Abstract
Nop15p is an essential protein that contains an RNA recognition motif (RRM) and localizes to the nucleoplasm and nucleolus. Cells depleted of Nop15p failed to synthesize the 25S and 5.8S rRNA components of the 60S ribosomal subunit, and exonucleolytic 5′ processing of 5.8S rRNA was strongly inhibited. Pre-rRNAs co-precipitated with tagged Nop15p confirmed its association with early pre-60S particles and Nop15p bound a pre-rRNA transcript in vitro. Nop15p-depleted cells show an unusually abrupt growth arrest prior to substantial depletion of ribosomal subunits. Following cell synchronization in mitosis, Nop15p-depleted cells undergo nuclear division with wild-type kinetics, activate the mitotic exit network and disassemble their mitotic spindle. However, they uniformly arrest at cytokinesis and fail to assemble a contractile actin ring at the bud neck. In dividing wild-type cells, segregation of nucleolar proteins to the daughter nuclei occurs after separation of the nucleoplasm. In these late mitotic cells, Nop15p was partially delocalized from the nucleolus to the nucleoplasm, consistent with a specific function in cell division in addition to its role in ribosome synthesis.
Keywords: cell division/nucleolus/pre-rRNA/ribosome synthesis/Saccharomyces cerevisiae
Introduction
Most steps in ribosome synthesis take place within the nucleolus, a specialized subnuclear structure. During ribosome synthesis, a complex processing pathway converts a large pre-rRNA to the mature 18S, 5.8S and 25S/28S rRNAs. In addition, the mature rRNA sequences within the pre-RNA undergo extensive covalent nucleotide modification and assemble with ∼80 ribosomal proteins. More than 140 non-ribosomal proteins that are required for ribosome synthesis have been identified by genetic and biochemical approaches in Saccharomyces cerevisiae (see Kressler et al., 1999; Venema and Tollervey, 1999; Warner, 2001; Fatica and Tollervey, 2002).
Far fewer human ribosome synthesis factors have been characterized, but the product of the open reading frame (ORF) NNP18/NOPP34/hNIFK was shown to be associated with pre-ribosomal particles (Fujiyama et al., 2002). NIFK was reported to localize to the nucleolus (Takagi et al., 2001) and was identified in a proteomic analysis of purified human nucleoli (Andersen et al., 2002). However, NIFK also localized to regions of condensed mitotic chromosomes, where it may interact with the forkhead-associated (FHA) domain of the cell proliferation marker pKI-67, and was proposed to function in mitosis (Sueishi et al., 2000; Takagi et al., 2001). A database search with this sequence clearly identified YNL110c as the probable yeast homologue (32% identity, 55% similarity over 174 amino acids). YNL110c/NOP15 was reported to be essential for viability in systematic analyses (De Antoni et al., 1997; Capozzo et al., 2000). Spores carrying a ynl110Δ mutation either failed to bud or were arrested after the first cycle, producing an abnormal bud either smaller or larger than wild-type. This led to the suggestion that Ynl110p/Nop15p functions during the cell cycle (Capozzo et al., 2000).
Recent papers have described other factors that function in both ribosome synthesis and activities required for cell cycle progression. Net1p/Cfi1p regulates release and activation of the Cdc14p phosphatase, thus triggering mitotic exit (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999), but also promotes transcription of the pre-rRNA by RNA polymerase I (Shou et al., 2001). Yph1p/Nop7p and Noc3p are associated with the origin of replication complex (ORC) and play key roles in the initiation of DNA replication (Du and Stillman, 2002; Zhang et al., 2002). However, both Nop7p and Noc3p are also components of pre-60S particles and each is required for pre-rRNA processing, 60S subunit synthesis and export to the cytoplasm (Harnpicharnchai et al., 2001; Milkereit et al., 2001; Adams et al., 2002; Oeffinger et al., 2002). Another 60S synthesis factor Sda1p (Baßler et al., 2001; Harnpicharnchai et al., 2001; Ihmels et al., 2002) is required to pass the ‘Start’ checkpoint at the G1–S boundary (Buscemi et al., 2000; Zimmerman and Kellogg, 2001) and one allele was also reported to lead to defects in the actin cytoskeleton. Moreover, a high-throughput screen implicated several other ribosome synthesis factors and ribosomal proteins in determining the timing of Start, and therefore the size of the cell at division (Jorgensen et al., 2002).
