation and Virus Infection Assay.
Pseudovirions were prepared essentially as described previously (Kaletsky et al., 2007). In brief, 293T cells were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum. Pseudotypes were produced by transfecting 293T cells with 10 μg of an HIV-luciferase vector, pNL-luc, and 30 μg of a plasmid encoding the Ebola glycoprotein or SARS Spike proteins (Simmons et al., 2005; Kaletsky et al., 2007). Vesicular stomatitis virus glycoprotein (VSV-G) pseudovirions were prepared as above using 6 μg of a VSV-G-encoding plasmid. Virions were concentrated by ultracentrifugation at 40,000 rpm in a SW41 rotor (Beckman Coulter, Fullerton, CA) through a 20% sucrose cushion for 1 h at 4°C. The pellets were resuspended overnight in phosphate-buffered saline at 4°C. To measure infection, 293T cells were pretreated for 1 h with inhibitors or Z-Val-Phe-CHO (MDL28170; Sigma, St. Louis, MO) as a positive control (Simmons et al., 2005). The media were removed and replaced with the same inhibitors at double the final concentration. An equal volume of pseudotypes was then added, and cells were spin-infected at 1200g for 2 h at 4°C. After spin infection, the cells were incubated for 6 h, and the medium was replaced with fresh medium without drug. Cell lysates were assayed for luciferase activity (Promega) at 40 h after infection.
Labeling Active Cathepsin L in HEK 293T using DCG-04.
The oxocarbazate CID 23631927, the thiocarbazate CID 16725315, MDL28170 (Calbiochem), and cathepsin L inhibitor III (Z-Phe-Tyr(t-Bu)-diazomethylketone; Calbiochem) were dissolved in DMSO to give 10 mM stock solution. Biotin-Lys-C5 alkyl linker-Tyr-Leu-epoxide (DCG-04) was prepared as described previously (Greenbaum et al., 2000). HEK 293T cells were seeded into six-well tissue culture plates (BD Biosciences Discovery Labware, Bedford, MA) at a concentration of 106 cells/well. Inhibitors were added to the well in a 1:1000 dilution to give a final concentration of 10 μM and incubated with the cells for 3 h. The adherent cells were then scrapped, pelleted, and lysed in 100 mM sodium acetate and 0.25% (v/v) Triton X-100, pH 5.5, and subjected to three freeze-thaw cycles, followed by sonication (30 s; Branson Ultrasonics Corporation, Danbury, CT). After centrifugation (13,200 rpm, 10 min, 4°C), the supernatant was incubated with 5 μM cysteine protease probe DCG-04 (Greenbaum et al., 2000) at 4°C overnight. The incubated sample (120 μg of total protein per lane) was mixed with NuPAGE 4× lithium dodecyl sulfate sample buffer (Invitrogen, Carlsbad, CA) and 10× sample reducing agent (Invitrogen) and denatured at 95°C for 3 min. SDS-polyacrylamide gel separation was performed with Xcell SureLock system (Invitrogen) on 10% Bis-Tris gel (Invitrogen) at 125 V for 1.5 h after transfer to a polyvinylidene difluoride membrane at 30 V (1.5 h). The membrane was blocked in casein solution (Vector Laboratories) for 1 h and then incubated with avidin and biotinylated horse-radish peroxidase, followed by washing (4×) and detection with SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA) in an image reader (LAS-3000; Fujifilm, Tokyo, Japan). Band intensity was quantified using the software Multi Gauge 3.0 (Fujifilm). To confirm the position of the Cat-L band, the membrane was later incubated with Western blot stripping buffer (Thermo Fisher Scientific) for 10 min and redetected with anti-Cat-L rabbit polyclonal antibody (Calbiochem) in 1:2000 dilution for 1 h. Goat anti-rabbit IgG peroxidase conjugate (Calbiochem) was used as the secondary antibody (1:8000 dilution). Protein loading was again checked using mouse anti-β-Actin monoclonal antibody (Sigma) with 1:2000 dilution followed by 1:5000 diluted goat anti-mouse IgG peroxidase conjugate (Thermo Fisher Scientific).
