Summary
Adults maintain tissue-specific stem cells via niche signals. A leading paradigm for niche function is the Drosophila testis, where a small cluster of cells called the hub produce locally available signals allowing only adjacent cells to self-renew. We show here that the principle signaling pathway implicated in this niche, chemokine activation of STAT1,2, does not primarily regulate self-renewal of germline stem cells (GSCs), but rather governs GSC adhesion to hub cells. In fact, GSC renewal does not require hub cell contact, as GSCs can be renewed solely by contact with the second resident stem cell population, somatic cyst stem cells (CySCs), and this involves BMP signaling. These data suggest a modified paradigm whereby the hub cells function as architects of the stem cell environment, drawing into proximity cellular components necessary for niche function. Self-renewal functions are shared by the hub cells and the CySCs. This work also reconciles key differences in GSC renewal between the Drosophila testis and ovary niches, and highlights how a niche can coordinate the production of distinct lineages by having one stem cell type rely on a second.
Results
The critical self-renewal event in the Drosophila testis is thought to be activation of STAT in GSCs, caused by secretion of the cytokine Unpaired from the hub (Figure S1A, C). For instance, depletion of stat from all testis cells causes loss of both stem cell populations, the GSCs and CySCs, while misexpression of unpaired leads to self-renewal of both lineages away from the niche1,2. However, we recently found that germ cell-intrinsic STAT activation could not renew germ cells away from the niche3. Instead, intrinsic STAT activation in somatic cells renewed not only CySCs, but also permitted GSC renewal away from the niche3. This suggested that STAT-activated CySCs produce a germline self-renewal signal, and that the previously observed loss of GSCs upon global stat depletion was secondary to CySC loss. To test this, we examined stat-depleted testes by temperature inactivation of statts, but rescued the CySCs by restoring somatic expression of stat (Figure S1B, D–E; Table 1, Tj Gal4, c587 Gal4 or EyaA3 Gal4 entries). As expected, CySCs persisted because stat had been restored to this lineage specifically (Figure 1A, Zfh-1, green, marks CySCs), but now stat-depleted germline cells were maintained indefinitely in most testes (57/64 or 89% still had spermatogonia eight days after stat depletion; Figure 1C; see Table 1 for quantitation)4. In particular, amplifying spermatogonia and spermatocytes were observed at all stages of differentiation in their normal organization emanating from the testis apex near the hub, shown by normal Bam accumulation, branching fusomes, continual cycling through S phase, and spermatocyte Ptb accumulation (Figure 1D–G). These testes appeared to harbor as many (or even more) differentiating germ cells than wild-type testes. This is in striking contrast to the unrescued situation, where most testes had lost all germ cells to differentiation by eight days, and 0/37 had spermatogonia present (Figure 1B, C; Table 1).
Table 1. stat in the CySC lineage prevents loss of stat-depleted germ cells.
Global stat depletion by temperature shift (#1,4,7,10, 13) resulted in progressive germ cell loss to differentiation. Rescue of stat in the CySCs (#2,5,8,11,14,16) prevented germ cell loss in most testes, but these testes did not maintain GSCs at the hub. CySC expression of UAS stat alone did not cause the observed GSC loss (#3,6,9,12,15). Similar results were obtained with other stat alleles in combination with statF and other CySC drivers (#17–24). Stat rescue in the hub by Upd Gal4 (#25) did not rescue germ cells. Stat rescue in germ cells by Nos Gal4 VP16 (#26–28) resulted in modest germ cell loss, although in these testes, some CySCs always remained (see text). Germline stat RNAi (#29) caused GSCs to be lost from the hub, but these testes maintained germ cells.
