SUMMARY
In skeletal muscle development, the myogenic regulatory factors myf5 and myoD play redundant roles in the specification and maintenance of myoblasts, whereas myf6 has a downstream role in differentiating myocytes and myofibers. It is not clear whether the redundancy between myf5 and myoD is within the same cell lineage or between distinct lineages. Using lineage tracing and conditional cell ablation in mice, we demonstrate the existence of two distinct lineages in myogenesis: a myf5 lineage and a myf5-independent lineage. Ablating the myf5 lineage is compatible with myogenesis sustained by myf5-independent, myoD-expressing myoblasts, whereas ablation of the myf6 lineage leads to an absence of all differentiated myofibers, although early myogenesis appears to be unaffected. We also demonstrate here the existence of a significant myf5 lineage within ribs that has an important role in rib development, suggested by severe rib defects upon ablating the myf5 lineage.
INTRODUCTION
Transient condensations of paraxial mesoderm, known as somites, are the major source of skeletal muscle in vertebrates. Somites differentiate into dorsolateral dermomyotome (source of skeletal muscle and dermis) and ventromedial sclerotome (source of ribs and vertebrae). Myoblasts are proliferating progenitors of the myogenic lineage that differentiate into myocytes, and several myocytes fuse to form mature multinucleated myofibers (Buckingham et al., 2003; Parker et al., 2003). Myogenesis involves complex transcriptional networks in which the basic helix-loop-helix domain containing myogenic regulatory factors (MRFs) myf5, myoD, myf6 (mrf4), and myoG is downstream to the paired domain transcription factors pax3 and pax7 (Parker et al., 2003; Pownall et al., 2002).
Relatively normal myogenesis in myf5−/− and myoD−/− mice, absence of myogenesis due to loss of myoblasts in myf5−/− myoD−/− double null mice, and lack of differentiated myofibers in myoG−/− mice collectively suggest a genetic hierarchy in which myf5 and myoD have an upstream, redundant role in specification and maintenance of myoblasts, whereas myoG has a downstream role in differentiating myocytes and myofibers (Braun and Arnold, 1995; Braun et al., 1992; Hasty et al., 1993; Nabeshima et al., 1993; Rudnicki et al., 1992, 1993). However, the basis of functional redundancy between myf5 and myoD has been a matter of debate. Whereas the serial lineage model assumes expression of both genes within the same lineage, the parallel lineage model purports the existence of distinct lineages. Similar to myoG, initial knock out studies implicated myf6 in terminal differentiation (Braun and Arnold, 1995; Patapoutian et al., 1995; Zhang et al., 1995). A subsequent study, however, showed that myf5−/− myoD−/− double mice null lacking myoblasts were also deficient in myf6, and restoring myf6 expression in these mice partially rescued embryonic myogenesis, thereby calling to question the position of myf6 in the genetic hierarchy (Kassar-Duchossoy et al., 2004).
The lateral part of the dermomyotome is thought to play a role in the development of distal rib (Hirao and Aoyama, 2004; Kato and Aoyama, 1998). Severe rib anomalies in myf5−/− as well as myf6−/− mice suggested a role for myf5 and myf6 in rib morphogenesis (Braun et al., 1992; Yoon et al., 1997), but their relative involvement was unclear. Rib anomalies associated with myf6−/− were proposed to be the result of cis effects of the myf6−/− null on the linked myf5 locus (Kaul et al., 2000; Yoon et al., 1997). However, a conditionally generated myf5−/− mutant, developed subsequently, showed no rib defects, and the authors concluded that the previously documented rib anomalies were due to long-range cis effects of the neo cassette on a putative distal gene (Kaul et al., 2000). Although the above-described observations appear to rule out a direct role of the myf5 gene itself on rib formation, the significance of the myf5 cell lineage in rib development is not known.
In this study, we use conditional cell ablation and lineage tracing in mice to demonstrate that myf5 is expressed in only a subset of developing myoblasts. Early ablation of myf5-expressing cells during embryogenesis is compatible with myogenesis, sustained by a myf5-independent myoblast lineage. Surprisingly, such a redundancy does not exist for the myf5 lineage in developing ribs, and loss of this lineage leads to a severe mutant rib phenotype. We also demonstrate an expansion in the myoD-expressing myoblast population upon ablating myf5-expressing cells, suggesting remarkable adaptability of the myogenic program to developmental insults. However, ablating the genetically downstream myf6-expressing cells has the opposite effect of preserving normal rib morphogenesis, while eliminating differentiated myofibers, ascertaining that myf6 plays a predominant role in differentiated cells of skeletal muscle.
