During the past two decades, magnetic resonance imaging (MRI) has become a powerful technique in clinical diagnosis and biological molecular imaging.[1–4] A signficant advantage of MRI is the ability to acquire tomographic information of whole animals with high spatial resolution and soft tissue contrast. In addition, images are acquired without the use of ionizing radiation (e.g., X-ray and CT) or radiotracers (e.g., PET and SPECT) permitting long term longitudinal studies. Since spatial resolution increases with magnetic field strength, the ability to track small cell populations has been realized.
MRI contrast agents are frequently utilized to permit the visual differentiation of cells and tissues that are magnetically similar but histologically distinct. Paramagnetic gadolinium [Gd(III)] complexes are the most widely used contrast agents, as Gd(III) reduces the longitudinal relaxation time (T1) of local water protons due to its high magnetic moment and symmetric S-state. Areas enriched with Gd(III) exhibit an increase in signal intensity and appear bright in T1-weighted images. Furthermore, chelation of the Gd(III) ion (required to decrease latent toxicity) provides a means for chemical modification with targeting or bioactive moieties and cell transduction domains.
Recent advances in design and amplification strategies have produced a wide variety of bioactivatable contrast agents for investigating biologically important events such as ion fluctuation, enzyme activity, peroxide evolution, and temperature variation.[5–12] However, the majority of these agents are incapable of penetrating cells and therefore are of limited use in cell tracking experiments.
Recent results suggest that Gd(III) contrast agents have shown promise in cell tracking and fate-mapping experiments. For example, tracking stem cells in adult rat brains post stroke and monitoring β-islet cell transplantation has demonstrated potential.[13–15] However, there are few examples of MR probes with the essential characteristics of high Gd(III) loading for enhanced contrast coupled with facile cell uptake and long-term cell retention. Herein, we report a multimodal, cell permeable, Gd(III) enriched polyvalent DNA gold nanoparticle (DNA-Gd(III)@AuNPs) conjugate for cellular MR imaging. This conjugate takes advantage of high cellular uptake, excellent stability, and high Gd(III) loading of polyvalent DNA-AuNPs.[16,17] These are properties not shared by all nanostructures and are a result of the dense loading of the oligonucleotides on the surface of the DNA-AuNPs and their ability to bind to proteins, which facilitates endocytosis.[18,19] In addition to gene regulation, DNA-AuNPs have been used in detection systems for DNA, proteins, metal ions, small molecules, and intracellular siRNA.[18],[20–31]
These Gd(III) enriched DNA-AuNP conjugates represent a new class of MR contrast agent with the capability of highly efficient cell penetration and accumulation that provides sufficient contrast enhancement for imaging small cell populations with μM Gd(III) incubation concentrations. Moreover, these conjugates are labelled with a fluorescent dye permitting multimodal imaging to confirm cell uptake and intracellular accumulation, and providing a means for histological validation.[32]
NP conjugates were prepared by reacting citrate stabilized gold nanoparticles with thiol-labeled 24-mer poly dT oligonucleotides. DNA oligos were synthesized on a solid support with post modification carried out in solution. The poly dT contained five conjugation sites (hexylamino labelled dT groups conjugated with a cross linker, azidobutyrate N-hydroxysuccinimideester) for covalently attaching Gd(III) complexes through click chemistry. Click chemistry has proven to be an efficient method for preparing Gd(III)-based MR contrast agents with high synthetic yields and increased relaxivity.[33]
After purification by RP-HPLC, the DNA-Gd(III) conjugates were characterized by MALDI-MS, which confirmed formation of the conjugates. The DNA-Gd(III) conjugates were then immobilized on citrate stabilized AuNPs following literature procedures used to make the analogous Gd(III)-free NPs to yield DNA-Gd(III)@AuNPs (Scheme 1).[34] Excess DNA-Gd(III) was removed by repeated centrifugation and re-suspension of the NPs until the supernatant was free of Gd(III). When suspended in aqueous solution, the NP conjugates appear deep red in color due to the plasmon resonance of the Au at 520 nm, and they are stable for months at room temperature (see supporting information). Cy3-labelled DNA oligos were synthesized in order to make Cy3-DNA-Gd(III)@AuNPs for fluorescence microscopy and flow cytometry to confirm cell uptake and labelling efficiency, respectively.
Scheme 1.