Results
Kinetics of Human Cathepsin L Inhibition.
CID 23631927 was found to inhibit cathepsin L with an IC50 of 6.9 ± 1.0 nM when assayed immediately after mixing with the enzyme. Upon preincubation with cathepsin L for a period of 1, 2, or 4 h, the tetrahydroquinoline oxocarbazate demonstrated a time-dependent reduction of IC50 from 2.3 ± 0.1 nM (1 h) to 1.2 ± 0.1 nM (2 h) to 0.4 ± 0.1 nM (4 h) (Fig. 1C), demonstrating a slow on-rate of inhibition.
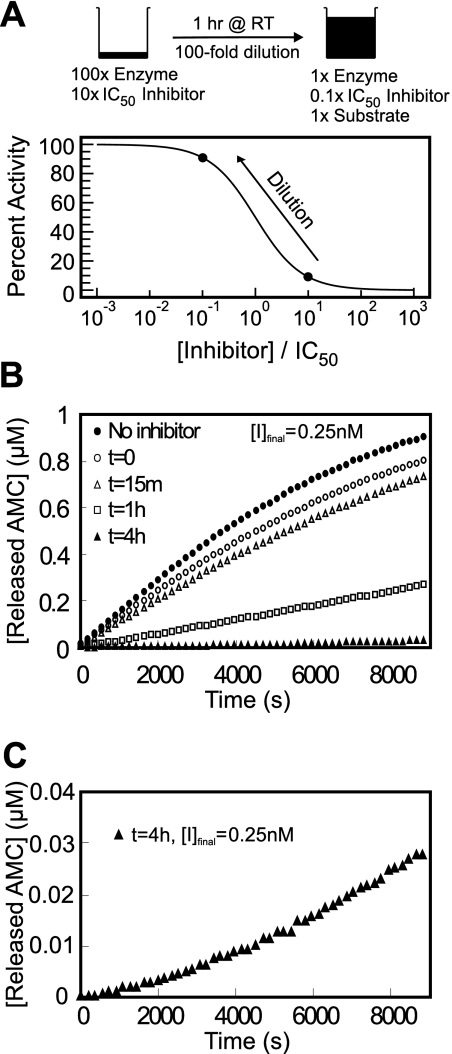
Inhibitor reversibility was evaluated using a preincubation/dilution assay (Copeland, 2005). After preincubation of the enzyme with inhibitor at 10 × IC50, most of the enzyme (>90%) is associated with the inhibitor as shown in Fig. 2A. Upon 100-fold dilution with buffer when the reaction was initiated with substrate, a total of 28% of enzymatic activity returned after 6000 s for the 1-h preincubated reaction. For the case of 4-h preincubation, the substrate cleavage rate after 8820 s was 8.9 times greater than the initial hydrolysis rate (Fig. 2C), indicating that the enzyme-inhibitor complex was very slowly reversible.
Fig. 2.
A, dilution protocol for determination of reversibility: 870 ng/ml cathepsin L and 25 nM inhibitor were combined and incubated at room temperature. B, reversibility data for CID 23631927 after 0 (○), 15 min (△), 1 h (□), and 4 h (▲) of preincubation with cathepsin L and upon 100-fold dilution into assay buffer containing 1 μM Z-Phe-Arg-AMC as substrate, and the final inhibitor concentration was 0.25 nM. A full enzyme-substrate reaction without inhibitor (●) served as a positive control. C, enlarged reaction progress curve with 4-h preincubation of cathepsin L and CID 23631927 to clearly demonstrate the recovering rate of enzymatic cleavage of the substrate.
