Abstract
This study reports an integrated microfluidic system capable of isolation, counting, and sorting of hematopoietic stem cells (HSCs) from cord blood in an automatic format by utilizing a magnetic-bead-based immunoassay. Three functional modules, including cell isolation, cell counting, and cell sorting modules are integrated on a single chip by using microfluidic technology. The cell isolation module is comprised of a four-membrane-type micromixer for binding of target stem cells and magnetic beads, two pneumatic micropumps for sample transport, and an S-shaped channel for isolation of HSCs using a permanent magnet underneath. The counting and sorting of HSCs are performed by utilizing the cell counting and sorting modules. Experimental results show that a separation efficiency as high as 88% for HSCs from cord blood is achieved within 40 min for a sample volume of 100 μl. Therefore, the development of this integrated microfluidic system may be promising for various applications such as stem cell research and cell therapy.
INTRODUCTION
Hematopoietic stem cells (HSCs) have the therapeutic potential to differentiate into red blood cells, white blood cells, and platelets, which are extremely useful for cell therapy.1 HSCs are commonly recognized to express the specific antigen CD34 on the cell surface in clinics. They can be derived from mobilized peripheral blood (PB), bone marrow (BM), and cord blood (CB). Especially, the presence of HSCs in CB is an attractive cell source that is readily available and can be collected noninvasively with a lower frequency of Cytomegalovirus infection. In addition, HSCs from CB for transplantation have been widely used and are capable of decreasing human leukocyte antigen (HLA) mismatches from a related family donor.2 Therefore, there exists a great need to isolate the HSCs from CB. The amount of HSCs available for transplantation in clinical usage has to be counted precisely for efficacy issues and to maximize the amount recovered from the CB. However, HSC isolation requires a relatively time-consuming and multiple-step process, costly and bulky equipment, and experienced personnel to achieve reasonable separation efficiency.3 Therefore, there still remains a critical need to develop a compact, fast, cost-effective, and automatic platform for HSC isolation, counting, and sorting.
Recently, biomicroelectromechanical systems (bio-MEMSs) have been demonstrated as a promising technology for cell isolation and counting. They have advantages including automation, miniaturized device size, and a short processing time, when compared to their large-scale counterparts. There are several bio-MEMS approaches for cell isolation including electrokinetic,4 dielectrophoretic,5, 6 hydrodynamic,7, 8 acoustic,9 laminar flow control,10 and a combination of gravity and hydrodynamic forces.11 However, the systems mentioned above always require complicated fabrication processes or costly, bulky peripheral equipment. Furthermore, even though these devices can be used for cell isolation, they require precise fluid control to achieve reasonable separation efficiency.
Recently, many types of magnetic particles have been developed for cell isolation, including purification processes and immunoassays which can be effectively used in the separation of cells.12, 13, 14 Especially, a “laboratory-on-a-chip,” integrating several functional modules on a single chip, has been realized.15 Several magnetic-bead-based immunoassays implemented in microfluidic systems have been reported for biomedical applications.16, 17 The isolation of biosamples can be realized using magnetic beads coated with antibodies specific to target pathogens.18 Advantages including lower cost, less labor-intensive operation, and less sample∕reagent consumption in these miniature systems also facilitate the development of these instruments for biodetection and analysis.