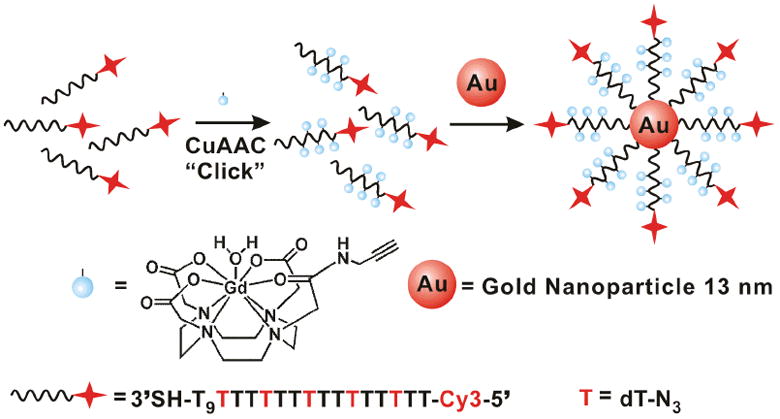
Schematic illustration of the synthesis of Cy3-DNA-Gd(III)@AuNP conjugates.
The relaxation efficiency of these newly synthesized MR contrast agent conjugates were determined by taking the slope of a plot of the measured 1/T1 as a function of Gd(III) concentration. The resultant relaxivity, r1, of the Gd(III) complex after conjugation to DNA was determined to be 8.7 mM−1s−1 at 37 °C in water at 60 MHz (1.41 T). This represents a two-fold increase over the unconjugated Gd(III) complex (3.2 mM−1s−1, Table 1). This doubling in relaxivity is consistent with Soloman–Bloomberg–Morgan theory where decreases in rotational correlation time, τr, result in increases in r1.[1,19]
Table 1.
Relaxivities (r1s) of Gd(III) complexes and conjugates at 60 MHz and 600 MHz.
|
r1(mM−1s−1) |
||
|---|---|---|
| 60 MHz (1.41 T)a | 600 MHz (14.1 T)b | |
| DOTA-Gd(III) | 3.2c | 2.2 |
| DNA-Gd(III) | 8.7 | NM |
| 13 nm DNA-Gd(III)@AuNP/ionic | 16.9 | 5.1 |
| 13 nm DNA-Gd(III)@AuNP/particle | 5779 | 1275 |
| 30 nm DNA-Gd(III)@AuNP/ionic | 20.0 | NM |
| 30 nm DNA-Gd(III)@AuNP/particle | 13,120 | NM |
Measured in pure water at 37°C.
Measured in cell media at 25°C.
Data taken from reference[1].
NM = not measured.
It is important to note that the relaxivity of Gd(III) increases further when DNA-Gd(III) is immobilized on the surface of AuNPs through gold thiol linkages. We have examined two different sizes of AuNPs (13 and 30 nm) and found that the ionic relaxivity [per Gd(III)] was 16.9 mM−1s−1 for 13 nm DNA-Gd(III)@AuNPs and 20.0 mM−1s−1 for 30 nm DNA-Gd(III)@AuNPs.
The degree of conjugation of chelates to the AuNP surface, was determined by calculating the Gd(III) to Au ratio following ICP-MS where 13 nm DNA-Gd(III)@AuNPs have 342 ± 1 Gd(III) per NP and 30 nm DNA-Gd(III)@AuNPs have 656 ± 20 Gd(III) per NP. These calculations are based on the assumption that there are 67,500 Au atoms per 13 nm AuNP, and 800,589 Au atoms per 30 nm AuNP (numbers were determined by geometric arguments and the crystal structure of bulk gold). Taking into account the loading of Gd(III) per particle, the 13 nm DNA-Gd(III)@AuNPs exhibited a relaxivity of approximately 5779 mM−1s−1 per particle.
Previously, three research groups have shown increased relaxivities of Gd(III) complexes by attaching thiol derivatized Gd(III) chelates on AuNPs.[35],[36] Moriggi et al. reported the highest relaxivity, namely 50 mM−1s−1 [per Gd(III)] at 60 MHz and 25 °C. The higher relaxivity of this conjugate is a result of two water molecules (as opposed to 1) coordinated to each Gd(III) ion, and the increased rigidity due to the nature of the attachment of the chelates to the particle surface.[36]
T1-weighted MR images of the DNA-Gd(III)@AuNPs in solution phantoms were acquired at 3 T and 14.1 T at 25 °C (see supporting information). The images clearly show that at each concentration [60 μM, 40 μM, 20 μM Gd(III)], DNA-Gd(III)@AuNPs appear significantly brighter than DOTA-Gd(III) samples at the same concentration at both field strengths. T1 analysis at 14.1 T reveals a 52% reduction in T1 for DNA-Gd(III)@AuNPs [60 μM Gd(III)] versus a 31% reduction for DOTA-Gd(III). The image-based r1 (at 14.1 T) of DNA-Gd(III)@AuNP is 5.1 mM−1s−1 whereas the r1 of DOTA-Gd(III) is 2.1 mM−1s−1 (Table 1).