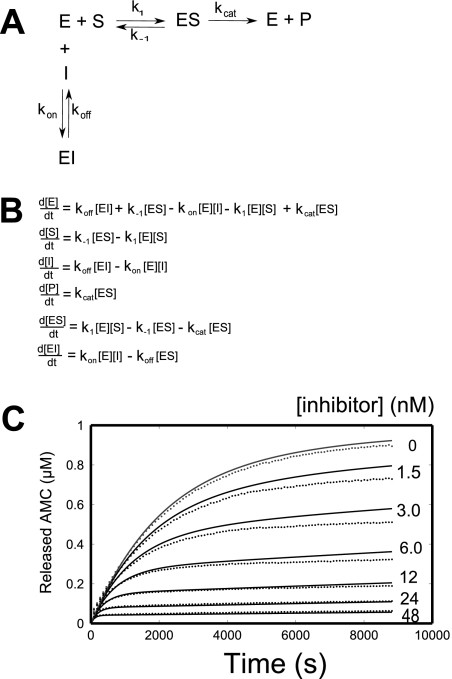
Transient kinetic data analysis was performed on the inhibition data for the oxocarbazate CID 23631927. Given the close structural resemblance to the thiocarbazate CID 16725315, the same inhibition model (Fig. 3, A and B) was used for both inhibitors, as reported previously (Shah et al., 2008). MATLAB code was developed for the optimization, and the fitted parameters for both inhibitors are presented in Table 1 for comparison. The slow on-rate of inhibition is clearly apparent in Fig. 3C, as product formation rate slowly dropped in the assay. The best fit parameters for CID 23631927 were k1 = 2.27 × 106 M−1 · s−1, k−1 = 0.29 s−1, kcat = 3.78 s−1, kon = 153,000 M−1 · s−1, and koff = 4.40 × 10−5 · s−1. The five parameters, k1, k−1, kcat, and koff do not vary substantially between the thiocarbazate (CID 16725315) and the oxocarbazate (CID 23631927) tests. For the two slow on-rate inhibitors, the fitted kinetic parameter kon for CID 23631927 was rate controlling, displaying a rate almost five times greater than that for CID 16725315, resulting in a calculated inhibition constant for the oxocarbazate of Ki = 0.29 nM. This calculated Ki value corresponds well with the IC50 value (0.4 nM) displayed after 4-h preincubation of cathespin L with CID 23631927.
Fig. 3.
A, single-step mechanism for simple, reversible, slow binding inhibition. B, ordinary differential equations governing the single-step mechanism of inhibition. C, nonlinear regression for transient dynamics of CID 23631927 using code written in MATLAB to fit in a five-parameter inhibition kinetic model. Reaction progress curves (●) are shown for 8.7 ng/ml human cathepsin L enzyme and 1 μM Z-Phe-Arg-AMC substrate with varying concentrations of CID 23631927 inhibitor.
TABLE 1.
Comparison of kinetic parameters and inhibition potency data between CID 16725315 and CID 23631927
| Thiocarbazate CID 16725315 | Oxocarbazate CID 23631927 | |
|---|---|---|
| k1 | 3.09 × 106 M−1·s−1 | 2.27 × 106 M−1·s−1 |
| k−1 | 0.32 s−1 | 0.29 s−1 |
| kcat | 4.31 s−1 | 3.78 s−1 |
| kon | 3.27 × 104 M−1·s−1 | 1.53 × 105 M−1·s−1 |
| koff | 3.75 × 10−5 s−1 | 4.40 × 10−5 s−1 |
| Ki = koff/kon | 1.15 nM | 0.29 nM |
| Human Cat L. IC50 | ||
| 0 h | 56 ± 4 nM | 6.9 ± 1.0 nM |
| 4 h | 1.0 ± 0.5 nM | 0.4 ± 0.1 nM |
| SARS IC50 | 273 ± 49 nM | |
| Ebola IC50 | 193 ± 39 nM | |
| Inhibition of intracellular Cat L. | 2% | 38% |
| Cat L/B Selectivity | 40–50 | 735 |
Cathespsin L/B Selectivity and Cytotoxicity.
