Abstract
We have designed, built, and evaluated a microfluidic device that uses deterministic lateral displacement for size-based separation. The device achieves almost 100% purity and recovery in continuously sorting two, four, and six micrometer microspheres. We have applied this highly efficient device to the purification of fungal (Aspergillus) spores that are spherical (∼4 μm diameter) with a narrow size distribution. Such separation directly from culture using unfiltered A. niger suspensions is difficult due to a high level of debris. The device produces a two to three increase in the ratio of spores to debris as measured by light scatter in a flow cytometer. The procedure is feasible at densities up to 4.4×106 spores∕ml. This is one of the first studies to apply microfluidic techniques to spore separations and has demonstrated that a passive separation system could significantly reduce the amount of debris in a suspension of fungal spores with virtually no loss of spore material.
INTRODUCTION
It is well known that microfabrication is enabling improved process integration and a shift toward using smaller quantities of reagents, but these technologies can also provide new sample processing tools for chemistry and biology, for example, those of West et al.1 or Yen et al.2 Microfabrication facilitates precise control of fluid and particle flow, making it possible to control and separate the suspended particles.3, 4 Much of the biological application of these separation methods have centered on blood, no doubt because of its medical significance, but some success with blood may be because blood typically flows through microchannels without clogging. Literature contains only isolated instances of work with other types of cells and particles, none of which were done in a really challenging medium. For example, live and dead yeast5 and E. coli6 have been separated by microfluidic dielectrophoresis (DEP). Micrococcus luteus, an airborne bacteria, was also recently separated by DEP.7 An inertial microfluidic device was recently shown to separate E. coli from red blood cells8 and immunomagnetic separation can be used for smaller numbers of organisms.9 Capillary electrophoresis was recently used to separate conidia (spores) from various filamentous fungi including Aspergillus,10 but there are few microfluidic systems that demonstrated application to fungal biology.
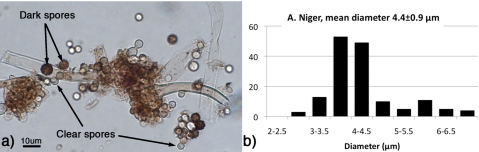
A commonly found mold Aspergillus is an industrially important organism used for producing a variety of molecules including citric acid, and some strains can cause infections. It is desirable to purify the Aspergillus spores for applications in immunological, molecular or spore coat analysis, proteomics, and for commercial sale of the spores where new methods achieving high throughput and low cost are needed. This requires separating mature spores from other parts of the organisms present in the culture, such as immature spores, foreign contaminants, and culture materials [Fig. 1a]. This can be difficult or impossible using conventional techniques, such as membrane filtration, centrifugation, and flow cytometry. DEP is a promising approach as it may be able to differentiate viable spores from other fungal material although volume throughput may be an issue.11, 12, 13 Inertial microfluidics, which can separate particles by size, see Ref. 4, could certainly produce sufficient volume without clogging but may not have the required size resolution and may not remove submicron-sized particles. The Aspergillus spores being spherical and with a narrow size distribution [Fig. 1b)] lend themselves to sorting by deterministic lateral displacement,14, 15 which has the capacity for reasonable volume throughput and excellent size resolution.
Figure 1.
(a) DIC image of A. niger spore preparation. Spores are circular objects less than 10 μm in diameter. (b) Histogram of spore diameter. Objects that are not clearly identifiable as spores are not counted in the histogram.
Deterministic lateral displacement uses flow through an array of obstacles to alter the trajectory of particles above a certain hydrodynamic size. It takes advantage of the fact that the center of a small particle can be closer to a boundary than the center of a large particle. Along any surface there then exists a boundary layer of fluid containing particles below a certain size. This boundary layer is fundamental to deterministic lateral displacement and, it is worth mentioning, to the work of Yamada and Seki.16, 17 In deterministic lateral displacement, particles encounter rows of obstacles, each one shifter laterally with respect to the previous row. This shift allows a small amount of this boundary layer of fluid to cross the column of obstacles. Particles below the critical size follow this fluid while particles larger than the critical size follow the slightly tilted column of obstacles. More complete descriptions of the method exist in numerous other papers18, 19
The aim of this paper is to describe and characterize a high quality microfluidic device with excellent separation efficiency measured by microspheres for separating and purifying a suspension of fungal spores in a complex, unfiltered culture. It is designed to separate an Aspergillus niger spore preparation into three groups for small (0–3.5 μm), medium (3.5–5 μm), and large (5–10 μm) particles. Figure 1a shows a micrograph characterizing the size of the spores and Fig. 1b shows the size distribution. In this paper we describe the device, show its excellent performance when tested with microspheres, and show the results for purification of A. niger spores.