| Genotype | Days after stat depletion | GSCs at hub/testis | Percent of testes with: |
Testes Scored | ||
|---|---|---|---|---|---|---|
| GSCs at hub | Spermatogonia | Spermatocytes | ||||
| 1. statF/stat85c9 | 0 | 9.3 | 100% | 100% | 100% | 15 |
| 2. Tj Gal4/UAS stat; statF/stat85c9 | 0 | 4.6 | 100% | 100% | 100% | 25 |
| 3. Tj Gal4/UAS stat; stat85c9/+ | 0 | 6.9 | 100% | 100% | 100% | 35 |
| 4. statF/stat85c9 | 2 | 2.6 | 73% | 100% | 100% | 30 |
| 5. Tj Gal4/UAS stat; statF/stat85c9 | 2 | 0 | 0% | 99% | 99% | 80 |
| 6. Tj Gal4/UAS stat; stat85c9/+ | 2 | 5.4 | 100% | 100% | 100% | 32 |
| 7. statF/stat85c9 | 4 | 0.01 | 1% | 36% | 100% | 73 |
| 8. Tj Gal4/UAS stat; statF/stat85c9 | 4 | 0.05 | 4% | 97% | 99% | 105 |
| 9. Tj Gal4/UAS stat; stat85c9/+ | 4 | 3.0 | 88% | 100% | 100% | 26 |
| 10. statF/stat85c9 | 8 | 0 | 0% | 0% | 8% | 37 |
| 11. Tj Gal4/UAS stat; statF/stat85c9 | 8 | 0 | 0% | 89% | 94% | 64 |
| 12. Tj Gal4/UAS stat; stat85c9/+ | 8 | 2.4 | 90% | 100% | 100% | 20 |
| 13. statF/stat85c9 | 16 | 0 | 0% | 0% | 0% | 63 |
| 14. Tj Gal4/UAS stat; statF/stat85c9 | 16 | 0 | 0% | 80% | 70% | 66 |
| 15. Tj Gal4/UAS stat; stat85c9/+ | 16 | 2.31 | ND1 | 57% | 61% | 49 |
| 16. Tj Gal4/UAS stat; statF/stat85c9 | 24 | 0 | 0% | 63% | 63% | 8 |
| Other stat alleles: | ||||||
| 17. statF/stat397 | 8 | 0 | 0% | 0% | 0% | 9 |
| 18. Tj Gal4/UAS stat; statF/stat397 | 8 | 0 | 0% | 90% | 92% | 52 |
| 19. statF/stat06346 | 8 | 0 | 0% | 0% | 7% | 28 |
| 20. Tj Gal4/UAS stat; statF/stat06346 | 8 | 0 | 0% | 88% | 100% | 16 |
| Other Gal4 Drivers: | ||||||
| 21. c587 Gal4; UAS stat; statF/stat85c9 | 8 | 0 | 0% | 77% | 79% | 42 |
| 22. c587 Gal4; UAS stat; statF/stat85c9 | 16 | 0 | 0% | 74% | 67% | 27 |
| 23. EyaA3 Gal4/UAS stat; statF/stat85c9 | 8 | 0 | 0% | 98% | 98% | 42 |
| 24. EyaA3 Gal4/UAS stat; statF/stat85c9 | 16 | 0 | 0% | 82% | 41% | 17 |
| 25. Upd Gal4; UAS stat; statF/stat85c9 | 8 | 0 | 0% | 0% | 0% | 29 |
| 26. Nos Gal4 VP16/UAS stat; statF/stat85c9 | 0 | 10 | 100% | 100% | 100% | 10 |
| 27. Nos Gal4 VP16/UAS stat; statF/stat85c9 | 8 | 3.1 | 84%2 | 92% | 99% | 90 |
| 28. Nos Gal4 VP16/UAS stat; statF/stat85c9 | 16 | 0.7 | 23%2 | 29% | 52% | 56 |
| stat RNAi: | ||||||
| 29. VDRC 10690 stat; nos Gal4 VP163 | - | 0.7 | 41% | 96% | 100% | 80 |
Expanded Fas3-positive somatic cell population in many testes made identification of hub and the number of GSCs attached to it impossible. GSCs/testis count includes only those testes without an expanded Fas3-positive population.
Testes with germline stat rescue maintained some CySCs, thus germ cell loss was modest (see text).
stat RNAi flies were aged 7 days at 29° after eclosion.
Figure 1. Germline stat depletion does not compromise self-renewal.