The IC50 of CID 23631927 against human liver cathepsin B was measured to be 5.072 μM ± 0.883 μM (data not shown) with no preincubation, resulting in a cathepsin L/B selectivity ratio of 735, considerably higher than the 40- to 50-fold selectivity previously reported for the thiocarbazate CID 16725315 (Shah et al., 2008). Prior work with tert-butyl N-[(2S)-1-[2-[2-(2-ethylanilino)-2-oxoethyl]sulfanylcarbonylhydrazinyl]-3-(1H-indol-3-yl)-1-oxopropan-2-yl]carbamate (PubChem SID 26681509) indicated that the selectivity was not a function of time (Shah et al., 2008). It is noteworthy that CID 23631927 proved to be nontoxic to human aortic endothelial cells up to 100 μM (data not shown). Thus, the oxocarbazate inhibitor tert-butyl N-[(2S)-1-[2-[2-(3,4-dihydro-2H-quinolin-1-yl)-2-oxoethoxy]carbonylhydrazinyl]-3-(1H-indol-3-yl)-1-oxopropan-2-yl]carbamate (SID 46493575) is one of the most potent and selective inhibitors of human cathepsin L reported to date.
Inhibition of Virus Pseudotype Infection.
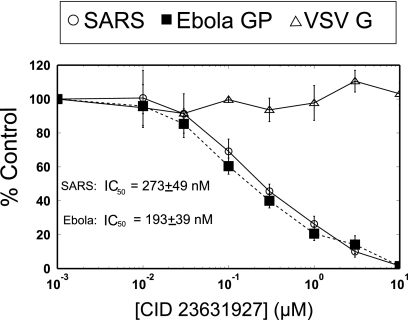
CID 23631927 inhibited entry of both SARS-CoV (IC50 = 273 ± 49 nM) and Ebola virus (IC50 = 193 ± 39 nM) pseudotypes in cellular assays. VSV-G, which does not rely upon cathepsin L activity to infect host cells, was used as a control and was not inhibited by CID 23631927 (Fig. 4). Although CID 23631927 revealed potency in blocking both SARS-CoV and Ebola virus pseudotype entry into 293T cells, CID 16725315 was found to have little inhibition for either SARS-CoV entry (IC50 > 10 μM) or Ebola viral entry (IC50 > 10 μM) (data not shown). This difference was postulated to be the result of differing cell permeability properties of the compounds, thus motivating the assay of intracellular cathepsin L inhibition exploiting a labeling probe.
Fig. 4.
CID 23631927 inhibition of SARS-CoV and Ebola virus pseudotype infections. CID 23631927 inhibits entry into cells of the SARS-CoV pseudotype at IC50 = 273 ± 49 nM and the Ebola virus pseudotype at IC50 = 193 ± 39 nM. Vesicular stomatitis virus glycoprotein pseudovirions (△) were used as a control and were not inhibited.
Inhibition of Intracellular Cathepsin L.
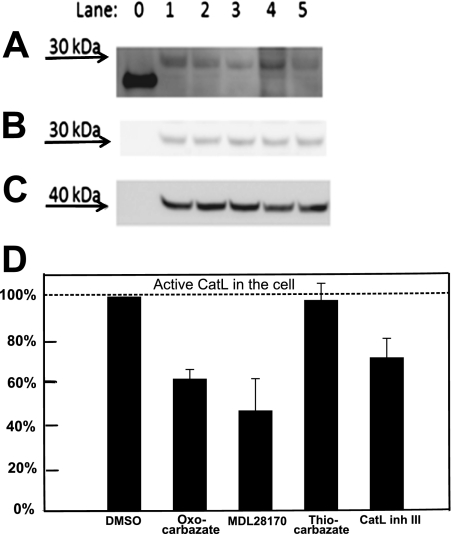
Cells were incubated with four different cathepsin L inhibitors at 10 μM for 3 h to permit time for each compound to permeate through cell membranes and interact with cathepsin L via inhibition of the active site of the enzyme. The chemical probe DCG-04, an epoxide electrophile with biotin tag (Greenbaum et al., 2002), labeled the active cathepsin L at the catalytic site. Any intracellular cathepsin L that had been inhibited during the 3-h incubation would thus be unable to bind the probe DCG-04, especially given the slow reversibility of the carbazate inhibitors. In the Western blot (Fig. 5A), band intensity was quantified with Multi Gauge 3.0 (Fujifilm) and correlated with the active cathepsin L activity remaining in the cell. The cathepsin L band position was verified in Fig. 5B, and protein loading in each lane was checked using β-actin (Fig. 5C). The quantified results were normalized against lane 1 (cells treated with DMSO only) and presented in Fig. 5D. The oxocarbazate inhibitor (CID 23631927) caused a 38% reduction in the active intracellular cathepsin L level compared with lane 1. The commercially available cysteine protease inhibitor MDL28170 revealed an IC50 against cathepsin L of 2.5 nM and IC50 against SARS in the range of 100 nM (Simmons et al., 2005). This positive control employing MDL28170 in lane 3 led to a 53% reduction of cathepsin L activity compared with lane 1. The thiocarbazate inhibitor (CID 16725315), however, did not affect cathepsin L level in the cell, as illustrated in lane 4, potentially indicating that the poor cellular uptake of this inhibitor in turn correlates with the poor activity in the viral entry assay. As a positive control, the cathepsin L inhibitor III (Calbiochem), a diazomethyl ketone (lane 5), led to a 29% reduction of cathepsin L activity compared with lane 1.