EXPERIMENTAL
Spore size measurement
Spore suspensions were fixed in 4% paraformaldehyde and imaged using a 60× objective on a differential interference contrast (DIC) microscope fitted with a calibrated digital camera. The diameters were determined by a three-point spherical fit using NIKON ELEMENTS software.
Microdevice design and construction
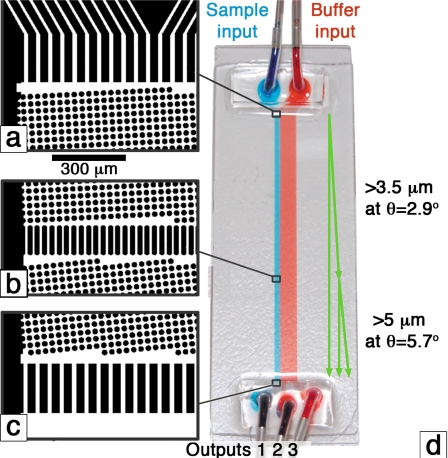
Deterministic lateral displacement separates micron-sized particles using fluid flow through a periodic array of obstacles. The array of obstacles is characterized by the distance between obstacles, the gap, and a tilt angle of the array with respect to the fluid flow direction, θ [Fig. 2d]. One can adjust the gap or the tilt and place multiple arrays in sequence to separate particles of one size range from particles of another size range.
Figure 2.
[(a)–(c)] Images from the photolithographic mask design. Black objects become 30 μm high PDMS features. Fluid flows through the white regions. (d) Image of the device carrying blue and red food dyes. The device is built on a standard glass slide (75×25 mm2). The green arrows show the idealized paths for particles of different sizes.
The device shown in Fig. 2d has an 820 μm wide input for the suspension while the right input, for filtered buffer, is 5180 μm wide. There are two arrays: The first is 33.7 mm long, which has a gap on the lithographic mask of 10.5 μm and a tilt of 2.86° giving a target critical diameter of 3.5 μm; the second is 16.9 cm long, which has a gap on the mask of 10.5 μm and a tilt of 5.7° giving a target critical diameter of 5 μm. Figures 2a, 2b, 2c show images of the beginning and end of both sections. This is the first applied device to use the procedure of Inglis20 for edge correction and the array is cubic rather than the more popular rhombic style. Both of these modifications have been made for a high performance device with a minimum wafer footprint.
Ideally particles in the sample stream that are less than 3.5 μm follow the fluid flow (blue dye) and are directed to output 1. Particles between 3.5 and 5 μm move at the array tilt angle in the first section, but not in the second so they are directed to output 2. Particles larger than 5 μm move at the array tilt angle in both sections and travel to output 3. The size range of 3.5–5 μm was chosen because it corresponds to the peak of the spore size distribution [Fig. 1b]. The actual gap depended on fabrication conditions and for all experiments here, it was 9.2 μm (by electron microscope), giving critical diameters of 4.6 and 3.1 μm.
The device was designed and built using standard soft lithographic techniques. A mask was designed (L-Edit, Tanner EDA) and manufactured, then transferred to 30 μm thick GM-1060 SU-8 photoresist (Gersteltec, Switzerland) using a mask aligner fitted with a 360 nm long pass filter. This gave polydimethylsiloxane (PDMS) pillars with vertical sidewalls. We formed a thin (250 μm) layer of PDMS, (RTV615, Momentive Performance Materials) between a glass slide and the SU-8 master. After curing the PDMS at 75 °C for 2 hours, the glass slide-PDMS device was separated from the SU-8 master by prying with a razor blade. It was then nonpermanently sealed to a second glass slide. This second glass slide had 1 mm diameter sand-blasted holes in it and short sections of 0.74 mm inner diameter (ID) silicone tubing connected to the holes. The glass-PDMS-glass device was immersed in water containing 2 g∕l Pluronic F108 (BASF), a surfactant to assist in wetting. The bath was placed in a vacuum desiccator overnight to enable complete wetting of the microfluidics.