Flow cytometry using immunoassays has been extensively explored in recent years. For example, human antiviral antibodies bound to polymer beads were detected by using fluorescent labeling.19 The population of CB mononuclear cells (MNCs) that can be recovered using magnetic beads coated with antimouse immunoglobulin was of high purity and viability.20 However, conventional flow cytometry requires a large amount of biosamples and bulky, expensive, precision optical equipment. Therefore, there is an increasing demand for a compact and inexpensive cellular analysis system. In order to solve these problems, microflow cytometers have been developed for biological applications.21, 22, 23 Different techniques have been developed to miniaturize these devices for detection of bioparticles such as resistive pulse sensing methods,24, 25 electrical microimpedance measurements,26 fluorescent optical scheme based on light scattering techniques,27 and laser-induced fluorescence techniques.28 Among them, the bead-based flow cytometry technique is a well-known method for biomedical diagnosis, virus detection, and other cell based analysis.29, 30 Basically, neighboring sheath flows with a higher velocity were used to squeeze the sample flow into a narrow stream so that fluorescence-labeled cells can be optically detected and sorted into the target collection chambers.29 A microflow cytometer integrated with embedded, etched, optical fibers for detection of particles and cells has been reported.30 A device combining the use of fluorescence detection and resistive pulse sensing enhanced by a metal-oxide-semiconductor field-effect transistor (MOSFET) has been used to accomplish absolute number counting and to calculate the relative percentage of T-cells.31 Although precise detection of cells can be realized, the whole procedure is still time-consuming and costly due to the lengthy and labor-intensive sample preparation process. Thus there is also a need to develop a microfluidic system to isolate cells with the capability to perform sample pretreatment processes. Therefore, an automatic microfluidic system that integrates the two major functions of sample pretreatment and magnetic-bead-based flow cytometry for isolation, counting, and sorting of HSC from CB has been developed in this study. Microfluidic components, such as micropumps and microvalves, are used to perform the entire process automatically. By an immunoassay technique, the antibody-conjugated magnetic beads labeled with fluorescence dyes can be used to identify a surface antigen specific for HSC and bind with the target cell. Subsequently, the cell-magnetic-bead complexes can be optically detected, counted, and finally sorted into the target collector through the feedback signal. The total process time can be as short as 40 min. Consequently, the integrated microfluidic system provides a fast, compact, and reliable approach to isolate and count the HSCs.
MATERIALS AND METHODS
Experimental procedure
A new microfluidic system is proposed in this study for stem cell applications by integrating cell isolation, counting, and sorting steps into an automatic process. The target cells are specifically caught by the antibody-conjugated magnetic beads by utilizing the affinity between the antibodies and the surface antigens of cells.32 Then, a magnetic field is used for isolation of the stem cells. Figure 1 shows the experimental procedure including incubation, isolation, counting, and sorting using magnetic beads. Superparamagnetic beads with a diameter of 4.5 μm (Dynabeads® Pan Mouse IgG, Dynal Biotech, Norway) coated with fluorescein isothiocyanate (FITC)-labeled capture antimouse IgG antibody, are mixed with CB in the four-membrane-type micromixer for 10 min [Fig. 1a]. The target cells can be specifically captured by the antibody-conjugated magnetic beads. The cell-magnetic-bead complexes are isolated using a permanent magnet that generates the required magnetic field (about 430 mT) and is attached onto the bottom of the microfluidic chip. Then, a wash buffer [×1 phosphate buffer saline (PBS)] is transported by the pneumatic pumps and is used to flush out nonbonded substances into the waste chamber [Fig. 1b]. Next, the purified cell-magnetic-bead complexes are then transported into the cell counting module, which can focus the complexes into a narrow stream using a sheath flow to inject them into the microfluidic channel. Subsequently, a photomultiplier tube (PMT) and a laser source are used to excite and detect the fluorescent signal bound to the magnetic beads [Fig. 1c]. Finally, the cell-bound magnetic beads are sorted and collected into the appropriate chambers by the downstream active microvalves by using the feedback signals from the detection module [Fig. 1d].
Figure 1.
The experimental procedure performed on the integrated microfluidic chip: (a) Mixing cells with magnetic beads; (b) purifying the target cells through a washing process; (c) fluorescent detection while the cell-magnetic-bead complexes pass through the optical detection region; (d) sorting of cell-magnetic-bead complexes utilizing microvalves.
Chip design and experimental setup
A schematic illustration of the magnetic-bead-based microfluidic system is shown in Fig. 2, which is made up of two polydimethylsiloxane (PDMS) (Sylgad 184A∕B, Dow Corning Corp., USA) layers and a glass layer. The chip is comprised of three functional modules including cell isolation, cell counting, and cell sorting modules. The cell isolation module is composed of a loading∕mixing chamber, a four-membrane-type micromixer, a wash buffer loading chamber, two pneumatic micropumps, and an S-shaped channel. The four-membrane-type micromixer includes four thin-film PDMS air chambers and three connecting air channels. The compressed air is injected into the air inlet and passes into the connecting channels to cause the four thin-film membranes to be deflected sequentially by controlling the driving frequency of an electromagnetic valve (EMV) controller. This EMV controller integrates a pressure regulator, a microcontroller (8051 microcontroller, model AT89C51 24 PC, ATMEL, USA), 12 EMVs (SD70M-6BG-32, SMC, Japan), an air compressor (MDR2-1A∕11, Jun-Air Inc., Japan), and a graphical user interface developed using VISUAL BASIC software (Visual Basic 2005, Microsoft, USA), which can be used to control the pressure, frequency, and duration for EMVs activation.