While this work was in progress, Nop15p was identified as a component of a pre-60S particle, and a strain expressing a haemaglutinin (HA)-tagged form of Nop15p showed some reduction in 60S subunit accumulation (Harnpicharnchai et al., 2001). Here we show that Nop15p is required for pre-rRNA processing, but also has a specific function in cell cycle progression.
Results
Cells depleted of Nop15p are defective in 60S subunit synthesis and pre-rRNA processing
The predicted ORF for YNL110c/NOP15 contains 220 amino acids (25.4 kDa), and analysis with PSORT (Nakai and Horton, 1999) identified one canonical RNA recognition motif (RRM; residues 92–168) and two putative nuclear localization signals (NLS), consistent with a role in nuclear RNA processing.
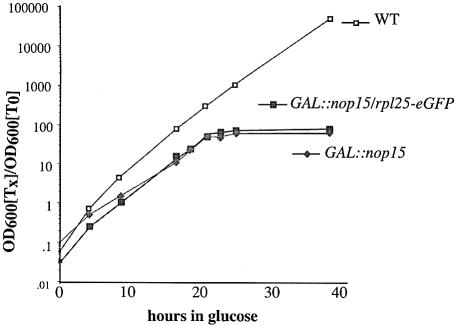
To examine the function of Nop15p, its expression was placed under the control of a repressible galactose promoter using a one-step PCR technique in strain YDL401 (see Materials and methods). Growth of the GAL::nop15 strain (YMO11) was not clearly different from that of the isogenic wild-type strain (YDL401) on permissive, galactose medium, but was progressively slowed following transfer to repressive, glucose medium (Figure 1). Mild growth impairment was initially seen, commencing ∼6 h after transfer, with much more stringent growth inhibition after 20 h. Both the sharp inflection point in the growth curve and the complete growth inhibition are unusual features for strains in which ribosome synthesis factors are depleted under GAL repression, which is never complete (see below).
Fig. 1. Depletion of Nop15p leads to abrupt growth arrest. Growth curves of GAL::nop15 strains following transfer to glucose medium, with and without expression of Rpl25p-eGFP. Strains were pre-grown in RGS medium and transferred to glucose medium for the times indicated. Strains were maintained in exponential growth by dilution with pre-warmed medium. Cell densities measured by OD600 are shown corrected for dilution. Wild-type (open squares); GAL::nop15; (filed diamonds); GAL::nop15; rpl25-GFP (filled squares).
To determine whether depletion of Nop15p inhibits rRNA synthesis, metabolic labelling with [H3]uracil was performed 16 h after transfer to glucose minimal medium (Figure 2). In the GAL::nop15 (YMO14) strain, synthesis of mature 25S and 5.8S rRNA was almost completely inhibited (Figure 2C and D). The 27SA pre-rRNA was accumulated, while the 27SB and 7S pre-rRNAs were lost. In contrast, synthesis of the 20S pre-rRNA and 18S rRNA remained robust. Synthesis of the 5S rRNA and tRNA was also little affected (Figure 2D).