Operation with microbeads
We prepared a mixture of 2.1, 4.2, and 5.7 μm diameter fluorescent polystyrene beads (Polysciences and Bangs Laboratories) at a number ratio of 2:1:2 in de-ionized water containing 1 g∕l F108. The total concentration of beads was 12×106∕ml. The “buffer” was 0.2 μm filtered de-ionized water. Prior to running, the microdevice was flushed with the buffer for 15 min at a pressure of 5 psi (35 kPa), and the bead mixture was placed in an ultrasonic bath for 30 min to break up aggregated beads. After the flush, the mixture of beads was connected to the input and run for 20 min at 5 psi. Short (4 cm) tubes connected the three outputs to polystyrene flow cytometry tubes with caps modified to hold the silicone tubing in place. The device geometry dictates that the sample will be diluted by 7.3 times (the buffer stream is 7.3 times wider than the sample stream).
Within 3 h the samples were taken to a BD FACScalibur flow cytometer where 10 000 events were collected for the input and each output sample. The experiment was repeated on four different days. For statistical analysis, single bead events, as opposed to aggregates and other outlying events, were counted. These were determined using polygon regions on the scatter and fluorescence dot plots.
Spore production and handling
A lawn of A. niger NCPF 2275 (recently renamed A. brasiliensis) was grown on Dichloran Rose Bengal Chloramphenicol agar (Oxoid PP2233) by inoculating from a frozen working stock and incubating the agar for 5 days at 37 °C. After incubation the spores were harvested by wiping the surface of the agar with a sterile cotton swab, which was then rinsed in 0.9% saline solution to retrieve the spores. The spore suspension is then stored at 4 °C until used (less than 2 weeks). All experiments involving viable Aspergillus spores were conducted within a physical containment level 2 laboratory. Harvesting large numbers of spores from agar plates was done within a class 2 biological safety cabinet in order to contain any potential airborne particles created during the process.
Operation with Aspergillus spores
The spore suspension was diluted in phosphate buffered saline (PBS) containing 0.1% Tween 20 (Sigma Aldrich) to a concentration of approximately 4.4×106 spores∕ml. The Tween was added to reduce the aggregation of spores. Prior to running, the microdevice was flushed with 0.2 μm filtered PBS for 15 min at a pressure of 10 psi (69 kPa). The spore suspension was then connected and run at 5–10 psi for 15–20 min. A 20 cm, 0.64 mm ID silicone tube connected the spore suspension to the device. Some large particles and aggregates settled in this tube before reaching the device where they would have contributed to clogging. As described earlier, output tubes were connected to flow cytometry tubes. These tubes were taken to a FACScalibur flow cytometry system for analysis within 3 hours after the run. The experiment was repeated on three different days using five different devices. The total output from the device is 0.59 μl∕min kPa, about 800 μl in 20 min at 10 psi.
RESULTS
Microbeads
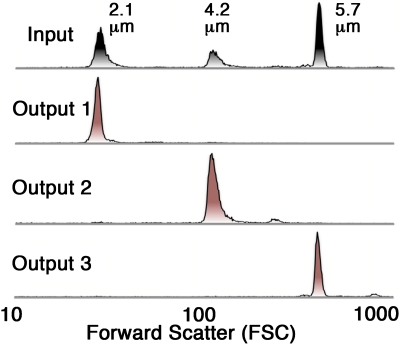
The input bead mixture comprised of 2.1, 4.2, and 5.7 μm beads. The 2.1 μm diameter particles were chosen to represent small debris, while the 4.2 μm particles could be regarded as equivalent to the target spores and the 5.7 μm particles simulated large spores and debris. Figure 3 shows forward scatter histograms for the input bead mixture and the three outputs. It clearly shows that nearly all of the 2.1 μm beads flow into output 1, nearly all the 4.2 μm beads flow to output 2 and nearly all the 5.7 μm beads flow to output 3.
Figure 3.