Germline stat depletion (A, D–G; Tj Gal4/UAS HA-stat; statF/stat85c9) or global stat depletion (B; statF/stat85c9) eight days after stat temperature inactivation. Hub (asterisk and Fas3, white); germ cells (vasa, magenta); DNA (blue). (A) Testes with germline stat depletion maintained CySCs (Zfh-1, green), and germ cells. (B) Testes with global stat depletion exhibited germ cell loss (lack of magenta cells) and CySC loss (Zfh-1, green; note, a few CySCs always persisted near the hub). (C) Global stat depletion led to progressive germ cell loss, first the more undifferentiated spermatogonia (blue; this category includes GSCs/single germ cells) followed by spermatocytes (orange), while most testes with germline stat depletion maintained spermatogonia (blue) indefinitely. Similar results were obtained with three CySC lineage Gal4 drivers and three strong stat alleles with the statts allele (Table 1). (D–G) Stat-depleted germ cells continually progressed through differentiation, shown by normal expression of the spermatogonial differentiation marker Bam (D, green), the appearance of branching fusomes through germ cell cysts (E, α-spectrin, green), synchronously cycling germ cell cysts (F, Edu pulse-labeled cells, green), and expression of the spermatocyte marker Ptb (G, green). Bar:10μm.
The continual production of normally differentiating germ cells indicated that stat-depleted germ cells must still be functioning as GSCs. In normal testes, GSCs are closely apposed to the hub. Surprisingly, when germline stat was depleted, germ cells lost contact with the hub (Figure 2A vs. B), and the nearest germ cells were now located several cell tiers away from the hub. The first few tiers of these displaced germ cells remained in contact with CySCs as judged by Zfh-1 expression, and these germ cells retained the hallmarks of GSCs, including expression of M5-4, appearance of dot spectrosomes, and cycling as individual cells (Figure 2C–E). While these markers are not unique to GSCs, but shared by gonialblasts (the differentiating daughters of GSCs), the appearance of these characteristics along with the continual, robust production of normally differentiating germ cells demonstrates that the displaced, stat-depleted germ cells still self-renew as GSCs.
Figure 2. stat depleted GSCs become displaced from the hub.
Germ cells (vasa, magenta), Hub (asterisk or Fas3, white). (A) Wild-type GSCs are tightly associated with the hub. (B) Two days after germline stat depletion (Tj Gal4/UAS HA-stat; statF/stat85c9), GSCs became displaced from the hub. (C–F) 8 days after germline stat depletion, germ cells nearest the hub accumulate M5-4 lacZ (C, white, arrows), have spherical or dumbbell-shaped spectrosomes (D, α-spectrin, green, arrows; D′ shows α-spectrin alone), and undergo mitosis as individual cells (E, phosphohistoneH3, green, arrow; a mitotic CySC is also present), all consistent with GSC identity. M5-4 lacZ (C, white) also accumulates in the hub (arrowheads), but not in CySCs (Tj, green), indicating CySCs do not take on hub fate. (F) stat-depleted GSCs fail to reliably orient mitotic spindles away from the hub (centrosomin reveals centrosomes, green). Two mitotic germ cells outlined, one oriented (arrow) and one non-oriented (arrowhead). (G) With germline stat depletion, the hub became surrounded by CySCs (Zfh-1, green). (H) Testes with Gal4-driven stat in the CySC lineage (Tj Gal4/UAS stat; stat85c9/+) undergo modest GSC loss, but still maintain GSCs at the hub (arrows, Zfh-1, green). (I) Germline RNAi knockdown of stat (nos gal4 VP16 VDRC 106980) caused GSCs to be displaced from the hub, and the hub became surrounded by CySCs (Zfh-1, green). Bar:10μm.
The only hallmark of GSCs that was disrupted under conditions of stat-depletion was the reliably oriented plane of cell division. Normally, GSCs divide perpendicular to the hub surface5, providing for a mechanism of asymmetric stem cell division, with one daughter cell invariantly pushed away from renewing influences, allowing its differentiation program to initiate. Such oriented divisions are directly dependent on tethering of one centrosome in the GSC to its interface with the hub6,7. While centrosome staining of wild-type mitotic GSCs shows invariant orientation6, the division plane of stat-depleted, displaced GSCs became randomized (Figure 2F shows one oriented mitotic GSC (arrow) and one non-oriented mitotic GSC (arrowhead); ratio of oriented:non-oriented mitotic GSCs was 1:1.7, n=16 GSCs; see Methods). Thus, this alternate niche configuration likely does not precisely parse the stem cell daughters into one renewing and one differentiating cell, demonstrating the niche’s capacity to function in a less regimented, population mode for self-renewal, where the production of differentiating daughter cells is stochastic8. We do not know whether GSCs in this alternate niche configuration completely bypass the other characteristics of asymmetric division, including stem cell inheritance of the mother centrosome and the mitotic checkpoint associated with centrosome attachment7,9.