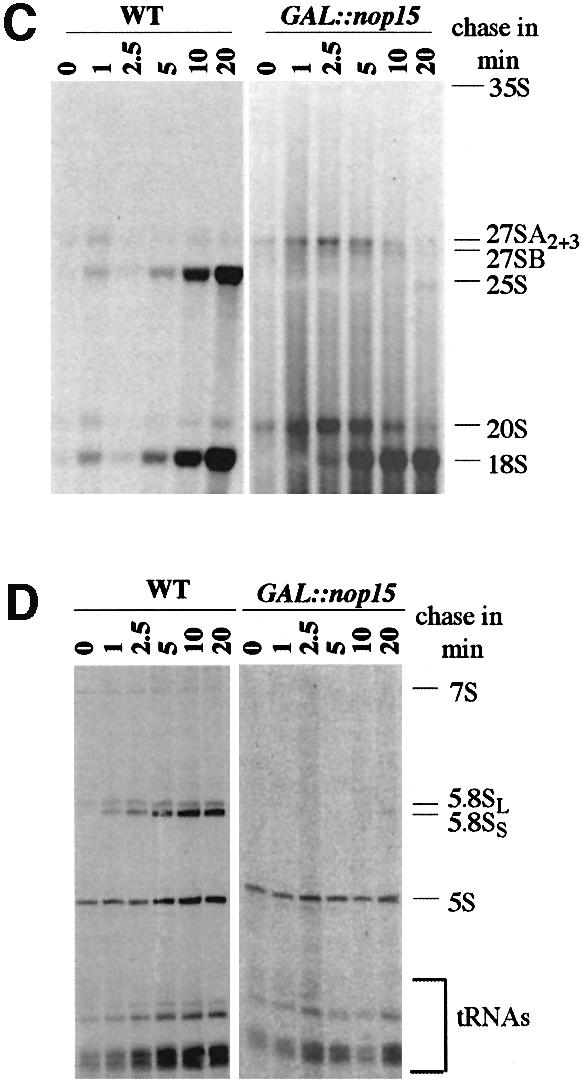
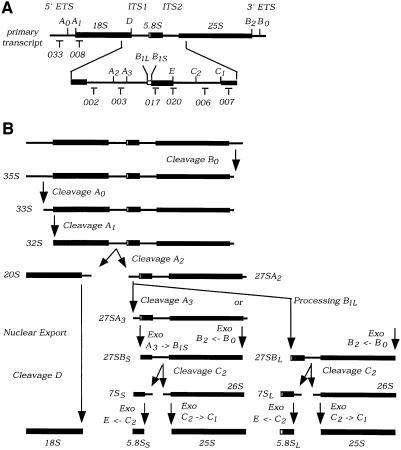
Fig. 2. Pulse–chase analysis of rRNA synthesis. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S and 25S, which are separated by the two internal transcribed spacers ITS1 and ITS2 and flanked by the two external transcribed spacers 5′ETS and 3′ETS. The positions of oligonucleotide probes utilized in northern hybridization and primer extension analyses are indicated. (B) Pre-rRNA processing pathway. For further discussion of the processing pathway and enzymes, see Kressler et al. (1999), Venema and Tollervey (1999), Warner (2001) and Fatica and Tollervey (2002). (C and D) Pre-rRNA was pulse-labelled with [3H]uracil for 2 min at 30°C and chased with a large excess of unlabelled uracil for the times indicated. Labelling was performed for the GAL::nop15 strain and a wild-type strain 16 h after transfer to glucose medium. (C) High molecular weight RNA separated on a 1.2% agarose/formaldehyde gel. (D) Low molecular weight RNA separated on an 8% polyacrylamide/urea gel.
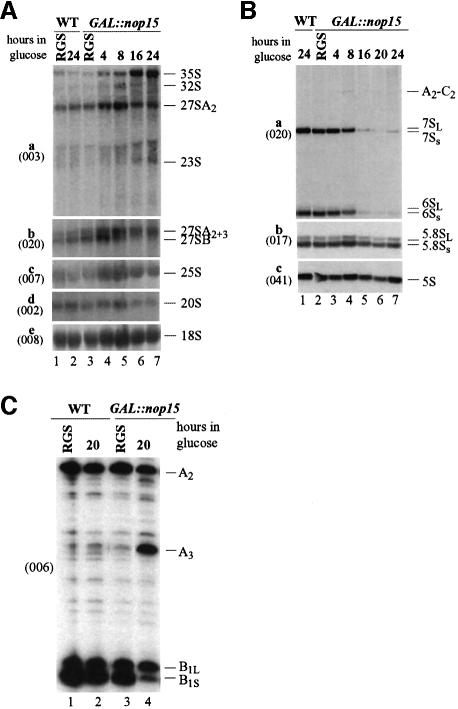
The effects of Nop15p depletion on pre-rRNA processing were assessed in more detail by northern hybridization and primer extension (Figure 3). Depletion of Nop15p resulted in some accumulation of the 35S primary transcript but had little impact on levels of the 27SA2 or 20S pre-rRNAs (Figure 3A). This indicates that the early cleavages at sites A0, A1 and A2 are kinetically delayed but still occur with good efficiency. In contrast, the levels of the 27SB pre-rRNAs were mildly reduced after 8 h and strongly reduced by 16 h after transfer to glucose medium. Levels of the 27SA2 + A3 pre-rRNAs, both of which are detected by oligo 020 (Figure 3A, b), were mildly increased following Nop15p depletion. This is in contrast to the depletion of 27SA2 (Figure 3A, a), suggesting that the 27SA3 pre-rRNA was accumulated. This was confirmed by primer extension (Figure 3C), which showed little alteration in the level of 27SA2, as shown by the primer extension stop at site A2, and strong accumulation of 27SA3, shown by the stop at site A3. The level of 27SBS was clearly reduced, whereas the level of 27SBL appeared unaffected, as shown by the primer extension stops at B1S and B1L, respectively. The exonucleases Rat1p and Xrn1p normally digest the pre-rRNA from site A3 to site B1S (Henry et al., 1994), and the loss of 27SBS is therefore a consequence of defective processing of 27SA3.