RESULTS
Alleles of myf5 and myf6 Cre Drivers: Cis Effect of Selection Cassette
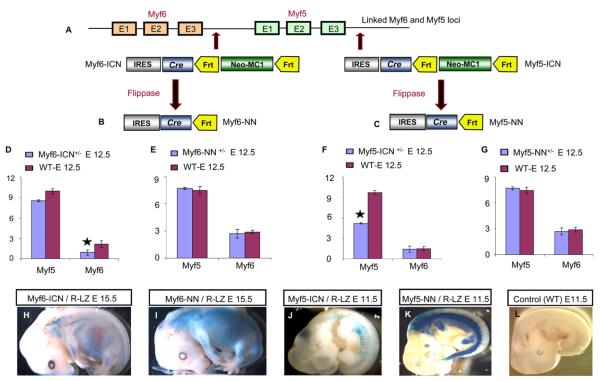
myf5 and myf6 are linked genes on mouse chromosome 10. myf5-cre and myf6-cre mice (Figure 1A) were previously generated in our laboratory and harbor an IRES-CRE-FRT-NEO-MC1-FRT sequence in the 3′UTR of the respective genes (Haldar et al., 2007; Keller et al., 2004). Previous reports suggested the cis effects of the selection cassette on the linked myf5 and myf6 loci as a cause of rib anomalies (Kaul et al., 2000). Although myf5-ICN and myf6-ICN mice (“ICN” denotes IRES-CRE-NEO-MC1) lack any rib phenotype in the homozygous or heterozygous state, we hypothesized that the exogenous MC1 promoter could have a local cis effect. myf6-ICN and myf5-ICN mice were bred to flp-e mice (Rodriguez et al., 2000), thereby removing the MC1-Neo selection cassette (Figures 1B and 1C) and generating two alleles for each of these Cre drivers: myf5-ICN, myf5-NN, myf6-ICN, and myf6-NN (“NN” denotes “no neo”).
Figure 1. Alleles of myf5 and myf6 Cre Drivers.
(A) myf5 and myf6 genes are linked on chromosome 10. The 3′UTR of myf6 (in myf6-ICN)or myf5 (in myf5-ICN) harbors an encephalomyocarditis Internal Ribosomal Entry Site (IRES) that is linked to the Cre coding sequence and is followed by the Neomycin (neo)-resistance selection cassette with MC1 promoter. The selection cassette with MC1 promoter is flanked by two FRT sequences.
(B and C) Breeding with FLP-e mice leads to recombination between FRT sites, generating the “NN” alleles.
(D–G) Semiquantitative RT-PCR on total RNA from E12.5 embryos shows (D) reduced expression of myf6 in myf6-ICN+/− mice (star) but (E) normal expression in myf6-NN+/− mice compared to wild-type. Similarly, myf5 expression is (F) reduced in myf5-ICN+/− mice (star) but (G) normal in myf5-NN+/− mice compared to wild-type.
(H and I) β-gal staining revealed a reduced myf6 lineage in E15.5 (H) myf6-ICN/R-LZ embryos compared to (I) myf6-NN/R-LZ embryos.
(J and K) Similarly, the myf5 lineage in E11.5 (J) myf5-ICN/R-LZ embryos is reduced compared to (K) myf5-NN/R-LZ embryos.
(L) A control wild-type E11.5 embryo shows an absence of any β-gal staining.
Semiquantitative real-time RT-PCR and reporter-based lineage analysis revealed that the presence of the selection cassette in the 3′UTR of myf6 or myf5 downregulates expression of the respective genes. This is demonstrated by significantly reduced myf6 expression and by the myf6 lineage in myf6-ICN heterozygous mice (Figures 1D and 1H) compared to myf6-NN heterozygous mice (Figures 1E and 1I). Similarly, myf5 expression and its lineage are significantly reduced in myf5-ICN heterozygous mice (Figures 1F and 1J) compared to myf5-NN heterozygous mice (Figures 1G and 1K). The expression of the linked gene (myf5 expression in myf6-NN and vice versa) was not affected in the NN alleles, and expression of myf5 and myf6 in the heterozygous NN alleles was comparable to wild-type (Figures 1E and 1G). This underscores the importance of selection cassette removal in developmental studies and is the justification of our use of the NN allele in subsequent analysis.
The Myogenic Lineage of myf5
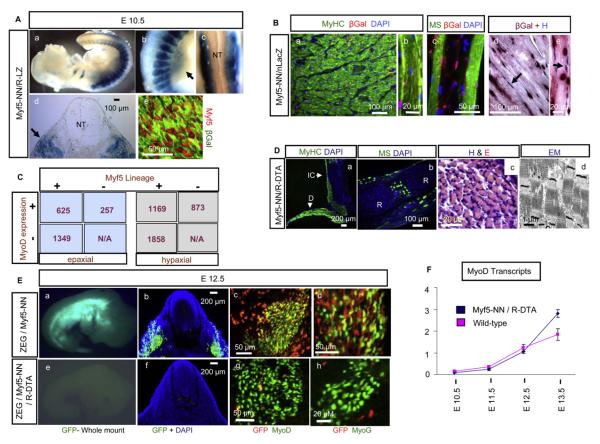
myf5 is involved in myoblast specification and maintenance (Parker et al., 2003; Pownall et al., 2002). myf5-NN mice were bred to ROSA-LacZ (R-LZ) reporter mice (Soriano, 1999). The resulting myf5-NN/R-LZ embryos revealed the expected distribution of the myf5 lineage in whole mount (Figures 2Aa–2Ac; Figures S1Aa and S1Ab; see the Supplemental Data available with this article online) and in sections (Figure 2Ad; Figures S1Ac and S1Ad). Cre-induced expression of β-galactosidase (β-gal) recapitulates the myf5 lineage, as suggested by consistent colocalization of the Myf5 protein with β-gal on sections (Figure 2Ae).
Figure 2. The myf5 Lineage in Myogenesis.