In testes with stat-depleted GSCs, the CySCs were rescued due to lineage-specific restoration of stat expression, and these Zfh-1-positive cells now surrounded the hub (which is the source of the STAT-activating ligand). We noted that the stat-depleted, displaced GSCs were interspersed among Zfh-1-expressing cells (Figure 2G). These Zfh-1-expressing cells did not adopt hub character (Figure S2 and Figure 2C), suggesting that it was their CySC fate and not any influence of hub cells that was integral to the maintenance of GSCs under these conditions. In fact, stat-depleted GSCs could not be rescued by restoration of stat solely to the hub cells (Table 1, Upd Gal4 entry). The requirement for CySCs was further confirmed because stat-deficient GSCs were rescued, although to a lesser degree, by cyst lineage restoration of zfh-1, a target of STAT in CySCs (Table S1). This is consistent with previous findings that Zfh-1 controls a CySC signal that can support GSC renewal3.
If CySCs are required for GSC renewal, loss of the CySC population should lead to GSC loss, even if the GSCs retain stat. To test this, we specifically depleted stat in the CySC lineage by temperature shift of the statts allele accompanied by Gal4 stat rescue in germline cells (Table 1, Nanos Gal4 VP16 entry). Unshifted controls averaged 36 Zfh-1-positive cells/testis (n=10), while testes depleted for stat in CySCs for eight and sixteen days averaged only 14.2 (n=25) and 9.5 (n=56) Zfh-1-positive cells/testis respectively. Over the same period, we observed a gradual loss of GSCs. While unshifted controls had 10 GSCs/testis on average (n=10), testes eight and sixteen days after stat loss in CySCs harbored only 3.1 (n=90) and 0.7 (n=56) GSCs/testis on average, respectively. This result is consistent with a requirement for CySCs in GSC renewal. We had expected that CySC stat-depletion would cause all CySCs to differentiate. The retention of some CySCs may be due to the hypomorphic nature of the statts allele, or alternatively, a partially redundant mechanism for CySC maintenance.
Next, we investigated why stat-depleted GSCs became displaced from the hub. It is known that an intrinsic increase in STAT activation in CySCs causes these cells to outcompete GSCs for niche access10. Since our strategy to examine stat-depleted GSCs utilized Gal4-driven rescue of stat in CySCs, we considered whether higher than normal STAT levels in CySCs led to excess STAT activation and hence could account for the observed GSC displacement from the hub. However, this did not explain the displacement of GSCs in our experiment. As expected, testes expressing excess somatic stat but otherwise stat/+ indeed led to some displacement of GSCs (Table 1: Tj Gal4/UAS stat; stat85c9/+). However, even after eight days, most such testes still retained GSCs at the hub (90% of testes had GSCs, averaging 2.4 GSCs/testis; Figure 2H, Table 1). Only upon depletion of stat in the germline were all GSCs lost from the hub, and this occurred within two days (Table 1: Tj Gal4/UAS stat; statts/stat85c9). To confirm that intrinsic reduction of STAT in the germline led to GSC displacement, we knocked down stat by RNAi in the germline. This experiment involved no extra stat expression in CySCs, yet also led to detachment of most GSCs from the hub (Figure 2I). These testes similarly exhibited continual, robust production of normally differentiating germ cells, and displaced GSCs clustered next to CySCs, which now surrounded the hub (Table 1). Thus, STAT is required intrinsically in GSCs to promote attachment to the hub. We note that under the conditions of stat depletion that must be used with the statts allele, there could be some residual STAT function in germ cells. Thus, we cannot eliminate the possibility that STAT plays some direct role in self-renewal of GSCs. However, a principal role in adhesion is sufficient to explain earlier mosaic experiments1,2. We suggest that stat null GSCs were lost, not because stat was essential for self-renewal, but rather, because an attachment defect led to their displacement from the niche, and nearby wild-type germ cells de-differentiated11 to take their place.