Fig. 3. Analysis of pre-rRNA processing. (A) Northern analysis. Lanes 1 and 2, wild-type strain in RGS medium and 24 h after transfer to glucose; lanes 3–7, GAL::nop15 strain in RGS medium and after transfer to glucose medium for the times indicated. (B) Northern analysis. Lane 1, wild-type strain 24 h after transfer to glucose; lanes 2–7, GAL::nop15 strain in RGS medium and after transfer to glucose medium for the times indicated. RNA was separated on a 1.2% agarose/formaldehyde gel (A) or 8% polyacrylamide/urea gel (B). Probe names are indicated in parentheses on the left. (C) Primer extension using oligo 006, which hybridizes within ITS2, 3′ to site C2 (the 3′ end of the 7S pre-rRNA). Primer extension stops at sites A2, A3, B1S and B1L show levels of the 27SA2, 27SA3, 27SBS and 27SBL pre-rRNAs, respectively. Lanes 1 and 2, wild-type strain in RGS medium and 20 h after transfer to glucose medium; lanes 3 and 4, GAL::nop15 strain in RGS medium and 20 h after transfer to glucose medium.
Analysis of low molecular weight RNAs (Figure 3B) showed that the 7S and 6S pre-rRNAs were mildly depleted after 8 h and strongly depleted by 16 h after transfer to glucose medium (Figure 3Ba). In contrast to the 27SB pre-rRNAs, no clear differences were observed in the ratios of the long and short forms of the 7S pre-rRNA (7SL and 7SS), showing that the processing of 27SBL to 7SL is also inhibited (see Figure 2A). Little depletion of mature 18S, 25S or 5.8S rRNA was seen even at late time points. We conclude that Nop15p is required for exonucleolytic processing of the 27SA3 pre-rRNA to 27SBS. The alternative form, 27SBL, continues to be synthesized but fails to be converted to 7SL.
These results demonstrate that Nop15p is a bona fide pre-rRNA processing factor with specific functions in the synthesis of the 25S and 5.8S rRNAs. It is, however, striking that the mature rRNAs were not substantially depleted even 24 h after transfer to glucose medium. The mature rRNAs are stable in most backgrounds and must be depleted by growth, which is very slow in the Nop15p-depleted strain at late times.
To test for potential defects in 60S subunit export, the Rpl25p–enhanced green fluorescent protein (eGFP) reporter (Gadal et al., 2001) was expressed in the GAL::nop15 strain. No nuclear accumulation of Rpl25p-eGFP was observed on Nop15p depletion (data not shown). Expression of Rpl25p-eGFP synergistically inhibits growth in some mutants with ribosome assembly defects (Oeffinger et al., 2002) but did not alter the kinetics of growth inhibition in the GAL::nop15 strain (Figure 1).
Together, these observations indicate that growth arrest in Nop15p-depleted strains is not a consequence of depletion of the cytoplasmic ribosomes or defects in assembly or export of the 60S subunits.
Nop15p is associated with precursors to the 25S and 5.8S rRNAs
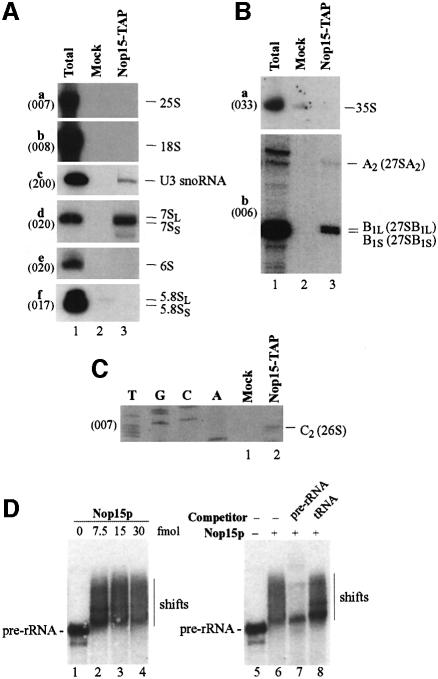
RNAs that co-precipitated with TAP-tagged Nop15p (Figure 4A–C, Nop15p-TAP lanes) were compared with a mock precipitation from the non-tagged parental strain YDL401 (Figure 4A–C, Mock lanes). Northern hybridization (Figure 4A) showed that the 7SL and 7SS pre-rRNA components of the pre-60S ribosomes co-precipitated with Nop15p-TAP, but were not detectably recovered in a mock precipitation from YDL401. The U3 snoRNA, a component of the 90S pre-ribosomes, was precipitated with lower efficiency. The 6S pre-rRNA and mature rRNAs were not co-precipitated detectably. High molecular weight pre-rRNAs were analysed by primer extension (Figure 4B and C). The 27SA2, 27SBL, 27SBS and 26S pre-rRNA components of pre-60S ribosomes were co-precipitated with Nop15p-TAP. In contrast, no co-precipitation of the 35S pre-rRNA was observed.