Forward scatter histograms of the input and sorted fractions for a single experiment using microspheres. The input sample consists of about 40% 2.1 μm, 20% 4.2 mm, and 40% 5.7 μm beads. 5000–10 000 events were counted in each run.
Aggregates can be seen as small bumps to the right of the main peaks. We are interested in reducing the amount debris or nonspore objects in the sample. From this perspective output 2 shows 96% removal of the small and large beads and 99% recovery of the 4.2 μm particles (Table 1). This is an excellent result, given that the data were obtained by analyzing the entire actual output. Earlier work by Huang et al.14 in 2004 showed very high resolution separations of 0.8, 0.9, and 1.0 μm particles; however, this was calculated based on images of beads flowing through the device, which represents a very small sample and shows only the best result. The more demanding off-chip analysis performed here indicates that this device performs as well or better than any previously reported work.
Table 1.
Percent and type of single beads in the input and each output. Values are the mean of four runs ± one standard deviation. Unclassified events are the percent of all events that were neither 2.1, 4.2, or 5.7 μm beads. They are believed to be aggregates.
| 2.1 μm | 4.2 μm | 5.7 μm | Unclassified events | |
|---|---|---|---|---|
| Input | 40±7 | 20±2 | 40±5 | 8±2 |
| Output 1 | 99±1 | 1±1 | 0.0±0.0 | 15±6 |
| Output 2 | 1.5±0.2 | 97±1 | 1.6±1 | 31±7 |
| Output 3 | 0.8±0.3 | 0.2±0.2 | 99±0.3 | 37±11 |
Spores
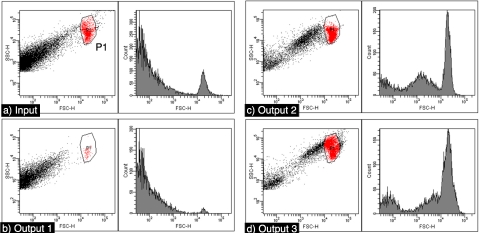
Operation with spores is more challenging than with beads because of an increased tendency to aggregate and the wide and continuous spectrum of differently shaped and sized particles in the spore preparation. Nevertheless, experiments with actual fungal cultures show a marked increase in the purity of the spores. Figure 4 shows the flow cytometry data from one run through our microdevice; it shows a clear improvement in the ratio of events within the polygon P1, to other debris. The polygon region is defined around the right uppermost cluster, which reliably appears in fresh spore preparations and is used to select for colony forming units. When 200 of these events were plated, 195 colonies formed, showing that a high percentage of events in P1 are viable spores. Table 2 shows the percent of events within P1, events with forward scatter less than ten were not included in the calculation. On average there is a twofold increase in the purity of spores in output 2 and a threefold increase in the purity of spores in output 3. The spore purity in output 3 is higher than in output 2 most likely because there is a larger amount of debris in the size range covered by output 2 relative to that covered by output 3. Figure 4 shows that there is still a significant amount of low light scattering particles in outputs 2 and 3. Control experiments, not shown, with 0.2 μm filtered buffer show an insignificant amount of detector noise or actual scattering events in the buffer.
Figure 4.
Dot plots and histograms for a single experiment with spores. Events within the polygon P1 are considered spores.
Table 2.
Summary of flow cytometry data from five runs.
| Sample name | Percentage within P1 gate |
|---|---|
| Input | 17±7 |
| Output 1 | 1.5±0.9 |
| Output 2 | 37±8 |
| Output 3 | 50±9 |
DISCUSSION
The fabricated device performs extremely well with microbeads and, in four runs averaged nearly 100% of the single bead events going to the intended, correct output. Defects and clogs in the post array cause beads to occasionally stop bumping, leading to contamination into the lower numbered outputs, that is, a 5.7 μm particle arriving in output 2, and 4.2 μm particle going to output 1. Clogs are usually caused by three or more beads trying to pass through a gap at the same time. This is rare, but nevertheless the clogs are usually permanent so the number of them increase with time. Defects are also rare but there are 156 818 posts in the array and a handful of those posts may not be vertical or may have dust particle or other defect on them. When a particle that is moving with the tilt angle hits such a post, it will change course and will move with the fluid flow for 44∕tan(θ) microns before resuming its course across streamlines.