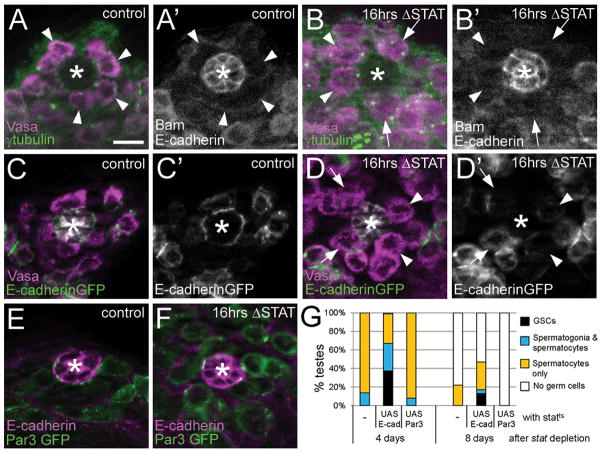
If STAT primarily regulates attachment, then adhesion defects should precede other signs of differentiation in GSCs. To ensure reliably oriented divisions, an elaborate cortical complex is postulated to be established in GSCs at the hub interface, linking one centrosome to the adherens junction (Figure 3A and C, respectively)6,7. Thus, in testes with global stat depletion, we examined these components in GSCs prior to overt signs of differentiation, scoring only GSCs that were still located next to the hub and that did not yet express the differentiation marker Bam (Figure 3A′, B′). At sixteen hours after stat depletion, there were on average 4.5 such undifferentiated GSCs per testis (n=26 testes) compared to 7.0 for controls (n=12). We found that 47% of such GSCs (n=86) exhibited defective centrosome placement (Figure 3B, arrows), compared to only 8% (n=64) in controls (Figure 3A). In addition, we expressed E-cadherin-GFP in germline cells, which selectively labeled the adherens junctions at the GSC-hub interface (Figure 3C′). In stat-depleted testes, E-cadherin-GFP became significantly delocalized in most GSCs, with 44% of GSCs (n=81) completely lacking enrichment at the hub interface compared to only 16% (n=102) in controls (Figure 3C, D; arrowheads and arrows show reduced or absent E-cadherin enrichment, respectively). We observed similar dispersal of Par3-GFP from the interface (Figure 3E, F; quantified in Table S2); although Par3 has not been postulated to be part of the cortical complex in GSCs, it fortuitously localized in prominent puncta at the hub-GSC interface, adjacent to E-cadherin. Thus, stat-depleted GSCs exhibit severe disruption of the hub-GSC interface, and this precedes overt differentiation. We also found that increasing germline E-cadherin expression moderately suppressed the germ cell loss phenotype (Figure 3G), suggesting that regulation of adhesion is a key role for STAT in the germline.
Figure 3. GSCs display hub adhesion defects prior to other signs of differentiation upon global stat depletion.
(A) Most control GSCs (statF/stat85c9, 18° permissive temperature) have one centrosome oriented toward the hub interface (arrowheads, γ-tubulin (green) shows centrosomes; vasa (magenta) shows germ cells). (B) Upon stat depletion (sixteen hours after upshift) many GSC centrosomes were misoriented (arrows), while others were not (arrowheads). These GSCs are Bam-negative (A′ and B′, white shows Bam with E-cadherin marking the hub). (C) In control GSCs (Nos Gal4 VP16/UAS E-cadherinGFP; stat85c9/+, 16hrs 29°), E-cadherinGFP (green) localizes to the hub interface (vasa, magenta; fas3, white). (D) Upon sixteen hour stat depletion (Nos Gal4 VP16/UAS E-cadherinGFP; statF/stat85c9) E-cadherinGFP was lost (arrows) or highly reduced (arrowheads) from the hub interface. E-cadherinGFP only (C′, D′). (E) Germline Par3GFP (green) colocalizes with E-cadherin (magenta) at hub-GSC interface in controls (Nos Gal4 VP16/UAS Par3GFP; stat85c9/+, 16hrs 29°). (F) Upon sixteen hour stat depletion (Nos Gal4 VP16/UAS Par3GFP; statF/stat85c9) Par3GFP (green) delocalized, with only modest interface enrichment remaining. Quantified in Table S2. (G) Germline UAS E-cadherin expression in statts testes delayed germ cell loss compared to statts alone or with UAS Par3 expression. Asterisk denotes hub. Bar:10μm.