Fig. 4. Nop15p associates with pre-rRNAs in vivo and in vitro. (A–C) Nop15p-TAP co-precipitates with pre-rRNAs. Lane 1, total RNA control (5 µg); lane 2, mock precipitate from a wild-type control strain; lane 3, precipitate from a strain expressing Nop15p-TAP. (A) Northern hybridization of high and low molecular weight RNAs separated on a 1.2% agarose/formaldehyde gel or 8% polyacrylamide/urea gel, respectively. (B and C) Primer extension analyses. Nop15p-TAP was immunoprecipitated from cell lysates using IgG–agarose, with release of bound RNA–protein complexes by cleavage of the protein A linker by TEV protease. RNA was recovered from the released material, and from a mock-treated, isogenic wild-type control strain. Oligonucleotides used are indicated in parentheses. (D) Nop15p binds to pre-rRNA in vitro. Gel mobility shift assay performed with an in vitro transcribed pre-rRNA fragment, extending from the 5′ region of ITS1 to the 3′ region of ITS2. Nop15p was expressed in E.coli as a GST fusion with a thrombin-sensitive linker and eluted from a glutathione–Sepharose column by cleavage with thrombin. Lanes 1–4, 10 fmol pre-rRNA incubated with 0–30 fmol Nop15p as indicated; lanes 5–8, competition experiment. The gel shift assay was performed in the presence of 200-fold molar excess of cold pre-rRNA (lane 7) and tRNA (lane 8). Reactions in lanes 6–8 contained 10 fmol RNA and 15 fmol Nop15p. Complexes were resolved by electrophoresis in native 6% acrylamide/bisacrylamide (80:1) gels containing 0.5× TBE.
These results show that Nop15p is associated with the early pre-60S ribosomes. The precipitation of the U3 snoRNA reveals some association with earlier pre-ribosomes. However, the lack of co-precipitated 35S indicates binding only to the later 32S pre-rRNA, upon which A2 cleavage occurs. Nop15p has an RRM and may bind directly to the pre-rRNA, probably immediately prior to cleavage at site A2, and remains associated with the pre-60S particle until after cleavage at site C2.
To test directly whether Nop15p exhibits RNA binding activity, a gel shift assay was performed (Figure 4D). A labelled pre-rRNA transcript was prepared, which extends from the 5′ region of ITS1 to the 3′ region of ITS2 and includes sites A2, A3, B1 and C2. Labelled transcript (10 fmol) was incubated with 0–30 fmol of recombinant Nop15p, which was expressed in Escherichia coli as a thrombin-cleavable GST fusion construct (see Materials and methods). Free and bound forms of the RNA were resolved by native gel electrophoresis (Fatica et al., 2002). Nop15p clearly retarded the migration of the RNA, although the bound material did not migrate as a discrete species. Binding was inhibited by cold competitor pre-rRNA transcript, but was not inhibited by a 200-fold molar excess of tRNA. We conclude that Nop15p has RNA binding activity. Further analyses will be required to determine whether this in vitro binding is sequence specific.
Cells depleted of Nop15p become non-viable and arrest with abnormal morphology
The association of Nop15p with pre-rRNAs and the processing defects seen following its depletion clearly demonstrate that Nop15p is a genuine ribosome synthesis factor. However, the sudden and dramatic growth arrest seen on depletion of Nop15p (Figure 1) is not expected for strains carrying a GAL-regulated allele of a ribosome synthesis factor and has not been seen in many other published analyses. GAL repression is not complete, and such strains generally continue growth at a reduced rate that can be supported by the residual ribosome synthesis. The growth arrest in the Nop15p-depleted strain therefore suggests a function in addition to ribosome synthesis, particularly as it occurs prior to substantial depletion of the mature rRNAs.