To determine the efficacy of cellular uptake, NIH/3T3 and HeLa cells, were labelled with increasing concentrations of DNA-Gd(III)@AuNPs or DOTA-Gd(III) for different amounts of time. Following contrast agent incubation, cells were rinsed with DPBS, counted and then percent viability was assessed via flow cytometry. Gd(III) and Au content were determined via ICP-MS of acid digested samples. The cellular uptake of DNA-Gd(III)@AuNPs was both time- and concentration-dependent (Figure 1 and 2). At all concentrations the Gd(III) uptake was > 50-fold higher for DNA-Gd(III)@AuNPs compared to DOTA-Gd(III). On average, cells take up 106–107 Gd(III) atoms per cell using only μM Gd(III) incubation concentrations.[37] Previous reports have suggested that at least 107–109 Gd(III) atoms per cell are necessary to produce detectable contrast enhancement.[38] These reports, however, used mM incubation concentrations of Gd(III) to reach that detection threshold due to the limited cellular uptake (lower transduction efficiency) and low relaxivity of the clinical agents used. Furthermore, compared with previous cell permeable contrast agents using either the small transduction molecule stilbene and oligomeric poly arginine conjugated DOTA-Gd(III), the DNA-Gd(III)@AuNP system exhibits the highest cellular uptake.[39]
Figure 1.
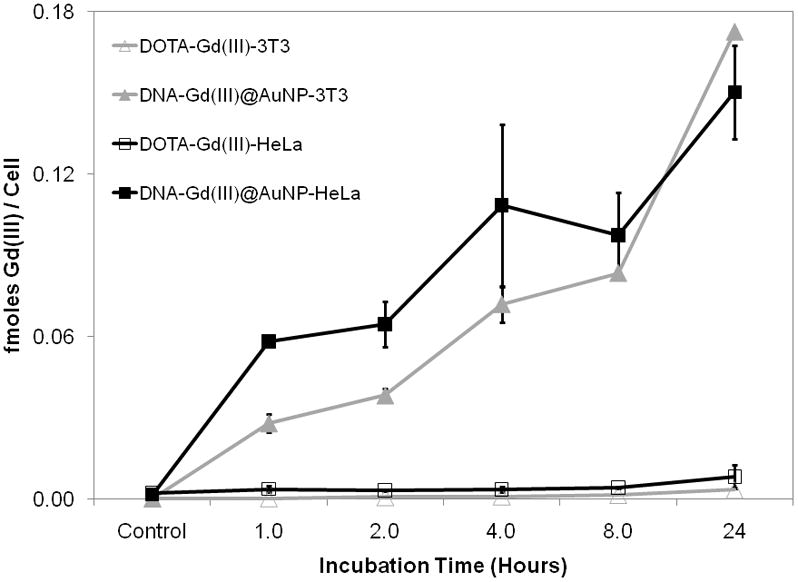
Time dependent cellular uptake of DNA-Gd(III)@AuNPs compared to DOTA-Gd(III) in NIH/3T3 and HeLa cells. Cells were incubated with 6.5 μM Gd(III) for both contrast agents. Error bars represent ± 1 standard deviation of the mean for duplicate experiments.
Figure 2.