Fig. 5.
Western blot to detect amount of intracellular active Cat-L after treatment with 10 μM inhibitors A, intracellular cysteine protease labeled by DCG-04 was detected at approximately 30 kDa in Western blot. Lane 0, papain (21 kDa), a purified cysteine protease served as a positive control for DCG-04 probe. Lane 1, treated with DMSO only without inhibitor; lane 2, oxocarbazate CID 23631927; lane 3, MDL28170; lane 4, thiocarbazate CID 16725315; lane 5, Cat-L inhibitor III. B, this band was subsequently confirmed to be cathepsin L by anti-cathepsin L rabbit polyclonal antibody. C, protein Loading in each lane was checked using β-actin (molecular mass, 42 kDa) as a control. D, statistical analysis of repeated Western blots was done using Multi Gauge 3.0 (Fujifilm). Data are presented as mean ± S.E. (n = 3).
Discussion
The oxocarbazate CID 23631927, designed based on the knowledge of the previously reported thiocarbazate chemotype (Myers et al., 2008b), has been demonstrated to be a subnanomolar (Ki = 0.29 nM), slow-on, slow-off inhibitor of human cathepsin L that blocks SARS-CoV and Ebola pseudotype virus entry in human cells (at IC50 values of 273 and 193 nM , respectively) with minimal in vitro toxicity. Given the >700-fold cathepsin L/B selectivity of the oxocarbazate inhibitor, we suggest that inhibition of cathepsin L alone is sufficient to block Ebola pseudotype virus entry in HEK 293T cells without the need to simultaneously block cathepsin B.
Individual kinetic parameters were estimated through transient kinetic analyses on the reaction progress curves at various inhibitor concentrations. Compared with CID 16725315, CID 23631927 had substantially higher kon values, resulting in a lower Ki. This improved rate of binding between CID 23631927 and cathepsin L may be attributed to the optimized tetrahydroquinoline structure, which has both stronger hydrophobic interactions in the P1′ subsite and a better hydrogen bond network with the receptor enzyme, as observed and predicted in the docked pose for the ligand-enzyme complex (Beavers et al., 2008). It is noteworthy that the oxocarbazate and thiocarbazate inhibitors are likely to be covalent reversible inhibitors, because replacement of the thiocarbazate sulfur in CID 16725315 results in a loss of activity (Myers et al., 2008a). It is quite possible that the mechanism of inhibition involves a tetrahydral intermediate by attack of the active site Cys25 residue on the thiocarbazate carbonyl or oxocarbazate carbonyl. Because it is difficult to predict temperature dependence of the IC50, we directly measured it and found a small shift from IC50 = 5.9 ± 0.6 nM (at 25°C, n = 3) to IC50 = 10.6 ± 0.9 nM (at 37°C, n = 3) (data not shown).
Endosomal cathepsins play important roles in mediating virus entry into host cells for SARS-CoV (Huang et al., 2006) and Ebola virus (Schornberg et al., 2006). In particular, Cathepsin L has been singled out as possibly playing a crucial role in triggering membrane fusion within endosomes via proteolysis (Simmons et al., 2005; Kaletsky et al., 2007). In the present study, cathepsin L inhibitor CID 23631927 was demonstrated to be effective in blocking virus entry in both SARS and Ebola pseudotype infection. This result validates the possibility of preventing the invasion and spread of such diseases using cathepsin L inhibitors and as such establishes this oxacarbazate as a viable candidate for further optimization.