The unavoidable size variation in the particles is responsible for some contamination in both directions. The 4.2 μm particles have a standard deviation of 0.14 μm, so assuming a normal distribution, 0.2% would be larger than 4.6 μm, accounting for all of the 4.2 μm beads found in output 3 but none of the 4.2’s in output 1. These 1% are presumed to be the result of clogs and defects. The 5.7 μm beads have a standard deviation of 0.43 μm. Assuming a normal distribution, 0.5% would be less than 4.6 μm. This accounts for one-third of the 5.7 mm beads found in output 2; there were no 5.7 μm beads in output 1. The other two-thirds, 1% of the 5.7 μm beads, are presumed to be the result of clogs and defects. We do not know the variance of the 2.1 μm particles, but assuming a standard deviation of 5% or 0.1 μm, particle size variation does not account for any of the 2.1 μm beads in output 2 or output 3. It is unclear how 2.1 μm particles were able to reach output 3 as great care was taken to avoid contamination. Diffusion is unlikely to play a role here as a 2.1 μm particle has a diffusion constant of 0.2 μm2∕s and traverses the array in a few seconds. So these particles would diffuse much less than one gap width, while traversing the arrays.
We emphasize that determining the enrichment factors in the microdevice outputs is based on the result of flow cytometry studies. For the spore experiments, it is the percent of all events falling within the specified polygon P1 on the forward∕side scatter plots. This region is associated with the presence of the spores, but it is not known what fraction of spores falls outside this region. We can be fairly certain that objects sorted in the microdevice have the correct size, but it is not clear what combination of spore size, granularity, and∕or opacity leads to a spore falling into P1. In principle, forward scatter reflects the particle size, however, the differing granularity and opacity are expected to influence the dot plots of forward and side scatter. We have shown evidence [Fig. 1a] that spores range from clear and smooth walled to opaque and rough walled, with both variants in significant proportion, somewhat contrary to an earlier report by Tokunaga et al.21 in 1973, who showed by electron microscopy that prior to release, the spores have a rough nodular surface.
This leads to the following question: How many of the events outside the polygon P1 are actually spores but with widely varying opacity∕granularity? One possibility is that the smooth walled, transparent spores form a loose cluster outside of P1. A second cluster to the left of P1 can clearly be seen in Fig. 4c, output 2. Perhaps these are the clear spores? For the experiment shown, a polygon around that region contains a 29.5% of events. Adding this to the events in P1 [Fig. 4c] accounts for 72% of all events in output 2. If these or other events are spores, then the purity of the microdevice product is very much underestimated here. Regrettably, simple microscopy does not provide large enough data sets to fully resolve this issue. Further study is therefore needed to correlate spore appearance with light scattering characteristics, as well as all-important viability. A significant amount of light scattering objects remain to be identified in the microdevice product.
The major limitation for the current device is the spore concentration. This is important for the industry where high concentrations are required for some applications, and regrettably, simple approaches such as postprocess centrifugation will cause unacceptable aggregation. Unfortunately, in the present device the input concentration cannot be made much higher as in-chip aggregation begins to clog the postarray and reduce the operable life of the device. Experiments with twice the spore density, around 10×106∕ml, caused discoloration of the postarray that was visible to the naked eye before the end of a 20 min run. A microscopic view showed that the discoloration was due to aggregates of spores and debris. At 4.4×106 spores∕ml, no significant aggregation of debris or spores is visible under microscope after the twenty minute run.
In the work presented here the volumetric throughput is quite small, only 5.5 μl of input sample∕min at 69 kPa, with about 400 spores∕s. We expect that this value may be increased by up to fivefold by increasing the driving pressure and tenfold by making a larger device with multiple parallel separation arrays.
CONCLUSIONS
In this work we have developed a device for the purification of Aspergillus spores by applying the principle of deterministic lateral displacement. The method takes advantage of the fact that the center of a small particle can be closer to a boundary than the center of a large particle. This fact is used to deflect the trajectory of particles above a particular hydrodynamic size as they flow through an array of obstacles. Aspergillus is an industrially important organism with round, micron-sized spores (mean diameter of 4.4 μm). We have designed the device to isolate spores by size from the debris laden spore suspension. The device was tested with polymer microbeads where it performed very well, recovering 99% of the target particles and eliminating 96% of smaller and larger particles. When tested with spore suspensions it provided a two- to threefold increase in spore purity, measured by light scatter.