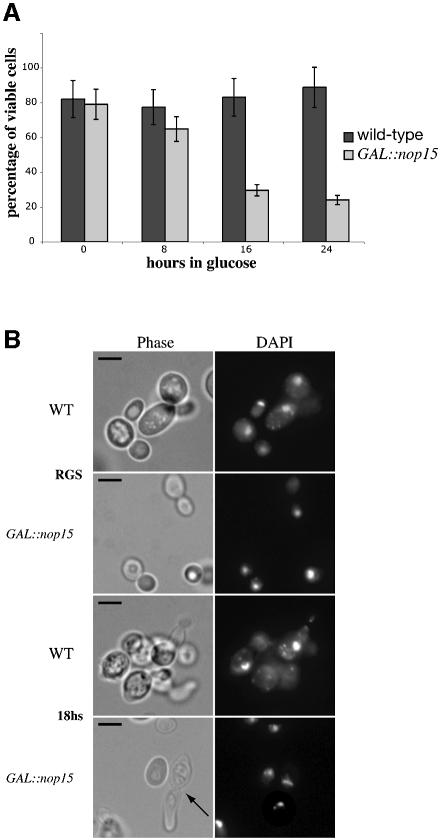
Cell viability was assessed during incubation in glucose medium by plating the cells on solid galactose medium. Cell numbers were calculated by counting in a haemocytometer slide. At 16 h after transfer to glucose medium, the viability of the Nop15p-depleted cells had dropped to ∼20%, 4-fold lower than the wild-type or the GAL::nop15 strain grown on galactose medium. (Figure 5A). In a similar analysis performed on a GAL::nop1 strain, viability dropped only much later in depletion (commencing after 60 h), and a slow decline in growth rate continued through 80 h of depletion (Tollervey et al., 1991). The inability of the cells to recover from depletion of Nop15p strongly suggested that reduced numbers of ribosomes is not the major cause of growth arrest.
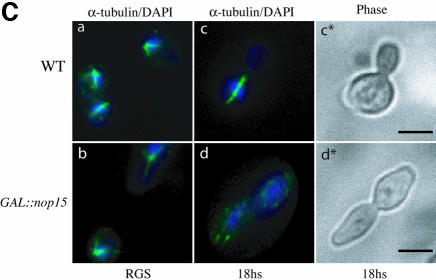
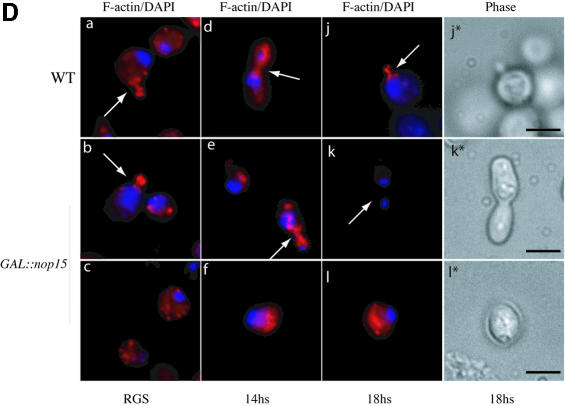
Fig. 5. Depletion of Nop15p leads to a cell cycle defect. (A) Percentage of cells from wild-type and GAL::nop15 strains that remain viable when plated on fresh, pre-warmed YRGS plates following incubation in glucose medium for the times indicated. Viability was determined by comparing the total number of cells plated with the number of colonies formed at each time point. (B) Cell morphology. (C) Staining with anti-tubulin. (D) Staining with rhodamine-conjugated phalloidin to detect F-actin. Wild-type and GAL::nop15 strains were examined during growth in RGS medium and following transfer to glucose medium for 14 or 18 h. The position of the nucleus was visualized by DAPI staining. In (C) and (D), the figures indicated with an asterisk show phase contrast images of cells in the corresponding immunofluorescence images. The bar represents 10 µm and arrows indicate the position of the bud neck.
Microscopic inspection of the GAL::nop15 strain 18 h after transfer to glucose medium showed 63% unbudded cells (from 600 cells counted), indicating an arrest in G1. However, 37% of the cells showed a distinctive and unusual elongated shape, which was not observed in galactose medium, demonstrating a defect in cell morphology (Figure 5B). These elongated cells contained duplicated, separated nuclei as shown by 4′,6-diamidino-2-phenylindole (DAPI) staining and had a marked constriction corresponding to the bud neck (indicated by an arrow in Figure 5B), where cytokinesis should have occurred. This morphology is distinct from that seen in cells defective in the mitotic exit network (MEN), which arrest with an intact, elongated mitotic spindle (Jimenez et al., 1998). To confirm this distinction, anti-tubulin staining was performed on wild-type and GAL::nop15 cells after 18 h in glucose medium to observe the organization of microtubules (Figure 5C). None of the elongated GAL::nop15 cells contained a mitotic spindle (from >500 cells examined), while the rounded cells arrested in G1 displayed an apparently normal interphase array of microtubules (Figure 5C, d). No cells examined showed morphology consistent with metaphase arrest.