(A) (Aa) The myf5 lineage (blue) in myf5-NN/R-LZ embryos comprises developing embryonic musculature at E10.5, which is predominantly in the (Ab and Ad) myotome but absent in the (Ac and Ad) neural tube. Migratory myogenic populations in the (Ab) limbs (arrow) and (Ad) nonmyogenic dermal precursors (arrows) are shown. (Ae) myf5 expression (red, anti-Myf5) occurs exclusively within the β-gal lineage marker (green, anti-β-gal) in myf5-NN/R-LZ embryos.
(B) The myf5 lineage (red, anti-β-gal) is present within (Ba and Bb) fast-type (green, anti-MyHC) myofibers and (Bc) slow-type (green, anti-MS) myofibers of adult myf5-NN/nLacZ intercostal muscle. (Bd and Be) Enzymatic detection of β-gal activity (red nuclei) reveals a similar distribution of the myf5 lineage, with hematoxylin as counterstain (blue nuclei). Arrows show myonuclei not derived from Myf5.
(C) The distribution of the myf5 lineage (with anti-β-gal) and MyoD expression (using anti-MyoD) was analyzed in epaxial (left, blue box) and hypaxial (right, gray box) regions by cell counting in E12.5 ZEG/myf5-NN embryos. The numbers in boxes are the numbers of nuclei counted. +, presence; −, absence.
(D) (Da) Fast-type (green, anti-MyHC) and (Db) slow-type (green, anti-MS) fibers seen in E18.5 myf5-NN/R-DTA intercostal musculature (D, diaphragm; IC, intercostals; R, ribs) that show normal morphology based on (Dc) H&E staining and (Dd) electron micrograph.
(E) The myf5 lineage (green) demonstrated in (Ea) whole mount and (Eb) sections (green, anti-GFP) in E12.5 ZEG/myf5-NN embryos. (Ec) MyoD (gree,n anti-MyoD) and (Ed) myogenin (green, anti-MyoG) are expressed within and in proximity to the myf5 lineage (red, anti-GFP). Absence of the myf5 lineage in ZEG/myf5-NN/R-DTA embryos is demonstrated in (Ee) whole mount (absence of green signal) and (Ef) sections (absence of green anti-GFP staining). (Eg) MyoD and (Eh) MyoG expression occurs independent of the myf5 lineage in ZEG/myf5-NN/R-DTA.
(F) Semiquantitative RT-PCR for myoD at E10.5, E11.5, E12.5, and E13.5 reveals increased numbers of MyoD transcripts in myf5-NN/R-DTA embryos compared to wild-type by E13.5.
myf5-NN mice were then bred to reporter mice conditionally expressing the nuclear-localized β-gal reporter gene from the CAG promoter (nLacZ mouse line, P.T. and M.R.C., unpublished data) to generate myf5-NN/nLacZ mice. Myoblasts differentiate into myocytes that fuse to form multinucleated myofibers. Therefore, if all myoblasts express myf5, then all myonuclei within myf5-NN/nLacZ should express β-gal. Surprisingly, we detected β-gal in only a fraction of adult myonuclei based on both immunohistochemistry, a more specific method (Figures 2Ba–2Bc), and enzymatic detection of β-gal activity, a more sensitive method (Figures 2Bd and 2Be). This was true for both fast-type (Figures 2Ba and 2Bb) and slow-type myofibers (Figure 2Bc). Intercostals and leg flexors of myf5-NN/nLacZ, used as representative musculature, were sectioned and labeled with anti-MyHC (skeletal muscle specific), anti-β-gal (myf5 lineage marker), and DAPI (non-specific nuclear marker). The number of β-gal-positive nuclei within MyHC-positive myofibers were counted and divided by the total number of nuclei (β-gal positive and/or DAPI positive) within MyHC-expressing myofibers. Based on this strategy, ~35% of myonuclei appeared to be a lineage of myf5 (2012 nuclei counted). Myonuclei are located peripherally, and there is a small probability of erroneously counting adjacent nonmyogenic connective tissue nuclei within the total pool of myonuclei used in the calculation. Another possible source of error could be the sensitivity of the technique. Considering these probable sources of error, we believe that the final contribution of myf5 to adult musculature could be as high as 50%. To rule out mosaic activity of the CAG promoter in myonuclei of nLacZ reporter mice, we bred the nLacZ mice to hprt-cre mice (Tang et al., 2002), which express Cre recombinase at the single-cell zygotic stage. All myonuclei of hprt-cre/nLacZ mice showed β-gal, ruling out promoter mosaicism as a potential cause of the observed partial myogenic contribution of the myf5 lineage (Figure S1B).