Figure 2.
Schematic illustration of the microfluidic chip for HSC detection including cell isolation, cell counting, and cell sorting modules.
Consequently, a gentle mixing effect can be successfully induced. More detailed information about this four-membrane-type micromixer can be found in our previous work.33 The dimensions of each quarter circular membrane are 9000 μm in diameter and 200 μm in height. Besides, two types of micropumps are used for the transportation of the biosample and buffer solution by controlling the driving frequency of EMV. Micropump 1 is activated by a controller system that coordinates three PDMS membranes connected by air channels,34 and generates a peristaltic effect to drive the solution forward. The width, length, and depth of each membrane are 2300, 4000, and 200 μm, respectively. Micropump 2 is activated by a rectangular-shaped PDMS membrane structure combined with a normally closed microvalve with a similar design to one in our previous study.35 The square-shaped PDMS-based membrane can be deflected upward and the liquid is transported forward in the microchannel by supplying compressed air. Then, the normally closed microvalve, which is a PDMS-based floating block structure, placed in the microchannel, is activated by pressure created by the deflection of the square membrane and is used to prevent a backflow. The waste buffer can be quickly delivered through the S-shaped channel without contamination. Note that only one PDMS membrane structure is used to simplify the original three-membrane structure, without the generation of backflow, with a new design for a normally closed microvalve. The dimensions of Micropump 2 are 6000 μm in length, 3000 μm in width, and 200 μm in height. The normally closed microvalve has dimensions of 3000 and 2000 μm in length and width, respectively. Then, in order to isolate the target cells, the sample is transferred to an S-shaped channel where a permanent magnet is attached. The magnetic-bead complexes are attracted to the magnet without being flushed away by the lower shear force generated in the S-shaped channel. The cross-sectional dimensions of the microchannel to the incubation chamber and the waste chamber are 2400 and 1200 μm in width, respectively, and 100 μm in height.
The cell counting module contains a flow-focusing region. Typically, cells can be squeezed to form a narrow stream by a sheath flow. Then the cells pass an incident laser which induces fluorescence for optical detection.8 The wavelength of the emitted fluorescent light ranges from 500 to 575 nm. By using a low pass filter and bandpass filter, the wavelength range has been successfully narrowed from 510 to 520 nm to increase the sensitivity. The widths of the sheath flow channel and the sample flow channel are 1600 and 400 μm, respectively. The cell sorting module utilizes two circular microvalves to control two microchannels, which can be closed or opened by activating the microvalves to guide the sample into the desired outlet. Note that the circular microvalve is 3000 μm in diameter.
Fabrication process
The microfluidic chip is fabricated using a MEMS process by constructing elastic PDMS structures on a glass substrate.34 Briefly, using a standard lithography and a two-step photoresist baking processes, a SU-8 master mold is formed to replicate the PDMS structure. A photograph of the microfluidic chip is represented in Fig. 3a. The dimensions of the chip are 60 mm in length and 32 mm in width. Scanning electron microscope (SEM) images of SU-8 molds of all the system components including a microchannel layer and an air channel layer were also showed in Fig. 3b. Detailed information regarding the microfabrication process can be found in our previous work.34
Figure 3.
(a) A photograph of the integrated microfluidic chip. The dimensions of this chip are measured to be 60×32 mm2; SEM images of SU-8 molds for [(b)(1) and (b)(2)] microchannel layer and (b)(3) the air channel layer.