A collection of ‘heat-inducible-degron’ strains has been described very recently (Kanemaki et al., 2003), in which the target protein is rapidly degraded following transfer to 37°C. In a strain expressing the degron-tagged Nop15p construct (generously provided by K.Labib, Paterson Institute), 35% of cells arrested with large buds and divided nuclei within 2 h of transfer to 37°C (data not shown), demonstrating that this phenotype does not require long incubation in restrictive conditions.
Bud neck constriction during cytokinesis involves an actin/myosin contractile ring (Bi et al., 1998). The ability of the Nop15p-depleted cells to form this ring was assessed using rhodamine-conjugated phalloidin to visualize the organization of polymerized or filamentous actin (F-actin). In the wild-type (Figure 5D, a, d and j), F-actin was observed in cortical patches and was concentrated around the bud neck in dividing cells (indicated by arrows in Figure 5D). The same organization was observed for the GAL::nop15 strain in permissive RGS medium (Figure 5D, b and c) and 14 h after shift to glucose medium (Figure 5D, e and f). After 18 h in glucose medium, the rounded GAL::nop15 cells arrested in G1 showed randomly distributed F-actin patches (Figure 5D, l). However, none of the elongated cells (from >500 cells examined) contained detectable F-actin at the bud neck (arrow in Figure 5D, k), indicating that actin is depolymerized in these cells.
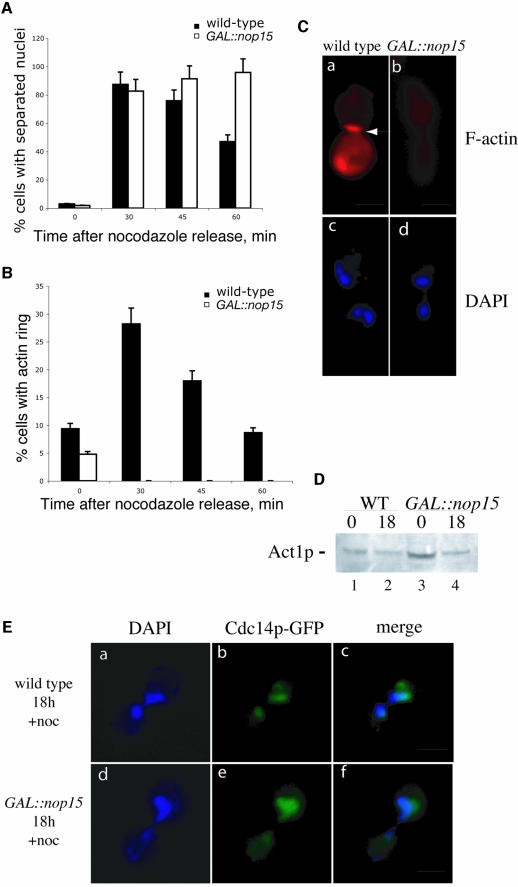
To analyse this cell cycle defect better, wild-type and GAL::nop15 cells were examined following synchronization. Cell division was arrested at the spindle assembly checkpoint by nocodazole treatment (Tolliday et al., 2002), which inhibits microtubule assembly, preventing spindle formation and nuclear division. Nocodazole was added 15 h after the shift to glucose medium, at which time the GAL::nop15 cells are still cycling, and both wild-type and mutant uniformly arrested as large-budded cells. After 3 h incubation, the nocodazole was washed out and nuclear separation and actin ring formation were monitored microscopically. In both wild-type and Nop15p-depleted cells, mitosis resumed with similar kinetics, as shown by the high efficiency of nuclear separation 30 min after release from nocodazole arrest (Figure 6A and C). However, while the wild-type cells carried on through cytokinesis, the Nop15p-depleted cells uniformly arrested as large budded cells (Figure 6A). Wild-type cells exhibited a peak of actin ring formation 30 min after release from nocodazole arrest, with ∼29% of cells showing an actin ring (Figure 6B and C), in good agreement with previous reports (Tolliday et al., 2002). In striking contrast, no Nop15p-depleted cells with an F-actin ring were detected 30, 45 or 60 min after nocodazole release (from >500 cell examined) (Figure 6B and C).