The myf5 Lineage in Embryonic Myogenesis
myf5-NN mice were bred to transgenic ZEG reporter mice, which express green fluorescent protein (GFP) in response to Cre (Novak et al., 2000). Based on GFP and MyoD expression in E12.5 ZEG/myf5-NN embryos, we identified three populations of cells: cells expressing GFP only (myf5 lineage not expressing myoD), cells expressing myoD only (myoD-expressing cells not derived from myf5), and cells expressing GFP and MyoD (Figure 2C; Figures S2Aa and S2Ad). This suggests the existence of myf5-independent (MyoD-positive, GFP-negative) and myf5-expressing (GFP-positive) myoblasts. Approximately 71% of the total MyoD expression in the epaxial region (Figure 2C, left, blue boxes) and 57% in the hypaxial region (Figure 2C, right, gray boxes) occurred within the myf5 lineage, suggesting either a predominant role or an earlier appearance of the myf5 lineage in the epaxial compared to the hypaxial region. Approximately 32% of the myf5 lineage showed MyoD expression in the epaxial region, and 39% showed expression in the hypaxial region (Figure 2C), suggesting the presence of either nonmyogenic myf5 lineages or MyoD-negative myoblasts. Most of the MyoD-negative myf5 lineages are located superficially in the epaxial domain (Figure S2Aa, arrow), are negative for MyHC (Figure S2Ab, arrow), and probably represent nonmyogenic dermal precursors (Hadchouel et al., 2000). Differentiating myocytes expressing MyHC (Figures S2Ab and S2Ae) and MyoG (Figures S2Ac and S2Af) were found within and outside the myf5 lineage in epaxial and hypaxial regions, suggesting that both the myf5 and myf5-independent lineages contribute to embryonic musculature. Moreover, Myf5 and MyoD proteins do not always colocalize at various embryonic stages (Figure S2B), further supporting the notion of distinct myoblast lineages.
The myf5 Lineage Is Dispensable in Myogenesis
Our results suggested that all myonuclei are not derived from myf5-expressing cells. We then asked if the myf5 lineage is required for myogenesis. myf5-NN mice were bred to ROSA-DTA (R-DTA) mice conditionally expressing the highly potent diphtheria toxin (DTA) from the ROSA locus (Wu et al., 2006). Surprisingly, despite ablating the myf5 lineage, myogenesis was preserved, as demonstrated by the presence of myofibers of both the fast type (Figure 2Da) and the slow type (Figure 2Db). Muscles were present in all anatomical regions surveyed (Figures S3Aa–S3Ad), and myofibers had normal morphology on sections (Figure 2Dc). However, these pups do not survive after birth due to a fatal rib anomaly (discussed later). The functional integrity of the musculature was demonstrated by spontaneous and stimulus-induced mobility in myf5-NN/R-DTA full-term embryos delivered by C-section, whereas ultrastructural integrity was demonstrated by electron microscopy (Figure 2Dd). It is noteworthy that whereas the myf5-NN/R-DTA mice die perinatally with severe rib cage defects, 90% of the myf5-ICN/R-DTA mice are viable, with a normal life span, and their musculature appears to be similar to that of myf5-NN/R-DTA mice based on histology (Figures S3Ba and S3Bb). Subtle downregulation of myf5 in myf5-NN mice (similar to the ICN allele), which is undetectable in the heterozygous state (analyzed in Figures 1G and 1K), may allow a small fraction of myoblasts to survive and proliferate to form musculature. This, however, seems unlikely since Myf5 expression in myf5-NN is unperturbed in even the homozygous state (Figure S3C).
Inefficient killing by DTA could account for ongoing myogenesis in myf5-NN/R-DTA mice. To rule this out, we generated ZEG/myf5-NN/R-DTA embryos and observed the absence of the fluorescent myf5 lineage within these embryos (Figures 2Ee and 2Ef) compared to ZEG/myf5-NN (Figures 2Ea and 2Eb) embryos at E12.5. In sections, occasional cells expressing GFP (the myf5 lineage) are observed very rarely within ZEG/myf5-NN/R-DTA embryos (Figures 2Eg and 2Eh), a frequency that is almost negligible when compared to littermate ZEG/myf5-NN embryos (Figures 2Ec and 2Ed); this finding reflects the interval between Myf5 expression and DTA-mediated cell death, which is ~12–24 hr (Wu et al., 2006). Further investigation into DTA-mediated myf5 lineage ablation revealed that, although the myf5 lineage is absent (Figures 2Ee and 2Ef), myf5 transcripts (Figure S4A) and proteins (Figures S4B and S4C) that are detectable at E11.5 are greatly reduced and are dramatically reduced further with increasing embryonic age such that by E13.5 Myf5 proteins are essentially undetectable by western blot (Figure S4B) and are barely detectable based on immunohistochemistry (Figure S4C). Further-more, a significant population of Myf5-expressing cells at E11.5 in myf5-NN/R-DTA mice is already undergoing apoptosis (Figure S4Ce). These results demonstrate that in vivo cell killing via DTA is associated with a time-lag between the onset of “DTA-inducing” gene expression (in this case, myf5) and DTA-induced apoptosis that has to be accounted for when analyzing lineage ablation by using this system.
Ongoing myogenesis within ZEG/myf5-NN//R-DTA embryos based on MyoD and MyoG expression (Figures 2Eg and 2Eh) suggests that embryonic myogenesis is not significantly perturbed in the absence of the myf5 lineage. Real-time RT-PCR analysis of myf5-NN/R-DTA embryos between E10.5 and E13.5 revealed significantly higher levels of myoD transcripts (Figure 2F) compared to wild-type by E13.5, suggesting an expansion of myf5-independent, myoD-expressing myoblasts to compensate for the loss of the myf5 lineage. Unlike myoD, there is no significant difference in myf6 expression within myf5-NN/R-DTA embryos at these stages (Figure S3D), suggesting that loss of the myf5 lineage does not affect the pool of differentiating myocytes, indicating efficient compensation by myf5-independent lineages.