Sample preparation
Superparamagnetic polystyrene beads with a diameter of 4.5 μm are used, which are coated with IgG groups in order to bind to specific antibodies. Isolation of HSCs is based on the expression of certain surface antigens or on the lack of expression of lineage specific antigens. The most commonly used surface marker for HSC selection is the transmembrane glycoprotein CD34.36 The primary antibody 1:20 dilution of FITC-conjugated CD34 (mouse IgG-FITC, abcame, U.K.) is first added into 100 μl of magnetic beads with a concentration of 4×108 beads∕ml and is shaken for 40 min using a large-scale shaker (INTELLI-MIXER, ELMI Ltd., Latvia) at room temperature. Next, the magnetic beads labeled with CD34-FITC and cells are loaded into the incubation chamber for the mixture to react. Since CB cells are not easily accessible for experiments, the combination of two leukemia cell lines, HL-60 (CD34- cells) and TF-1a (CD34+cells) are first used to simulate the MNCs to verify our designed chip. The isolation and detection of MNCs centrifuged from CB are then tested on the microfluidic chip by using the operating conditions obtained from the earlier simulated experiments. All of the cells used in this study (TF-1a, HL-60 and MNCs) are provided by the cell bank of the Bioeresource Collection and Research Center in Taiwan.
RESULTS AND DISCUSSION
Characterization of the microfluidic system
The four-membrane-type micromixer is designed for the delicate handling and incubation of the antibody-conjugated magnetic beads and cell samples, as demonstrated in our previous study.33 In this study, an air pressure of 5 psi, at a low frequency of 2 Hz, is applied to actuate the four-membrane-type micromixer to generate a gentle mixing effect by slow time-phased deflections of the four membranes. The reason for generating this gentle mixing effect is to mimic the mixing performance of a large-scale shaker such that the binding between the antibodies and antigens would not be broken and to prevent cell damage during the mixing step. The four-membrane-type micromixer is designed for gentle handling and incubation of magnetic beads and cells. The mixing index is used to quantify the mixing performance. The mixing efficiency is evaluated by measuring the concentration distribution along the cross section of the mixing chamber. First, 50 μl of blue ink and 50 μl of de-ionized water are loaded into the mixing chamber. Then mixing is performed to measure the mixing efficiency of the four-membrane-type micromixer. The mixing process is observed utilizing a high-speed charge-coupled device (CCD) (MC1311, Mikrotron, Germany). The four-membrane-type micromixer is actuated at a low pulsing frequency of 2 Hz with an applied pressure of 5 psi, which generates a slow, time-phased deflection of the four membranes to produce a gentle mixing effect. Experimental data indicate that the mixing can be completed within 2 min, which demonstrates the efficiency of this four-membrane-type micromixer. A normalized concentration profile across the mixing chamber is shown in Fig. 4, where Y+ is the normalized location across the mixing chamber and C+ is the normalized concentration. The data show that the mixing index of the micromixer is as high as 97% after mixing for 2 min. These gentle mixing results also demonstrate reliable cell capture without damage. More detailed information about the working principle of the mixer can be found in our previous work.33
Figure 4.
Normalized concentration profiles across the mixing chamber.
In this study, pneumatic micropumps are used to transport the cells and reagents in the microchannels. Figure 5a illustrates the relationship between the pumping rate and the driving frequency of the EMV for Micropump 1 at different pressure levels. The maximum pumping rate is measured to be 2.05 μl∕s at a driving frequency of 12 Hz and a pressure of 15 psi. Figure 5b shows the relationship between the pumping rate and the driving frequency of the EMV for Micropump 2 at different pressure levels. The maximum pumping rate for Micropump 2 is measured to be 2.26 μl∕s at a driving frequency of 15 Hz and a pressure of 15 psi. This micropump is used to prevent any backflow, which minimizes contamination. It can be clearly seen that the pumping rate of both Micropumps 1 and 2 increases with increased pressure. It is also observed that the pumping rate starts to drop after reaching the maximum pumping rate. This is because the time required to restore the membranes to their minimum deflection position is longer than the switching time of the EMV. The maximum frequency response limit of the PDMS membrane is measured to be 18 Hz.
Figure 5.
The relationship between the pumping rate and the driving frequency of the EMV at different pressure levels for (a) Micropump 1 and (b) Micropump 2.