ACKNOWLEDGMENTS
This work was funded by the Australian Research Council (Grant No. DP0880205) and was supported by a grant from the Fluorescence Applications in Biotechnology and Life Sciences Network. The authors are grateful for support from Professor Ewa Goldys and for helpful discussions with Varun KAS.
References
- West J., Manz A., and Dittrich P. S., Lab Chip 8, 1852 (2008). 10.1039/b811448j [DOI] [PubMed] [Google Scholar]
- Yen B. K. H., Gunther A., Schmidt M. A., Jensen K. F., and Bawendi M. G. A., Angew. Chem., Int. Ed. 44, 5447 (2005). 10.1002/anie.200500792 [DOI] [PubMed] [Google Scholar]
- Pamme N., Lab Chip 7, 1644 (2007). 10.1039/b712784g [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Lab Chip 9, 3038 (2009). 10.1039/b912547g [DOI] [PubMed] [Google Scholar]
- Li Y., Dalton C., Crabtree H. J., Nilsson G., and Kaler K. V. I. S., Lab Chip 7, 239 (2007). 10.1039/b613344d [DOI] [PubMed] [Google Scholar]
- Lapizco-Encinas B. H., Simmons B. A., Cummings E. B., and Fintschenko Y., Anal. Chem. 76, 1571 (2004). 10.1021/ac034804j [DOI] [PubMed] [Google Scholar]
- Moon H., Nam Y., Park J. C., and Jung H., Environ. Sci. Technol. 43, 5857 (2009). 10.1021/es900078z [DOI] [PubMed] [Google Scholar]
- Wu Z., Willing B., Bjerketorp J., Jansson J. K., and Hjort K., Lab Chip 9, 1193 (2009). 10.1039/b817611f [DOI] [PubMed] [Google Scholar]
- Qiu J., Zhou Y., Chen H., and Lin J., Talanta 79, 787 (2009). 10.1016/j.talanta.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Horká M., Růzička F., Kubescová A., Holá V., and Slais K., Anal. Chem. 81, 3997 (2009). 10.1021/ac900374v [DOI] [PubMed] [Google Scholar]
- Cheng I. -F., Chang H. -C., Hou D., and Chang H. -C., Biomicrofluidics 1, 021503 (2007). 10.1063/1.2723669 [DOI] [Google Scholar]
- Iliescu C., Tresset G., and Xu G., Biomicrofluidics 3, 044104 (2009). 10.1063/1.3251125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewpiriyawong N., Yang C., and Lam Y. C., Biomicrofluidics 2, 034105 (2008). 10.1063/1.2973661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. R., Cox E. C., Austin R. H., and Sturm J. C., Science 304, 987 (2004). 10.1126/science.1094567 [DOI] [PubMed] [Google Scholar]
- Davis J. A., Inglis D. W., Morton K. J., Lawrence D. A., Huang L. R., Chou S. Y., Sturm J. C., and Austin R. H., Proc. Natl. Acad. Sci. U.S.A. 103, 14779 (2006). 10.1073/pnas.0605967103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M. Y. J., Yamada M., and Seki M., Lab Chip 5, 778 (2005). 10.1039/b501885d [DOI] [PubMed] [Google Scholar]
- Yamada M. and Seki M., Anal. Chem. 78, 1357 (2006). 10.1021/ac0520083 [DOI] [PubMed] [Google Scholar]
- Inglis D. W., Davis J. A., Austin R. H., and Sturm J. C., Lab Chip 6, 655 (2006). 10.1039/b515371a [DOI] [PubMed] [Google Scholar]
- Heller M. and Bruus H., J. Micromech. Microeng. 18, 075030 (2008). 10.1088/0960-1317/18/7/075030 [DOI] [Google Scholar]
- Inglis D. W., Appl. Phys. Lett. 94, 013510 (2009). 10.1063/1.3068750 [DOI] [Google Scholar]
- Tokunaga M., Tokunaga J., and Harada K., J. Electron Microsc. 22, 27 (1973). [PubMed] [Google Scholar]