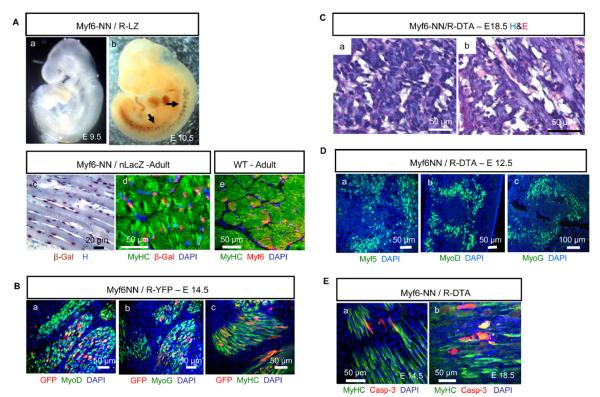
The myf6 Lineage Is Indispensable in Myogenesis
myf6 is believed to be genetically downstream of myf5 and to act in differentiated cells of the skeletal muscle lineage. The myf6 lineage is detectable by E10.5 (Figures 3Aa and 3Ab) and comprises a majority of, but not all, adult myonuclei, based on enzymatic (Figure 3Ac) and antibody-based (Figure 3Ad) detection of β-gal within myf6-NN/nLacZ mice. Significant Myf6 expression also occurs in adult musculature (Figure 3Ae). myf6-NN mice were bred to ROSA-YFP (R-YFP) reporter mice (Srinivas et al., 2001), and the myf6 lineage was found to comprise only a small fraction of cells expressing myoD, myoG, and myHC (Figures 3Ba–3Bc) within E14.5 myf6-NN/R-YFP embryos, suggesting that myf6 makes a minor contribution in early myogenesis. However, we observed a complete lack of differentiated myofibers in the newborn myf6-NN/R-DTA pups that were immobile and died soon after birth. On sections, basophilic clumps of cellular debris were observed in locations of skeletal musculature (Figures 3Ca and 3Cb), suggesting that skeletal muscles were formed prior to DTA-mediated killing. Robust expression of Myf5, MyoD, and MyoG within E12.5 myf6-NN/R-DTA embryos suggested that the absence of the myf6 lineage does not significantly compromise early embryonic myogenesis (Figures 3Da–3Dc).
Figure 3. The myf6 Lineage in Myogenesis.
(A) The myf6 lineage is undetectable at (Aa) E9.5 and is detectable at (Ab) E10.5 (arrows) based on β-gal staining (blue) of myf6-NN/R-LZ embryos. (Ac) The myf6 lineage (red nuclei) in the intercostal musculature of an adult myf6-NN/nLacZ mouse is detected by β-gal staining and hematoxylin counterstain (blue nuclei). (Ad) shows the same based on immunohistochemistry (red, anti-β-gal; green, anti-MyHC). (Ae) shows Myf6 expression (red, anti-Myf6) in adult intercostal muscles (green, anti-MyHC).
(B) Transverse sections through the epaxial region of myf6-NN/R-YFP embryos demonstrate that the myf6 lineage (red, anti-GFP) comprises only a fraction of (Ba) MyoD- (green, anti-MyoD), (Bb) MyoG- (green, anti-MyoG), and (Bc) MyHC-expressing (green, anti-MyHC) cells at E14.5.
(C) (Ca and Cb) E18.5 myf6-NN/R-DTA embryos show basophilic cellular debris in place of musculature (back muscles).
(D) Robust myogenesis within an E12.5 myf6-NN/R-DTA embryo demonstrated by expression of (Da) myf5 (green, anti-Myf5), (Db) myoD (green, anti-MyoD), and (Dc) myoG (green, anti-MyoG) in sections through the myotome.
(E) Apoptosis detected by expression of activated caspase-3 (red) within differentiated myofibers (green) at (Ea) E14.5 and (Eb) E18.5.
The number of apoptotic cells within myf6-NN/R-DTA embryos increased with embryonic age, as more and more differentiated cells of myogenic lineage express Myf6 and consequently DTA (Figures 3Ea and 3Eb), such that by E18.5 all differentiated myofibers were either already dead (extracellular patchy staining with MyHC) or dying (positive for activated caspase-3). These observations support the existing concept that myf6 plays a primary role in differentiated cells of the myogenic lineage and is not crucial in early myogenesis.