Cell isolation
Isolation of simulated cell samples
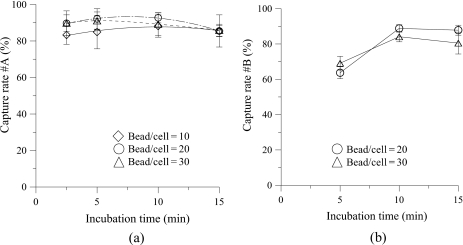
In this study, TF-1a cells and HL-60 cells are first used to verify the performance of the developed microfluidic chip. 100 μl of TF-1a cells with a concentration of 2×105 cells∕ml is first loaded into the incubation chamber. The magnetic beads are then added for incubation when the micromixer is activated. After the incubation step, the cell-magnetic-bead complexes and nonbinding cells are transported to the S-shaped channel by Micropump 1. Then a magnet with a magnetic field of 430 mT is used to attract the cell-magnetic-bead complexes such that the nonbinding cells can be washed out to the waste chamber when the washing buffer (PBS) is pumped by Micropump 2. The capture rate for the TF-1a cells is defined as 100% minus the ratio between the nonbinding cells collected from the waste chamber and the total number of loaded cells. Figure 6a shows the relationship between the incubation time and the capture rate of the TF-1a cells (CD34+). Three different ratios of the number of magnetic beads over the loaded number of the TF-1a cells (bead∕cell ratio), namely, 10, 20, and 30, are tested. For each case, four incubation times (2.5 min, 5.0 min, 10.0 min and 15.0 min, respectively) are tested. The experimental results show that a capture rate as high as 92.7% can be achieved with an incubation time of 10.0 min when a bead∕cell ratio of 20 is adopted. The results also indicate that the capture rate starts to decrease if a higher bead∕cell ratio and a longer incubation time are used. This is probably because nonbonded beads would collide more often with the magnetic-bead-cell complexes so that magnetic beads detach from the target cells, decreasing the overall binding effect. Note that a low capture rate is obtained when a bead∕cell ratio of 10 is used due to an insufficient binding effect. However, a capture rate higher than 80% is achieved for all cases.
Figure 6.
(a) The relationship between the capture rate of TF-1a cells (CD34+) and the incubation time for TF-la cells. A high capture rate of approximately 92.7% is achieved at a bead∕cell ratio of 20 for an incubation time of 10 min. (b) The relationship between the capture rate of TF-1a cells (CD34+) and the incubation time for a mixture of HL-60 and TF-1a cells. A capture rate of 88.8% can be obtained at a bead∕cell ratio of 20 with an incubation time of 10 min.
Next, a mixture of HL-60 cells (CD34-) and TF-1a cells (CD34+) to mimic conditions for MNCs is used. Since 1% of CD34+ cells are present in MNCs, 100 μl of a mixed cell solution with a concentration of 5×107 cells∕ml (HL-60 cells: TF-1a cells=100:1) is tested in the chip. The capture rate for the TF-1a cells is defined as the ratio of the TF-1a cells collected from target cell chamber to the total number of TF-1a cells initially loaded. Two bead∕cell ratios of 20 and 30 with incubation times of 5, 10, and 15 min are tested. The experimental results shown in Fig. 6b indicate that an optimized capture rate of 88.8% is obtained when the bead∕cell ratio is 20 with an incubation time of 10 min, which is a similar result to the former experiment using TF-1a cells. The optimum ratio of 20 can be explained by the surface area effect. The surface area of the TF-1a cell (with a radius of 20 μm) is 400π μm2 (4×10×10×π) while that of the magnetic bead (with a radius of 4.5 μm) is 20.25π μm2 (4×2.25×2.25×π), meaning that the surface area of TF-1a cells is about 20 times larger than that of the beads, which is consistent with the optimum binding ratio. This indicates the optimum operating conditions have been identified in the developed chip and will be applied for subsequent isolation of HSCs.
HSCs isolation
Since the size of a TF-1a cell is triple that of the HSC (with a radius of 13 μm) in MNCs, the quantity of magnetic beads is reduced in the following HSCs isolation test. Based on the optimal conditions determined from the experiment with TF-1a cells, four bead∕cell ratios (5, 10, 15, and 20) are tested to optimize the capture rate for HSCs in MNCs under four incubation times (2.5, 5.0, 10.0, and 15.0 min, respectively). The relationship between the incubation time and the captured number of HSCs is illustrated in Fig. 7. Since 1% of CD34+cells is present in MNCs and 100 μl of MNCs, with a concentration of 5×107 cells∕ml, is used, the calculated cell number is 5×104 cells. The experimental results show that captured HSC-magnetic-bead complexes in the target cell chamber is 4.4×104 at a bead∕cell ratio of 10 and an incubation time of 10 min, resulting in a capture rate of 88.0%, which is similar to that of TF-1a. Compared to the results for capturing TF-1a cells, fewer magnetic beads are used for maximizing the capture of HSCs due to the smaller size of HSCs. The surface area of a HSC is 169π μm2 (4×6.5×6.5×π), which is only about eight times larger than that of the magnetic bead (20.25π μm2), leading to a lower bead∕cell ratio than in the case of the simulated isolation. Besides, the binding effect decreases significantly when the bead∕cell ratio is higher than 10 (bead∕cell=15 and bead∕cell=20) due to the same reason previously described in the simulated conditions. As a result, the quantity of beads needs to be tuned properly to capture the target cell.