The design of CID 23631927 entailed substituting a tetrahydroquinoline anilide moiety for the 2-ethylphenyl anilide group in CID 16725315. This structural modification improved ligand-protein interaction in the S1′ pocket as demonstrated in molecular docking studies (Beavers et al., 2008). In addition, we hypothesize that entropic considerations contributed to increased potency. Using the absorption, distribution, metabolism, and excretion (ADME) property prediction software Qikprop 3.1 (Schrodinger LLC, New York, NY), we found CID 23631927 to have a predicted log value for the partition coefficient cLogP (octanol/water) of 5.296. This cLogP is higher than that of CID 16725315 (4.724). This estimate of cLogP is based on the condition that the inhibitor is uncharged, a condition that is likely to be upheld because the pKa = −3.6 (Hur and Guven, 2002) for the indole group of the oxocarbazate and thiocarbazate inhibitors. The predicted increase in hydrophobicity may result in higher permeability of CID 23631927 into cells, hence resulting in better potency in both the virus infection assay (Fig. 4) and intracellular inhibition of cathepsin L (Fig. 5). The poor activity of the thiocarbazate in the cell-based viral entry assay may be partially explained by the labeling assay in which thiocarbazate treatment had little effect in blocking intracellular cathepsin L activity. The t-benzyloxycarbonyl group on the inhibitors is a common amino protecting group; we have found that this group prevents self-cleavage of the thiocarbazate (Myers et al., 2008b). We note that this protecting group may be unstable under acidic conditions of the gastrointestinal tract and the oral availability of the compound, with or without the t-benzyloxycarbonyl protecting group, is unknown.
Taken together, the results of this study demonstrating the efficacy of CID 23631927 in blocking virus entry into human cells in conjunction with the structure of CID 23631927 offers both a new starting point for intervention in SARS and Ebola virus infection and a novel probe to explore the mechanism of virus entry.
Acknowledgments
We thank Dr. Rajesh Chandramohanadas for help in DCG-04 activity based probe labeling work.
This work was supported by the National Institutes of Health National Human Genome Research Institute [Grant U54-HG003915] (to S.L.D.) and the National Institutes of Health National Institute of Allergy and Infectious Diseases [Grants T32-AI55400 (to R.L.K.), U01-AI070369 (to P.B.), U54-AI057168 (to P.B.)].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.064261.
- SARS
- severe acute respiratory syndrome
- CoV
- coronavirus
- CID
- PubMed compound identification
- CID 16725315
- ([(S)-2-{N′-[(2-ethyl-phenylcarbamoyl)-methylsulfanylcarbonyl]-hydrazino}-1-(1H-indol-3-ylmethyl)-2-oxo-ethyl]-carbamic acid tert-butyl ester
- DCG-04
- biotin-Lys-C5 alkyl linker-Tyr-Leu-epoxide
- AMC
- 7-amido-4-methylcoumarin
- DMSO
- dimethyl sulfoxide
- VSV-G
- vesicular stomatitis virus glycoprotein
- HEK
- human embryonic kidney
- Cat-L
- cathepsin L
- SID
- PubChem substance identification
- SID 26681509
- tert-butyl N-[(2S)-1-[2-[2-(2-ethylanilino)-2-oxoethyl]sulfanylcarbonylhydrazinyl]-3-(1H-indol-3-yl)-1-oxopropan-2-yl]carbamate
- SID 46493575
- tert-butyl N-[(2S)-1-[2-[2-(3,4-dihydro-2H-quinolin-1-yl)-2-oxoethoxy]carbonylhydrazinyl]-3-(1H-indol-3-yl)-1-oxopropan-2-yl]carbamate.