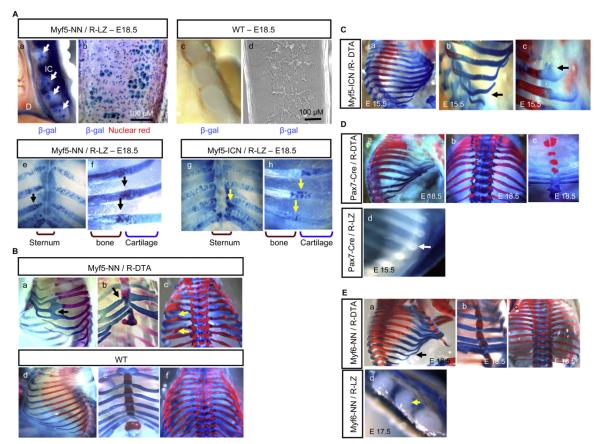
The myf5 Lineage in Rib Morphogenesis
Previous studies with targeted alleles of myf5 replaced with the β-gal reporter gene showed the presence of β-gal-expressing rib chondrocytes in null, but not in heterozygous, mice, leading the authors to suggest that the absence of myf5 leads to misincorporation of the myf5 lineage in ribs (Tajbakhsh et al., 1996). We observed significant myf5 lineage within ribs of both myf5-NN/R-LZ (Figures 4Aa, 4Ab, 4Ae, and 4Af) and myf5-ICN/R-LZ mice (Figures 4Ag and 4Ah), which comprised mostly chondrocytes and some cells in perichondrium (Figure 4Ab). The costochondral (Figures 4Af and 4Ah, arrows) and costosternal (Figures 4Ae and 4Ag, arrows) regions of ribs were particularly rich in the myf5 lineage and appeared smaller in myf5-ICN/R-LZ (Figure 4Ah, arrow) compared to myf5-NN/R-LZ mice (Figure 4Af, arrow) at the costochondral region. This apparent quantitative difference in the rib lineage of myf5 within the two alleles of myf5-cre mice (myf5-NN and myf5-ICN) translates into striking differences in the rib phenotype when these mice are bred to R-DTA mice. Whereas myf5-NN/R-DTA newborn pups have severely deformed ribs (Figures 4Ba–4Bc) and die immediately after birth, apparently due to their inability to breathe, myf5-ICN/R-DTA mice show normal rib cages, with few mild anomalies such as occasional waviness of floating ribs (Figures 4Ca–4Cc). The cartilaginous portion of the ribs shows fusions (Figure 4Ba, arrow) and loss of symmetric bilateral attachment to the sternum (Figure 4Bb, arrow), and the osseous portion shows occasional “knob-like” protrusions (Figure 4Bc, arrows) in the myf5-NN/R-DTA mice. These anomalies are detectable even at early embryonic stages, whereas heterozygous myf5-NN or R-DTA control mice show normal rib cages (Figures S5a–S5d).
Figure 4. Unique Role of the myf5 Lineage in Rib Morphogenesis.
(A) The myf5 lineage is detected by blue β-gal staining at E18.5 in ribs of myf5-NN/R-LZ, demonstrated in (Aa) whole mount (arrows) and (Ab) sections. (Aa) IC, intercostal muscles; D, diaphragm. The counterstain is nuclear red in (Ab). Control wild-type littermates show an absence of β-gal staining in (Ac) whole mount and in (Ad) sections. Rib lineages of Myf5 appear to be prominent at the (Ae and Ag) sternal end (arrows) and the (Af and Ah) costochondral region (arrows) and appear less often in the (Ah) “ICN” allele compared to the (Af) “NN” allele in costochondral regions.
(B) Abnormal rib cage of newborn myf5-NN/R-DTA mice characterized by (Ba) fusions (arrow) and (Bb) staggered asymmetric attachment to the sternum (arrow) at the cartilaginous part and by (Bc) abnormal protrusions in the bony part (arrow). (Bd)–(Bf) show comparative regions in a wild-type littermate.
(C) myf5-ICN/R-DTA mice have (Ca) normal rib cages and occasional mild anomalies such as (Cb) waviness of the floating ribs (arrow) and (Cc) occasional knobs in the costal regions (arrow).
(D) (Da–Dc) Ablating the pax7 lineage does not perturb rib morphogenesis. (Dd) The pax7 lineage is absent within ribs of E15.5 pax7-cre/R-LZ embryos (arrow).
(E) (Ea)–(Ec) Ablating the myf6 lineage does not perturb normal rib morphogenesis, except for (Ea) occasional mild waviness (arrow). (Ed) The myf6 lineage is absent within ribs of E17.5 myf6-NN/R-LZ embryos (arrow).
The rib defects seen upon myf5 lineage ablation could be due to extensive structural damage to developing somites. If this were true, then ablation of other somitic myotomal lineages, such as that of pax3 or pax7, should recapitulate the rib phenotype of myf5-NN/R-DTA. Whereas pax3-cre/R-DTA mice suffer extensive disruption of development not compatible with embryogenesis (data not shown), pax7-cre/R-DTA newborn pups show no rib cage anomalies (Figures 4Da–4Dc), and pax7 lineages were absent from ribs (Figure 4Dd). This indicates that nonspecific somitic damage is probably not the cause of rib anomalies upon myf5 lineage ablation. Intercostal musculature of myf5-NN/R-DTA mice appears to be normal based on histology and electron microscopy. Moreover, ablation of the myf6 lineage, which is absent in ribs (Figure 4Ed), eliminates all intercostal musculature while preserving rib development (Figures 4Ea–4Ec). These findings seem to rule out abnormal intercostal musculature as a cause of rib mispatterning in the absence of the myf5 lineage. Therefore, the myf5 lineage appears to play a direct role in rib development. Furthermore, electron microscopy of the mispatterned ribs of myf5-NN/R-DTA shows no obvious ultrastructural anomalies, suggesting the absence of any primary chondrocyte defect (Figures S5e and S5f).