Figure 7.
The relationship between the captured number of HSCs and the incubation time. An optimum capture rate of HSCs is measured to be 88.0%.
Cell detection
For cell counting, cell-magnetic-bead complexes can be hydrodynamically focused by the sheath flow (PBS), followed by detection when the cells pass through the detection region. First, a total number of 4×103 of TF-1a cell-magnetic-bead complexes is counted to evaluate the performance of the cell counting module. TF-1a cells bound with fluorescent-labeled magnetic beads, emitting the fluorescence when induced by a laser light, can be successfully detected, as shown in Fig. 8a(1). The optical detection of TF-1a cells is graphed in Fig. 8a(2). The cell sample flow rate and the sheath flow rate are 1.2 and 1.7 μl∕s at EMV frequencies of 6 and 10 Hz, respectively, under a pressure of 10 psi. Each signal peak represents the fluorescent response of a cell-magnetic-bead complex. A threshold of 0.5 mV is used to differentiate the signals from the noise. Note the sampling rate for signal acquisition is 2000 Hz. From this test, the counting efficiency is defined as the ratio of the number counted to the total number of loaded complexes and is measured to be 87.2%.
Figure 8.
[(a)(1) and (b)(1)] CCD images of the TF-1a cell-magnetic-bead complexes and HSC-magnetic-bead complexes concentrated in the chip; [(a)(2) and (b)(2)] the fluorescent signal from TF-a cells and HSCs.
After determining the counting efficiency of the cell counting module, the counting experiment for HSC-magnetic-bead complexes is then carried out in the developed chip. Figure 8b(1) shows the CCD images of HSCs bound with fluorescent-labeled magnetic beads. The optical detection of HSC is also graphed in Fig. 8b(2). Under the same conditions, the counting efficiency is measured to be 85.3%, which is similar to the TF-1 cell results. As a result, the final isolation efficiency (including the capture rate and the counting efficiency) for HSCs from MNCs is calculated to be 75.1% (88.0%×85.3%). From these results, HSCs can be isolated, counted, and sorted in the integrated microfluidic chip within a shorter period of time, with reasonable isolation efficiency.
Compared to the large-scale system, such as HSCs isolated from the CB, requiring 5 h (4.5 h for isolation and 30 min for counting and sorting) to perform the entire process, the microfluidic chip only requires 40 min (15 min for isolation and 25 min for counting and sorting) with a comparable yield. Although the total number of the cells processed in the chip is fewer than that in the large-scale system, fewer samples are required for counting and the entire process can be performed in an automatic manner in this chip. Furthermore, a high-throughput microfluidic chip with parallel channels can be designed to increase the number of the counted cells. Besides, an isolation efficiency of 88.8% can be obtained by using the developed chip, which is significantly higher than that obtained in the large-scale system (60.0%). Moreover, 75.1% of the final isolation efficiency (including the capture rate and the counting efficiency) is also higher than that of the large-scale system.
CONCLUSION
This study has successfully demonstrated an integrated microfluidic system for isolation, counting, and sorting of HSCs from CB by using antibody-conjugated magnetic beads. The single microfluidic chip is capable of biosample incubation, isolation, optical counting, and sorting automatically. TF-1a cells and HL-60 cells with similar characteristics to MNCs are first isolated to optimize the operating conditions for the developed system. Then, the isolation, counting, and sorting of MNCs has been performed successfully. The total operating time has been reduced to 40 min which is much faster than with the traditional system (5 h). Therefore, this integrated system provides a promising platform for stem cell research and cell therapy.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the financial support provided to this study by the National Science Council in Taiwan (Grant No. NSC 98-2120-M-006-001). This work was also partially supported by the Ministry of Education, Taiwan, R.O.C. under the NCKU Project for Promoting Academic Excellence & Developing World Class Research Centers and by the Ministry of Economic Affairs in Taiwan (Grant No. 99-EC-17-A-17-R7-0525). Besides, culture facilities in this work were provided by Professor Xi-Zhang Lin (Department of Medicine, National Cheng Kung University).