If cell-intrinsic Jak/STAT pathway activation does not primarily regulate GSC self-renewal, what does? The BMP pathway has been implicated in GSC renewal12–14. The ligands, gbb and dpp, are each expressed in CySCs (and in hub cells), and pathway activation has been observed in GSCs12–14. In testes with germline stat depletion, we observed strong BMP pathway activation in the displaced GSCs, shown by accumulation of phosphoSmad and the BMP target gene dad (Figure 4A, B). Interestingly, we noted that the levels of pSmad and Dad-lacZ were much higher in testes with stat-deficient displaced GSCs than in wild-type GSCs (wild-type not shown), possibly because higher levels of STAT in CySCs led to higher BMP ligand levels. Sustained zfh-1 expression in the cyst lineage, which generates ectopic GSCs far from the hub3, also led to strong BMP pathway activation in the ectopic GSCs (Figure 4C). Thus, Jak/STAT activation in CySCs leads to BMP ligand expression15, likely using zfh-1 as an intermediary.
Figure 4. Rescued GSCs are activated for and require the BMP pathway.
Germ cells (vasa, magenta), hub (asterisk). Eight days after germline stat depletion, the germ cells near the hub accumulated phospho-Smad (A, green, arrows; A′ pSmad alone) and Dad-lacZ (B, green, arrows; Zfh-1 in white; B′ Dad-lacZ alone). (C) Sustained zfh-1 expression in the cyst lineage (eyaA3 Gal4/UAS zfh-1; tubGal80ts; twelve days) generated excess GSCs (magenta)3 and these accumulated Dad-lacZ. DNA (blue). (D) Most testes with germline stat depletion retain spermatogonia (blue), but dampening the BMP pathway by coexpression of UAS sogCR1 (P<0.05 for 8d) or kekkon5 (P<0.0001 for 8d) reduced the proportion of testes with rescued germ cells. (E) Model for self-renewal in the testis niche. Bar:10μm.
In testes with germline stat depletion, we also found that dampening the BMP pathway by expression of the extracellular BMP antagonists sogCR1 or kekkon5 suppressed rescue of the germline (Figure 4D). Thus, BMPs are functionally important in CySC-supported GSC self-renewal. A primary role for BMPs in male GSCs makes the testis much more similar to the Drosophila ovary, where BMP pathway activation regulates GSC renewal via repression of the differentiation gene, bam16,17, as also occurs in the male germline18. A remaining difference, however, is that, in the male, BMP pathway activation is insufficient for induction of ectopic GSCs. Thus, there may be another assisting signal that also emanates from the CySCs. The Jak/STAT pathway is also similar between the two niches, with pathway activation primarily required in the somatic cell populations3,19,20. However, while the testis GSCs use Jak/STAT-regulated centrosome tethering to attach and orient the division plane, the ovary uses Jak/STAT-independent spectrosome localization to ensure an asymmetric outcome to stem cell divisions21.
One can formulate a revised model for this paradigmatic niche taking our data along with recent work (Figure 4E). The hub cells are architects of the stem cell environment, establishing close proximity of the three cell populations because STAT influences adhesion in nearby germline and somatic cells, probably through adherens junctions (this work)22 and integrin linkages10, respectively. STAT activation also governs self-renewal in the somatic lineage via zfh-1 and chinmo induction3,23. These CySCs then activate BMP, which is used to renew neighboring, hub-adherent germ cells. Thus, self-renewal activity relies not just on the hub, but also on one of the resident stem cell lineages. BMPs are expressed from both hub and CySCs12,13, possibly to generate sufficient quantity of BMP for renewal; alternatively, the ligand composition from hub versus CySCs may be qualitatively different. In either case, the parsing of different roles such as adhesion and self-renewal among multiple niche components is a departure from previous niche paradigms, and is likely to be utilized in the much more complex niche environments in mammals.