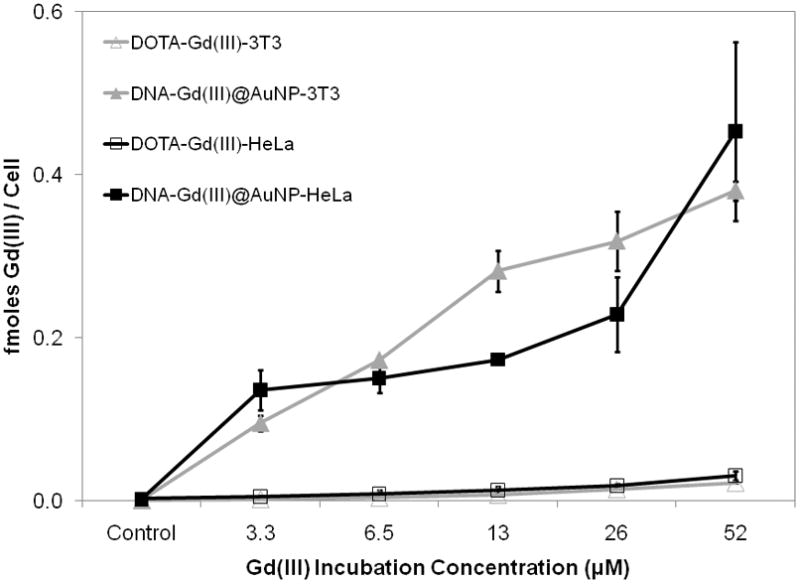
Concentration dependent cellular uptake of DNA-Gd(III)@AuNPs compared to DOTA-Gd(III) in NIH/3T3 and HeLa cells. Cells were incubated for 24 hours for both contrast agents. Error bars represent ± 1 standard deviation of the mean for duplicate experiments.
To demonstrate that μM Gd(III) incubation concentrations of DNA-Gd(III)@AuNP conjugates were sufficient to produce significant T1-weighted contrast enhancement of small cell populations, cells were labelled and imaged at 14.1 T. Specifically, NIH/3T3 cells were incubated with 5.0 μM or 20 μM [Gd(III) concentration] of DOTA-Gd(III) or DNA-Gd(III)@AuNP for 24 hours. T1-weighted MR images of cell pellets were acquired in 1.0 mm diameter glass capillaries, each containing approximately 106 cells (Figure 3). T1 analysis revealed a 43% and 29% T1 reduction with 20 μM and 5.0 μM DNA-Gd(III)@AuNP labelled cell pellets, respectively. Cell pellets incubated with DOTA-Gd(III) at either concentration showed no significant difference from control cell pellets. To our knowledge, these results represent the lowest reported incubation concentration of a Gd(III) complex or conjugate to produce greater than 40% reduction of T1 in cell pellets (see supporting information).[40]
Figure 3.
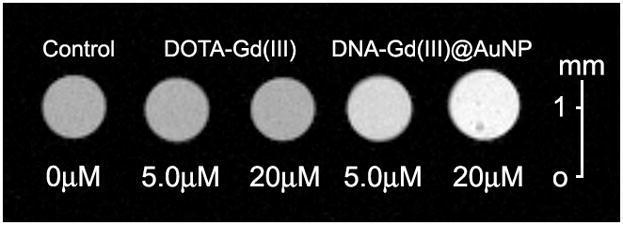
T1-weighted MR image of NIH/3T3 cells incubated with 20 μM and 5.0 μM [Gd(III) concentrations] DNA-Gd(III)@AuNP and DOTA-Gd(III) for 24 hours at 14.1 T (600 MHz) and 25 °C. (TE = 10.2 ms, TR = 750 ms, FOV = 10 × 10 mm2, slice thickness = 1.0 mm).
For comparison, MRI has been applied to tracking Gd(III) labelled β-islets for transplantation and stem cell migration with DO3AHP-Gd(III) with incubation concentrations ranging from 20–50 mM.[41] We are reporting a 1000-fold decrease in Gd(III) incubation concentration to obtain essentially the same contrast enhancement. We have found that efficient delivery and accumulation of Gd(III) complexes is critical for improving the detection limit for high resolution cellular imaging at high magnetic field.
The DNA-Gd(III)@AuNP conjugates are resistant to nuclease degradation which is important for long term cell tracking.[14] We have determined (via ICP-MS) that the ratio of Au to Gd(III), after cell internalization, remains constant for at least 24 hours. This implies that the DNA-Gd(III)@AuNP assembly did not undergo enzyme digestion over this time period which is consistent with previously published results using similar DNA-AuNP conjugates.[42]
To confirm the intracellular accumulation and uptake efficiency of the DNA-Gd(III)@AuNPs, bimodal AuNP conjugates were synthesized by conjugating Cy3 to the 5′ end of the DNA-Gd(III) strands [the ratio of optical to MR signal can be adjusted by altering the stoichiometry of the Cy3-labelled DNA-Gd(III) strands with non-labelled strands]. Specifically, NIH/3T3 and HeLa cells were labelled with 0.1–0.2 nM Cy3-DNA-Gd(III)@AuNPs for 24 hours, rinsed three times with DPBS, and imaged using a confocal laser scanning microscope (CLSM).