NOMENCLATURE
- Bio-MEMSs
Biomicroelectromechanical systems
- BM
Bone marrow
- CB
Cord blood
- CCD
Charge-coupled device
- DEP
Dielectrophoretic
- EMV
Electromagnetic valve
- FITC
Fluorescein isothiocyanate
- HLA
Human leukocyte antigen
- HSC
Hematopoietic stem cell
- MOSFET
Metal-oxide-semiconductor field-effect transistor
- MNCs
Mononuclear stem cells
- PB
Peripheral blood
- PBS
Phosphate buffer saline
- PDMS
Polydimethylsiloxane
- PMT
Photomultiplier tube
References
- Dhot P. S., Nair V., Swarup D., Sirohi D., and Ganguli P., Indian J. Pediatr. 70, 989 (2003). 10.1007/BF02723826 [DOI] [PubMed] [Google Scholar]
- Tomonari A., Takahashi S., Takasugi K., Ooi J., and Tsukada N., Int. J. Hematol. 84, 438 (2006). 10.1532/IJH97.06119 [DOI] [PubMed] [Google Scholar]
- Melnik K., Nakamura M., Comella K., Lasky L. C., Zborowski M., and Chalmers J. J., Biotechnol. Prog. 17, 907 (2001). 10.1021/bp010079r [DOI] [PubMed] [Google Scholar]
- Chien Y. S., Lin C. H., Kao F. J., and Ko C. W., Mater. Sci. Forum 505–507, 643 (2006). 10.4028/www.scientific.net/MSF.505-507.643 [DOI] [Google Scholar]
- Han K. H. and Frazier A. B., Lab Chip 8, 1079 (2008). 10.1039/b802321b [DOI] [PubMed] [Google Scholar]
- Pommer M. S., Zhang Y., Keerthi N., Chen D., Thomson J. A., Meinhart C. D., and Soh H. T., Electrophoresis 29, 1213 (2008). 10.1002/elps.200700607 [DOI] [PubMed] [Google Scholar]
- Yang S. Y., Hsiung S. K., Hung Y. C., Chang C. M., Liao T. L., and Lee G. B., Meas. Sci. Technol. 17, 2001 (2006). 10.1088/0957-0233/17/7/045 [DOI] [Google Scholar]
- Yang S. Y., Lien K. Y., Huang K. J., Lei H. Y., and Lee G. B., Biosens. Bioelectron. 24, 855 (2008). 10.1016/j.bios.2008.07.019 [DOI] [PubMed] [Google Scholar]
- Laurell T., Petersso F., and Nilsson A., Chem. Soc. Rev. 36, 492 (2007). 10.1039/b601326k [DOI] [PubMed] [Google Scholar]
- Wu Z., Liu A. Q., and Hjort K., J. Micromech. Microeng. 17, 1992 (2007). 10.1088/0960-1317/17/10/010 [DOI] [Google Scholar]
- Huh D., Bahng J. H., Ling Y., Wei H. H., Kripfgans O. D., Fowlkes J. B., Grotberg J. B., and Takayama S., Anal. Chem. 79, 1369 (2007). 10.1021/ac061542n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier T., Mohanty S., Drese K. S., Rampf F., Kim J., and Schonfeld F., Microfluid. Nanofluid. 7, 205 (2009). 10.1007/s10404-008-0376-3 [DOI] [Google Scholar]
- Estes M. D., Do J., and Ahn C. H., Biomed. Microdevices 11, 509 (2009). 10.1007/s10544-008-9257-5 [DOI] [PubMed] [Google Scholar]
- Ueda H., Kajikawa K. K., Furuse M., Fuchino S., and Ishiyama A., IEEE Trans. Appl. Supercond. 19, 2157 (2009). 10.1109/TASC.2009.2018441 [DOI] [Google Scholar]
- Choi J. W., Oh K. W., Thomas J. H., Heineman W. R., Halsall H. B., Nevin J. H., Helmicki A. J., Henderson H. T., and Ahn C. H., Lab Chip 2, 27 (2002). 10.1039/b107540n [DOI] [PubMed] [Google Scholar]
- Huang Y., Mather E. L., Janice L. B., and Madou M., Anal. Bioanal. Chem. 372, 49 (2002). 10.1007/s00216-001-1191-9 [DOI] [PubMed] [Google Scholar]