Methods
Fly Stocks and Crosses
The temperature-sensitive stat allele (statF, FBal0060556) was used in trans with a strong stat allele, either stat85c9, stat06346, or stat397 (FBal0130583, FBal0009559, FBal0130586, Erika Bach). Germ cell stages present in adult testes were tabulated before and at various time points after stat was inactivated by a shift from 18° (the permissive temperature) to 29° (the non-permissive temperature). Spermatogonia were identified by their smaller cell size and the presence of bright Hoechst staining. Included in this category were GSCs attached to the hub and individual or paired germ cells away from the hub. Spermatocytes were identified by increased cell size and much reduced Hoechst staining. Gal4 drivers were Traffic Jam Gal4 (Kyoto Stock Center, DGRC #104055), c587 Gal4 (Erika Matunis), or EyaA3 Gal43 for CySC expression, Nos Gal4 VP16 (Ruth Lehmann and Erica Selva) for germline expression, and Upd Gal4 (Doug Harrison) for hub expression. Rescue transgenes were UAS Stat-HA (FBtp0036673, Erika Bach, NYU, NY) and UAS zfh-1 (FBtp0016144, Bloomington Stock Center). Bam expression was visualized using a Bam-GFP reporter transgene (Dennis McKearin, UT Southwestern, Tx) and staining for GFP. Germline depletion of stat was also accomplished using an RNAi transgene (#106980; Vienna Drosophila Resource Center) crossed to Nos Gal4 VP16. We analyzed cortical components of the GSC at the interface with hub cells by crossing Nos Gal VP16 with UAS E-cadherin-GFP (also called UAS DEFL, Kyoto Stock Center) or UAS Par3GFP (Daniel St Johnston, MRC, UK) in stat mutant testes. Dad expression was visualized using the enhancer trap insertion P{lacZ}DadP1883 (FBal0094852), followed by lacZ immunostain. The BMP pathway was dampened by coexpressing either UAS sogCR1 (activated form of sog, FBtp0018246, Ethan Bier)24 or UAS kekkon5 (FBtp0018362, Joe Duffy)25 in the CySC lineage along with UAS stat in statts flies. These are both extracellular BMP ligand antagonists. Various time points after stat temperature inactivation, the types of germ cells present were scored, and compared to controls (the same genotype, lacking BMP antagonist expression). Significance was determined by the Chi-square test of independence (http://faculty.vassar.edu/lowry/odds2x2.html).
Immunostaining
Testes were dissected and fixed in 4% paraformaldehyde in Buffer B (16.7 mM KH2PO4/K2HPO4, 75 mM KCl, 25 mM NaCl, 3.3 mM MgCl2, pH 6.8) with 0.1% Triton X-100 for 20 minutes. Samples were blocked for one hour in 4% normal donkey serum in PBS with 0.1% Triton. Primary antibodies were incubated overnight in block, and were goat anti-Vasa (1:400, Santa Cruz sc-26877), mouse anti-Fasciclin III (1:50, Developmental Studies Hybridoma Bank), rabbit anti-Zfh-1 (1:5000, Ruth Lehmann), rabbit anti-Stat23 (1:1000, Erika Bach), mouse anti-HA (1:600, Covance), rabbit anti-GFP (1:1000, Molecular Probes), rabbit anti-alpha-spectrin (1:600, Dan Kiehart, Duke Univ., N.C.), rabbit anti-Ptb (1:200, David Standiford), mouse anti-lacZ (1:10,000, Promega), rabbit anti-phosphohistone H3 (1:500, Upstate Biotech), rabbit anti-centrosomin (1:1000, Thomas Kaufman, Bloomington, In), mouse anti-γtubulin (1:100, Sigma Clone GTU-88), rat anti-E-Cadherin (1:20, Developmental Studies Hybridoma Bank), mouse anti-phosphoSmad (1:1000, Carl-Henrik Heldin), and guinea pig anti-traffic jam (1:10,000, Dorothea Godt). Secondary antibodies conjugated to A488, Cy3, or Cy5 (Molecular Probes and Jackson Immunologicals) were used at 1:400. Testes were stained 5 minutes with Hoechst 33342 (Sigma) at 1.0 μg/ml to reveal DNA.
S Phase Labeling
To label cells in S phase, freshly-dissected testes were incubated for 30 minutes in 10μM Edu26 then fixed. Testes were stained as above with other antibodies, followed by visualization of the Edu label with the Click-iT EdU Alexa Fluor 647 Kit (Invitrogen) according to the manufacturer’s instructions.