DISCUSSION
Targeted loss-of-function analyses in mice demonstrated functional redundancy between myf5 and myoD in the specification and maintenance of myoblasts. However, it was not clear whether the redundancy is within the same cell lineage (serial lineage model) or between distinct lineages (parallel lineage model). One study attempted to address this in vitro by showing that ablation of either myf5- or myoD-expressing embryonic stem cells did not compromise their myogenic line of differentiation in culture (Braun and Arnold, 1996), whereas another study suggested distinct roles of myf5 and myoD in epaxial and hypaxial musculature, respectively (Kablar et al., 1997), raising the possibility of multiple myogenic lineages. Here, we demonstrate the existence of two distinct myogenic lineages: a myf5 lineage and a myf5-independent lineage (MyoD expressing), which seem to be the basis of functional redundancy between myf5 and myoD. The presence of such intercellular redundancy in terms of lineage commitment during embryogenesis reflects the evolution of the myogenic pathway and provides for an inbuilt safeguard against potential disruption of the myogenic program.
Myf5 and MyoD expression are not mutually exclusive, as suggested by widespread expression of MyoD within the myf5 lineage, nor are they coexpressed, as suggested by mostly distinct expression of Myf5 and MyoD. This suggests that, under normal circumstances, most of the myogenic lineage of myf5 eventually expresses MyoD, or vice versa. The presence of skeletal musculature in the absence the myf5 lineage, however, suggests that such sequential expression is not required for myogenesis. Compared to the final contribution of the myf5 lineage to adult myonuclei (about 50%), a higher fraction of myoblasts (71% in epaxial musculature and 57% in hypaxial musculature) comprises a lineage of myf5 at E12.5, suggesting that MyoD may not be expressed in all myoblasts. Therefore, myoD-independent myoblasts may exist in a manner similar to myf5-independent myoblasts that we describe here. However, we may have missed a significant proportion of the MyoD expression outside the myf5 lineage based on the reported variability of Myf5 and MyoD expression with the cell cycle (Kitzmann et al., 1998). Another possibility could be that more myf5-independent lineages contribute to myogenesis at later embryonic stages (after E12.5), thereby reducing the fraction of the final myf5 lineage in the adult musculature. In either scenario, it is not certain at this time whether the myf5-independent compartment is a single compartment or is heterogeneous based on MRF expression.
Cells expressing myf5 undergoe DTA-mediated apoptosis within myf5-NN/R-DTA embryos, precluding any cell-autonomous differences in the levels of myoD secondary to the absence of myf5. Therefore, an increased number of myoD transcripts in myf5-NN/R-DTA embryos reflects an increased number of myoD-expressing cells that probably compensate for the loss of the myf5 lineage to restore normal myogenesis. It would be interesting to know how the myogenic program maintains a balance between these distinct lineages and determines expansion or restriction of individual lineages in different scenarios. Another intriguing question is whether such distinct lineages also exist during satellite-cell-mediated postnatal myogenesis.
Skeletal muscle of myf5-NN/R-DTA appear to be indistinguishable from that of myf5-ICN/R-DTA based on histology. Therefore, myf5-NN/R-DTA mice, like myf5-ICN/R-DTA mice, would most likely have had a normal lifespan if not for the severe rib defects. It is remarkable that the lineage of myf5, an MRF, is dispensable in myogenesis but is required for rib development. Because a conditionally generated myf5 null mouse that lacks rib defects has been described, it is the myf5 cell lineage, not the myf5 gene, that is critical for rib development, which suggests that myf5 expression is not exclusive to the myogenic lineage. In this regard, it is noteworthy that lateral aspects of cervical vertebrae also show the presence of the myf5 lineage (Figure S5g). Transient expression of myf5 in presomitic mesoderm has been reported (Cossu et al., 1996), and it is possible that the presomitic cells transiently expressing myf5 are the primary source of the substantial nonmyogenic lineages of myf5, whereas somitic cells initiating myf5 expression are the primary source of myf5’s myogenic lineage. Whether somitic expression of myf5 occurs within the lineages of presomitic cells that had transiently expressed myf5 as well as the identity of the subset of myf5-expressing cells that are involved in rib development are currently under investigation.
In summary, we demonstrate here the existence of two independent lineages in myogenesis and show a role of the myf5 lineage in rib development by using lineage analysis in conjunction with lineage ablation, a strategy that could be effectively applied to analyze other developmental pathways.
EXPERIMENTAL PROCEDURES
Animal Handling and Genotyping
All studies involving animal subjects were approved by the University of Utah Institutional Animal Care and Use Committee and were conducted strictly in accordance with the relevant protocol. myf5-cre and myf6-cre mouse lines have been described elsewhere (Haldar et al., 2007; Keller et al., 2004). The nLacZ reporter line was generated by targeting the conditional, codon-optimized, and nuclear localization signal-containing β-gal gene to the polr2a locus, ~1 kb downstream of the polyA signal. The promoter driving nLacZ expression is the CAG promoter, a hybrid promoter comprised of cytomegalovirus immediate early enhancer and the chicken β-actin promoter (Tang et al., 2002). The specific details of this mouse line are available upon request and will be published elsewhere.
Real-Time RT-PCR
Semiquantitative real-time RT-PCR was performed by using total RNA from wild-type and mutant embryos extracted with TRIzol (invitrogen), purified, and treated with DNase. Real-time RT-PCR was performed on RNA samples (25 ng) by using the QuantiTect SYBR green RT-PCR kit (QIAGEN, Inc., Valencia, CA) and a DNA Engine Opticon 2 system (MJ Research, Waltham, MA). Results were normalized to Gapdh signals for each sample. Standards for Gapdh and all other primer sets showed similar slopes, indicating equivalent amplification kinetics. Please refer to the Supplemental Data for primer sequences.