- Gijs M. A. M., Microfluid. Nanofluid. 1, 22 (2004). [Google Scholar]
- Lien K. Y., Lin J. L., Liu C. Y., Lei H. Y., and Lee G. B., Lab Chip 7, 868 (2007). 10.1039/b700516d [DOI] [PubMed] [Google Scholar]
- McHugh T. M., Miner R. C., Logan L. H., Daniel P., and Stites D. P., J. Clin. Microbiol. 26, 1957 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S. R., Macardle P. J., and Zola H., J. Immunol. Methods 217, 121 (1998). 10.1016/S0022-1759(98)00111-2 [DOI] [PubMed] [Google Scholar]
- Huh D., Gu W., Kamotani Y., Grotberg J. B., and Takayama S., Physiol. Meas 26, R73 (2005). 10.1088/0967-3334/26/3/R02 [DOI] [PubMed] [Google Scholar]
- Yi C., Zhang Q., Li C. W., Yang J., Zhao J., and Yang M., Anal. Bioanal. Chem. 384, 1259 (2006). 10.1007/s00216-005-0252-x [DOI] [PubMed] [Google Scholar]
- Ateya D. A., Erickson J. S., P. B.Howell, Jr., Hilliard L. R., Golden J. P., and Ligler L. F. S., Anal. Bioanal. Chem. 391, 1485 (2008). 10.1007/s00216-007-1827-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton J. E., Jin P., Sexton L. T., Horne L. P., Sherrill S. A., Mino W. K., and Martin C. R., Small 3, 1424 (2007). 10.1002/smll.200700106 [DOI] [PubMed] [Google Scholar]
- Carbonaro A. and Sohn L., Lab Chip 5, 1155 (2005). 10.1039/b504827c [DOI] [PubMed] [Google Scholar]
- Sun T., Holmes D., Gawad S., Green N. G., and Morgan H., Lab Chip 7, 1034 (2007). 10.1039/b703546b [DOI] [PubMed] [Google Scholar]
- Wang Z., El-Ali J., Engelund M., Gotsæd T., Perch-Nielsen I. R., Mogensen K. B., Snakenborg D., Kutter J. P., and Wolff A., Lab Chip 4, 372 (2004). 10.1039/b400663a [DOI] [PubMed] [Google Scholar]
- Fu L. M., Yang R. J., Lin C. H., Pan Y. J., and Lee G. B., Anal. Chim. Acta 507, 163 (2004). 10.1016/j.aca.2003.10.028 [DOI] [Google Scholar]
- Chang C. M., Hsiung K., and Lee G. B., Mater. Sci. Forum 505–507, 637 (2005). [Google Scholar]
- Lin C. H. and Lee G. B., J. Micromech. Microeng. 13, 447 (2003). 10.1088/0960-1317/13/3/315 [DOI] [Google Scholar]
- Wang Y. N., Kang Y., Xu D., Chon C. H., Barnett L., Kalam S. A., and Li D., Lab Chip 8, 309 (2008). 10.1039/b713932b [DOI] [PubMed] [Google Scholar]
- Haukanes B. I. and Kvam C., Biotechnology 11, 60 (1993). 10.1038/nbt0193-60 [DOI] [PubMed] [Google Scholar]
- Lee Y. F., Lien K. Y., Lei H. Y., and Lee G. B., Biosens. Bioelectron. 25, 745 (2009). 10.1016/j.bios.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. H. and Lee G. B., J. Micromech. Microeng. 16, 341 (2006). 10.1088/0960-1317/16/2/019 [DOI] [Google Scholar]
- Yang Y. N., Lin H. I., Wang J. H., Shiesh S. C., and Lee G. B., Biosens. Bioelectron. 24, 3091 (2009). 10.1016/j.bios.2009.03.034 [DOI] [PubMed] [Google Scholar]
- Aroviita P., Teramo K., Hiilesmaa V., and Kekomäki R., Transfusion (Bethesda, Md.) 45, 613 (2005). 10.1111/j.0041-1132.2005.04268.x [DOI] [PubMed] [Google Scholar]