References
- Beavers MP, Myers MC, Shah PP, Purvis JE, Diamond SL, Cooperman BS, Huryn DM, Smith AB., 3rd (2008) Molecular docking of cathepsin L inhibitors in the binding site of papain. J Chem Inf Model 48:1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. (2005) Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HA, Riese RJ, Shi GP. (1997) Emerging roles for cysteine proteases in human biology. Annu Rev Physiol 59:63–88 [DOI] [PubMed] [Google Scholar]
- Copeland RA. (2005) Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists J. Wiley, Hoboken, NJ: [PubMed] [Google Scholar]
- Enserink M. (2003) Breakthrough of the year. SARS: a pandemic prevented. Science 302:2045. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Jones S, Klenk HD, Schnittler HJ. (2003) Ebola virus: from discovery to vaccine. Nat Rev Immunol 3:677–685 [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. (2000) Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol 7:569–581 [DOI] [PubMed] [Google Scholar]
- Greenbaum DC, Baruch A, Grainger M, Bozdech Z, Medzihradszky KF, Engel J, DeRisi J, Holder AA, Bogyo M. (2002) A role for the protease falcipain 1 in host cell invasion by the human malaria parasite. Science 298:2002–2006 [DOI] [PubMed] [Google Scholar]
- Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, Vasilieva N, Dermody TS, Harrison SC, Dormitzer PR, et al. (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem 281:3198–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur D, Guven A. (2002) The acidities of some indoles. J Mol Struct Theochem 583:1–18 [Google Scholar]
- Kaletsky RL, Simmons G, Bates P. (2007) Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol 81:13378–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehr R, Mintern JD, Herman AE, Lennon-Duménil AM, Mathis D, Benoist C, Ploegh HL. (2005) Cathepsin L is essential for onset of autoimmune diabetes in NOD mice. J Clin Invest 115:2934–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath ME. (1999) The lysosomal cysteine proteases. Annu Rev Biophys Biomol Struct 28:181–204 [DOI] [PubMed] [Google Scholar]
- Myers MC, Shah PP, Beavers MP, Napper AD, Diamond SL, Smith AB, 3rd, Huryn DM. (2008a) Design, synthesis, and evaluation of inhibitors of cathepsin L: Exploiting a unique thiocarbazate chemotype. Bioorg Med Chem Lett 18:3646–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MC, Shah PP, Diamond SL, Huryn DM, Smith AB., 3rd (2008b) Identification and synthesis of a unique thiocarbazate cathepsin L inhibitor. Bioorg Med Chem Lett 18:210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager CT, Dutch RE. (2005) Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol 79:12714–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo C, Joyce JA. (2008) Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci 29:22–28 [DOI] [PubMed] [Google Scholar]
- Potts W, Bowyer J, Jones H, Tucker D, Freemont AJ, Millest A, Martin C, Vernon W, Neerunjun D, Slynn G, et al. (2004) Cathepsin L-deficient mice exhibit abnormal skin and bone development and show increased resistance to osteoporosis following ovariectomy. Int J Exp Pathol 85:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Deveraux Q, Turk B, Sali A. (2004) Comprehensive search for cysteine cathepsins in the human genome. Biol Chem 385:363–372 [DOI] [PubMed] [Google Scholar]
- Schedel J, Seemayer CA, Pap T, Neidhart M, Kuchen S, Michel BA, Gay RE, Müller-Ladner U, Gay S, Zacharias W. (2004) Targeting cathepsin L (CL) by specific ribozymes decreases CL protein synthesis and cartilage destruction in rheumatoid arthritis. Gene Ther 11:1040–1047 [DOI] [PubMed] [Google Scholar]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. (2006) Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 80:4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Myers MC, Beavers MP, Purvis JE, Jing H, Grieser HJ, Sharlow ER, Napper AD, Huryn DM, Cooperman BS, et al. (2008) Kinetic characterization and molecular docking of a novel, potent, and selective slow-binding inhibitor of human cathepsin L. Mol Pharmacol 74:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102:11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D, Guncar G. (2003) Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr D Biol Crystallogr 59:203–213 [DOI] [PubMed] [Google Scholar]
- Turk V, Turk B, Turk D. (2001) Lysosomal cysteine proteases: facts and opportunities. EMBO J 20:4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B. (2007) Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr Pharm Des 13:387–403 [DOI] [PubMed] [Google Scholar]