The fluorescence micrographs show that the Cy3-DNA-Gd(III)@AuNPs localize in small vesicles in the perinucleur region, which is consistent with previous reports that show AuNP conjugates are taken up through an endocytic mechanism (Figure 4).[43] A second batch of cells was incubated under the same conditions and allowed to leach for 24 hours (media with contrast agent is replaced with fresh media after rinsing). During this time the cell number doubled, but the fluorescence signal persisted in essentially every cell.
Figure 4.
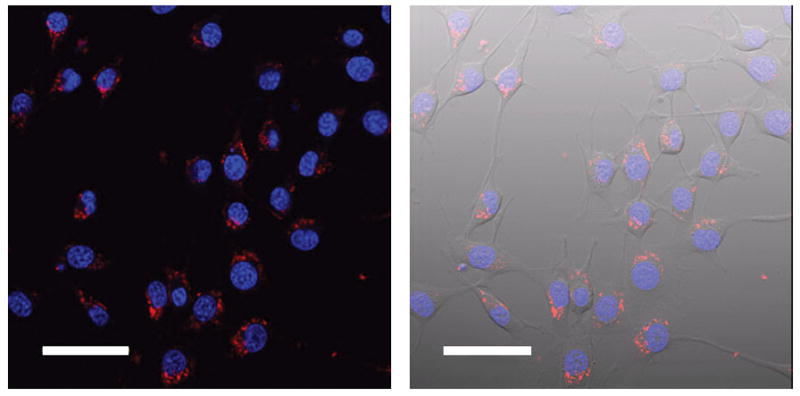
Confocal fluorescence micrographs of NIH/3T3 cells incubated with 0.2 nM particle concentration of Cy3-DNA-Gd(III)@AuNPs for 4.0 hours and a 24 hour leach in fresh media and 1 μM DAPI for 10 minutes. Left: merge of the blue (DAPI) and red [Cy3-DNA-Gd(III)@AuNPs] channels; Right: overlay, with transmitted light image. Scale bar = 50 μm
Cell labelling efficiency was evaluated using analytical flow cytometry and showed that at 0.3 nM Cy3-DNA-Gd(III)@AuNP incubation concentration, 80% of the cells were labelled after 4.0 hours. In both NIH/3T3 and Hela cells, labelling reached 100% after a 24 hour incubation (see supporting information). Importantly, we did not observe any evidence of cell toxicity or cell number variation under any of the conditions tested using DNA-Gd(III)@AuNPs or DOTA-Gd(III) (see supporting information).
In conclusion, we have demonstrated a new multimodal, cell permeable MR contrast agent based upon polyvalent DNA-AuNPs. These particles exhibit excellent biocompatibility and stability, high Gd(III) loading, a greater than 50-fold increase in cell uptake compared to the clinically available contrast agent [DOTA-Gd(III)], and relatively high relaxivity. Work is underway to investigate the utility of these agents for in vivo applications regarding cell tracking in animal models. A primary goal of these studies will be to determine the minimum number of cells that can be detected by this approach.
When modified with a fluorophore, the DNA-Gd(III)@AuNPs can be used as multimodal imaging agents where fluorescence microscopy showed that the particles localize in the peri-nuclear region inside cells. Since AuNPs serve as CT contrast agents, these DNA-Gd(III)@AuNP conjugates have promise as multimodal imaging probes for MR, fluorescence, and CT. The library of available probes for cancer and biological cellular imaging is growing and the strategy presented in this work represents a promising new addition.[20,44–47]
Supplementary Material
Acknowledgments
We acknowledge the financial support of the NCI/CCNE grant # CA119341 and NIH grant # R01EB005866 for financial support of this work. We thank E. Alex Waters for help with the 3T solution phantom images.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Ying Song, Department of Chemistry and The International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113 (USA), Fax: (+) 847-491-3832; (+1) 847-467-5123.
Xiaoyang Xu, Department of Chemistry and The International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113 (USA), Fax: (+) 847-491-3832; (+1) 847-467-5123.
Keith W. MacRenaris, Department of Chemistry and The International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113 (USA), Fax: (+) 847-491-3832; (+1) 847-467-5123
Xue-Qing Zhang, Department of Biomedical Engineering, Northwestern University, Evanston, IL 60208.