β-Galactosidase Staining
For whole-mount staining, samples were fixed in 1% PFA, 0.2% glutaraldehyde, 0.002 M MgCl2, 0.025 M EGTA, and 0.02% NP-40 in 1× PBS for 2 hr at 4°C, followed by overnight staining at room temperature in 0.005 M K3Fe(CN)6, 0.005 M K4Fe(CN)6, 0.002 M MgCl2, 0.01% Na-doc, 0.02% NP-40, and X-gal substrate in 1× PBS. Ribs were cleared in 1% KOH/20% glycerol until soft tissues disappeared.
Fixed and cryopreserved samples were also sectioned at 8–12 μm thickness and stained at room temperature overnight in β-gal staining solution (described above). Counterstaining was carried out with hematoxylin by following the established protocol.
Whole-Mount Skeletal Staining
Skeletal staining was carried out with Alizarin red and alcian blue by following the established protocol (Wellik and Capecchi, 2003).
Histology and Immunodetection
Hematoxylin and Eosin staining was carried out on 4–8 μm sections of tissue fixed in 4% PFA and embedded in paraffin by following standard protocols. Fluorescence-based immunohistochemistry was performed on 8–12 μm fixed and frozen sections. Cell counting was carried out with help from ImageJ software by using the ITCN plug in (Byun et al., 2006).
For western blot, embryonic samples from different stages of mouse development were homogenized in 200–500 μl lysis buffer (1% Triton X-100, 0.2% SDS, .001 M EDTA in 0.05 M Tri-HCl [pH 7.4]). Each sample (15 μg protein) was separated on a 8% SDS-PAGE gel, transferred onto a PVDF gel, and probed with primary antibodies, followed by horseradish peroxidase-conjugated secondary antibody. The signal was visualized by using the chemiluminescence ECL-Plus system.
Please refer to the Supplemental Data for information on antibodies used.
Electron Microscopy
Tissue samples were fixed by using 2% formaldehyde and 2% glutaraldehyde in 0.1 M PBS (pH 7.4) containing 1% osmium tetroxide, dehydrated through an ascending series of graded ethanols, and processed for embedment in Epon 812 resin. Ultrathin sections (0.75–0.5 μm) were mounted on copper grids and stained with uranyl acetate and lead citrate before observation under an electron microscope (Philips, TECNAI-T12).
Apoptosis Detection
Apoptosis was detected by using a rabbit polyclonal antibody directed against the activated, cleaved form of caspase-3 (Cell signaling). A TUNEL assay was performed by using a fluorescein In Situ Cell Death Detection Kit (Roche), and the assay was performed according to the manufacturer’s instructions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sen Wu for generating and making available the ROSA-DTA mouse line, and Gabriel Kardon, Eugenio Sangiorgi, Lara Carrol, Amir Pozner, and Anne Boulet for helpful comments and insights.
Footnotes
SUPPLEMENTAL DATA Supplemental Data include information on primer sequences and antibodies used and are available at http://www.developmentalcell.com/cgi/content/full/14/3/437/DC1/.
REFERENCES
- Braun T, Arnold HH. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. EMBO J. 1995;14:1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Arnold HH. Myf-5 and myoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J. 1996;15:310–318. [PMC free article] [PubMed] [Google Scholar]
- Braun T, Rudnicki MA, Arnold HH, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun J, Verardo MR, Sumengen B, Lewis GP, Manjunath BS, Fisher SK. Automated tool for the detection of cell nuclei in digital microscopic images: application to retinal images. Mol. Vis. 2006;12:949–960. [PubMed] [Google Scholar]
- Cossu G, Tajbakhsh S, Buckingham M. How is myogenesis initiated in the embryo? Trends Genet. 1996;12:218–223. doi: 10.1016/0168-9525(96)10025-1. [DOI] [PubMed] [Google Scholar]
- Hadchouel J, Tajbakhsh S, Primig M, Chang TH, Daubas P, Rocancourt D, Buckingham M. Modular long-range regulation of Myf5 reveals unexpected heterogeneity between skeletal muscles in the mouse embryo. Development. 2000;127:4455–4467. doi: 10.1242/dev.127.20.4455. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hirao A, Aoyama H. Somite development without influence of the surface ectoderm in the chick embryo: the compartments of a somite responsible for distal rib development. Dev. Growth Differ. 2004;46:351–362. doi: 10.1111/j.1440-169x.2004.00752.x. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kato N, Aoyama H. Dermomyotomal origin of the ribs as revealed by extirpation and transplantation experiments in chick and quail embryos. Development. 1998;125:3437–3443. doi: 10.1242/dev.125.17.3437. [DOI] [PubMed] [Google Scholar]
- Kaul A, Koster M, Neuhaus H, Braun T. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- Patapoutian A, Yoon JK, Miner JH, Wang S, Stark K, Wold B. Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development. 1995;121:3347–3358. doi: 10.1242/dev.121.10.3347. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Yoon JK, Olson EN, Arnold HH, Wold BJ. Different MRF4 knockout alleles differentially disrupt Myf-5 expression: cis-regulatory interactions at the MRF4/Myf-5 locus. Dev. Biol. 1997;188:349–362. doi: 10.1006/dbio.1997.8670. [DOI] [PubMed] [Google Scholar]
- Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.