Spindle orientation and centrosome attachment
In order to examine spindle orientation in stat-depleted GSCs, we examined the first tier of germ cells nearest to the hub eight days after stat inactivation. Mitotic germ cells were identified by centrosomin enrichment27, DNA compaction, and nuclear breakdown (revealed by loss of vasa exclusion from the nucleus). At this stage of the cell cycle, spindle orientation is reflected by centrosome position. Germ cells were scored as oriented if one centrosome was located within an arc inscribing approximately one quarter of the germ cell contour nearest the hub.
Centrosome attachment to the hub interface in GSCs sixteen hours after stat depletion was scored by γtubulin stain, which reveals centrosomes in all stages of the cell cycle. GSCs were scored as having an attached centrosome if one centrosome was visible at the GSC-hub interface (in some cells, centrosome separation might not yet have occurred). GSCs were scored as having unattached centrosomes if both centrosomes were visible and neither was anchored at the hub-GSC interface.
Quantitation of germline Par3-GFP mislocalization
Germline Par3-GFP localization was quantified by measuring the average intensity along an arc, drawn either along the hub-GSC interface or along each lateral edge of the GSC. In all cases, Z slices with the highest intensity were chosen. The two lateral edge measurements were averaged, and this value was used as the denominator in a ratio with the hub interface value (hub/lateral edges). The resulting ratios from all scored GSCs were averaged, and a p value calculated by a two tailed Students t test.
Supplementary Material
Acknowledgments
We thank members of the fly community, Bloomington Stock Center, Vienna Drosophila Resource Center, and the Developmental Studies Hybridoma Bank for reagents. We thank Erika Matunis for helpful comments on the manuscript, and Seth Donoughe for technical assistance. This work was supported by NIH R01GM8060804 and 060804-10S1 to S.D.
Footnotes
Author Contributions Experiments were conceived and analyzed by J.L.L. and S.D. J.L.L. performed the experiments and wrote the manuscript. S.D. edited the manuscript.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/naturecellbiology/
References
- 1.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–9. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 2.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–5. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 3.Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SR, et al. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J Cell Physiol. 223:500–10. doi: 10.1002/jcp.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy RW, Tokuyasu KT, Lindsley DL, Garavito M. The germinal proliferation center in the testis of Drosophila melanogaster. J Ultrastruct Res. 1979;69:180–90. doi: 10.1016/s0022-5320(79)90108-4. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–50. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–21. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287:1427–30. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 9.Cheng J, et al. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issigonis M, et al. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–6. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brawley C, Matunis E. Regeneration of Male Germline Stem Cells by Spermatogonial Dedifferentiation in Vivo. Science. 2004 doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 12.Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr Biol. 2003;13:2065–72. doi: 10.1016/j.cub.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 13.Kawase E, Wong MD, Ding BC, Xie T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131:1365–75. doi: 10.1242/dev.01025. [DOI] [PubMed] [Google Scholar]
- 14.Schulz C, et al. A Misexpression Screen Reveals Effects of bag-of-marbles and TGFbeta Class Signaling on the Drosophila Male Germ-Line Stem Cell Lineage. Genetics. 2004;167:707–23. doi: 10.1534/genetics.103.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Li Z, Cai Y. The JAK/STAT pathway positively regulates DPP signaling in the Drosophila germline stem cell niche. J Cell Biol. 2008;180:721–8. doi: 10.1083/jcb.200711022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–60. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–91. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 18.McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–51. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 19.Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell. 2005;9:501–10. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Onieva L, Fernandez-Minan A, Gonzalez-Reyes A. Jak/Stat signalling in niche support cells regulates dpp transcription to control germline stem cell maintenance in the Drosophila ovary. Development. 2008;135:533–40. doi: 10.1242/dev.016121. [DOI] [PubMed] [Google Scholar]
- 21.Deng W, Lin H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- 22.Voog J, D’Alterio C, Jones DL. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454:1132–6. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaherty MS, et al. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 18:556–68. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K, et al. Processing of the Drosophila Sog protein creates a novel BMP inhibitory activity. Development. 2000;127:2143–54. doi: 10.1242/dev.127.10.2143. [DOI] [PubMed] [Google Scholar]
- 25.Evans TA, Haridas H, Duffy JB. Kekkon5 is an extracellular regulator of BMP signaling. Dev Biol. 2009;326:36–46. doi: 10.1016/j.ydbio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–20. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li K, Kaufman TC. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 1996;85:585–96. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.