Prof. Chad A. Mirkin, Email: chadnano@northwestern.edu, Department of Chemistry and The International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113 (USA), Fax: (+) 847-491-3832; (+1) 847-467-5123
Prof. Thomas J. Meade, Email: tmeade@northwestern.edu, Department of Chemistry and The International Institute for Nanotechnology, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208-3113 (USA), Fax: (+) 847-491-3832; (+1) 847-467-5123
References
- 1.Merbach AE, Toth E, editors. The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. 1. Wiley; New York: 2001. [Google Scholar]
- 2.Aime S, Cabella C, Colombatto S, Geninatti Crich S, Gianolio E, Maggioni F. J Magn Reson Imaging. 2002;16:394. doi: 10.1002/jmri.10180. [DOI] [PubMed] [Google Scholar]
- 3.Hu X, Norris DG. Annu Rev Biomed Eng. 2004;6:157. doi: 10.1146/annurev.bioeng.6.040803.140017. [DOI] [PubMed] [Google Scholar]
- 4.Winter Patrick M, Caruthers Shelton D, Wickline Samuel A, Lanza Gregory M. Curr Cardiol Rep. 2006;8:65. doi: 10.1007/s11886-006-0013-2. [DOI] [PubMed] [Google Scholar]
- 5.Caravan P. Chem Soc Rev. 2006;35:512. doi: 10.1039/b510982p. [DOI] [PubMed] [Google Scholar]
- 6.Major JL, Meade TJ. Acc Chem Res. 2009;42:893. doi: 10.1021/ar800245h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aime S, Delli Castelli D, Geninatti Crich S, Gianolio E, Terreno E. Acc Chem Res. 2009;42:822. doi: 10.1021/ar800192p. [DOI] [PubMed] [Google Scholar]
- 8.Duimstra JA, Femia FJ, Meade TJ. J Am Chem Soc. 2005;127:12847. doi: 10.1021/ja042162r. [DOI] [PubMed] [Google Scholar]
- 9.Major JL, Parigi G, Luchinat C, Meade TJ. Proc Natl Acad Sci U S A. 2007;104:13881. doi: 10.1073/pnas.0706247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W-h, Fraser SE, Meade TJ. J Am Chem Soc. 1999;121:1413. [Google Scholar]
- 11.Caravan P, Cloutier Normand J, Greenfield Matthew T, McDermid Sarah A, Dunham Stephen U, Bulte Jeff WM, Amedio John C, Jr, Looby Richard J, Supkowski Ronald M, Horrocks William De W, Jr, McMurry Thomas J, Lauffer Randall B. J Am Chem Soc. 2002;124:3152. doi: 10.1021/ja017168k. [DOI] [PubMed] [Google Scholar]
- 12.Kalman FK, Woods M, Caravan P, Jurek P, Spiller M, Tircso G, Kiraly R, Bruecher E, Sherry AD. Inorg Chem. 2007;46:5260. doi: 10.1021/ic0702926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modo M, Mellodew K, Cash D, Fraser SE, Meade TJ, Price J, Williams SCR. Neuroimage. 2004;21:311. doi: 10.1016/j.neuroimage.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Modo M, Bulte JW, editors. Molecular and Cellular MR Imaging. CRC Press; FL: 2007. [DOI] [PubMed] [Google Scholar]
- 15.Biancone L, Simonetta GC, Cantaluppi V, Giuseppe MR, Russo S, Scalabrino E, Esposito G, Figliolini F, Beltramo S, Paolo CP, Giuseppe PS, Aime S, Camussi G. NMR in biomedicine. 2007;20:40. doi: 10.1002/nbm.1088. [DOI] [PubMed] [Google Scholar]
- 16.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science (Washington, DC, U S) 2006;312:1027. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 17.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:308. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 19.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 21.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 22.Taton TA, Mirkin CA, Letsinger RL. Science (Washington, D C) 2000;289:1757. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 23.Cao YC, Jin R, Nam JM, Thaxton CS, Mirkin CA. J Am Chem Soc. 2003;125:14676. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 24.Han MS, Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2006;128:4954. doi: 10.1021/ja0606475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JS, Han MS, Mirkin CA. Angew Chem, Int Ed. 2007;46:4093. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Han MS, Mirkin CA. Angew Chem, Int Ed. 2007;46:3468. doi: 10.1002/anie.200605249. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Georganopoulous DG, Hill HD, Mirkin CA. Anal Chem. 2007;79:6650. doi: 10.1021/ac070867g. [DOI] [PubMed] [Google Scholar]
- 28.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J Am Chem Soc. 2009;131:2072. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowman JMC, Ballard ET, Ackerson CJ, D’Antonio J, Feldheim Daniel L, Margolis D, Melander C. J Am Chem Soc. 2008;130:6896. doi: 10.1021/ja710321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Lu Y. Angew Chem, Int Ed. 2007;46:7587. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]
- 31.Agasti SS, Chompoosor A, You CC, Ghosh P, Kim CK, Rotello VM. J Am Chem Soc. 2009;131:5728. doi: 10.1021/ja900591t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frullano L, Meade TJ, Biol J. Inorg Chem. 2007;12:939. doi: 10.1007/s00775-007-0265-3. [DOI] [PubMed] [Google Scholar]
- 33.Song Y, Kohlmeir EK, Meade TJ. J Am Chem Soc. 2008;130:6662. doi: 10.1021/ja0777990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959. [Google Scholar]
- 35.(a) Park JA, Reddy PAN, Kim HK, Kim IS, Kim GC, Chang Y, Kim TJ. Bioorg Med Chem Lett. 2008;18:6135. doi: 10.1016/j.bmcl.2008.10.017. [DOI] [PubMed] [Google Scholar]; (b) Debouttiere PJ, Roux S, Vocanson F, Billotey C, Beuf O, Favre-Reguillon A, Lin Y, Pellet-Rostaing S, Lamartine R, Perriat P, Tillement O. Adv Funct Mater. 2006;16:2330. [Google Scholar]; (c) Alric C, Taleb J, Le Duc G, Mandon C, Billotey C, Le Meur-Herland A, Brochard T, Vocanson F, Janier M, Perriat P, Roux S, Tillement O. J Am Chem Soc. 2008;130:5908. doi: 10.1021/ja078176p. [DOI] [PubMed] [Google Scholar]
- 36.Moriggi L, Cannizzo C, Dumas E, Mayer CR, Ulianov A, Helm L. J Am Chem Soc. 2009;131:10828. doi: 10.1021/ja904094t. [DOI] [PubMed] [Google Scholar]
- 37.Note that on average the cells internalize approximately 105 Gd(III)-conjugates/cell, which is 2 orders of magnitude higher than citrate-stabilized AuNPs of the same size.
- 38.The nanoparticle concentration is over two orders of magnitude lower since each particle contains approximately 50 strands of DNA-Gd conjugates.
- 39.(a) Allen MJ, MacRenaris KW, Venkatasubramanian PN, Meade TJ. Chem Biol. 2004;11:301. doi: 10.1016/j.chembiol.2004.03.003. [DOI] [PubMed] [Google Scholar]; (b) Endres PJ, MacRenaris KW, Vogt S, Allen MJ, Meade TJ. Mol Imag. 2006;5:485. [PubMed] [Google Scholar]
- 40.Biancone L, Crich Simonetta G, Cantaluppi V, Romanazzi Giuseppe M, Russo S, Scalabrino E, Esposito G, Figliolini F, Beltramo S, Perin Paolo C, Segoloni Giuseppe P, Aime S, Camussi G. NMR in biomedicine. 2007;20:40. doi: 10.1002/nbm.1088. [DOI] [PubMed] [Google Scholar]
- 41.Crich SG, Biancone L, Cantaluppi V, Duo D, Esposito G, Russo S, Camusi G, Aime S. Mag Reson Med. 2004;51:938. doi: 10.1002/mrm.20072. [DOI] [PubMed] [Google Scholar]
- 42.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:308. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chithrani BD, Chan WCW. Nano Lett. 2007;7:1542. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 44.Smith AM, Duan H, Mohs AM, Nie S. Adv Drug Delivery Rev. 2008;60:1226. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alivisatos P. Nat Biotechnol. 2004;22:47. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 46.Xia YN. Nat Mater. 2008;7:758. doi: 10.1038/nmat2277. [DOI] [PubMed] [Google Scholar]
- 47.Kim BYS, Jiang W, Oreopoulos J, Yip CM, Rutka JT, Chan WCW. Nano Lett. 2008;8:3887. doi: 10.1021/nl802311t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