Fig. 6. Cytokinesis and actin ring formation are inhibited in synchronized, Nop15p-depleted cells. Wild-type and GAL::nop15 cells were shifted to glucose medium for 15 h and then treated with nocodazole for 3 h, arresting the cells in late M phase. Cells were examined before nocodazole washout (0 min samples) and at times points following release from arrest. (A) Efficiency of nuclear separation. (B) Efficiency of actin ring formation. (C) Typical wild-type and GAL::nop15 cells visualized 30 min after nocodazole release. F-actin was visualized by staining with rhodamine-conjugated phalloidin (panels a and b). Nuclei were visualized by DAPI staining (panels c and d). (D) Western blot analysis of Act1p in wild-type and GAL::nop15 cells. Whole-cell extracts were prepared and equal amounts of protein were separated by SDS–PAGE. Act1p was detected by western blotting using primary anti-actin antibody (I-19) and secondary peroxidase-conjugated rabbit-anti goat antibody. (E) Release of Cdc14p from nucleolus to nucleoplasm during late mitosis. The release of Cdc14p–GFP during mitotic exit was observed in wild-type (panel b) and GAL::nop15 (panel e) cells 30 min after release from nocodazole arrest. Nuclei were visualized by DAPI staining (panels a and d). The bar represents 10 µm.
The total level of actin present in the cells was assessed by western blotting (Figure 6D), which revealed no clear difference between the mutant and wild-type. The ability of the Nop15p-depleted cells to activate the MEN was assessed by examining the localization of Cdc14p–GFP (Figure 6E) [kindly provided by E.Schiebel (Hofken and Schiebel, 2002)]. In both the wild-type and Nop15p-depleted cells, Cdc14p–GFP was predominantly nucleoplasmic, indicating that Nop15p is not required for its release from nucleolar sequestration.
The Nop15p-depleted cells arrested correctly at the spindle assembly checkpoint and subsequently were able to undergo nuclear separation, activation of the MEN and disassembly of the mitotic spindle with the same efficiency as the wild-type, showing that they do not have a non-specific defect in cell cycle progression. However, the Nop15p-depleted cells uniformly arrested at cytokinesis and failed to form an actin ring at the bud neck. We conclude that Nop15p is required, directly or indirectly, for the recruitment of actin to the developing bud neck. The consequent failure to form a normal contractile ring presumably leads to the observed failure to undergo cytokinesis.
The nucleolus segregates late and Nop15p becomes partially delocalized during nuclear separation
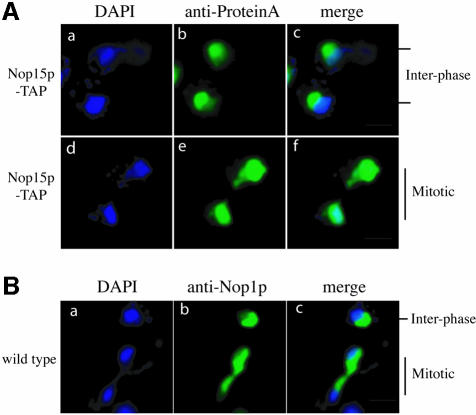
Nop15p was reported previously to be localized to the nucleolus, based on its exclusion from the nucleoplasmic region of the nucleus (Harnpicharnchai et al., 2001). This was confirmed by the co-localization in indirect immunofluorescence of Nop15p-TAP with a DsRed fusion with the well characterized nucleolar protein Nop1p, the yeast homologue of human fibrillarin (data not shown) (Gadal et al., 2001). In interphase cells, Nop15p-TAP was largely excluded from the nucleoplasm, which was decorated by DAPI staining Figure 7A, a–c). The localization of Nop15p was dramatically altered in the mitotic cells, filling the entire nuclear volume, as shown by the co-localization of the protein A tag with the DAPI-stained region (Figure 7A, d–f). A second striking observation was that in late mitotic cells in which the nucleoplasm of the daughter nuclei was well separated as shown by DAPI staining (Figure 7A, d), Nop15p-stained material formed a bridge between the nuclei (Figure 7A, e).
Fig. 7. The nucleolus segregates late during yeast cell division. The localization of the nucleolus was determined in wild-type interphase and mitotic cells. (A) The nucleolar marker Nop1p localized with anti-Nop1p antibody. (B) The localization of Nop15p-TAP. The nucleoplasm was visualized by DAPI staining. The bar represents 10 µm.
To determine whether the late segregation of Nop15p is a general feature of nucleolar proteins, we also examined the behaviour of Nop1p (Figure 7B). In both interphase and mitotic cells, Nop1p was largely excluded from the nucleoplasm. In mitotic cells, segregation of Nop1p to the daughter nuclei also occurred after substantial separation of the nucleoplasm.
We conclude that the nucleolus was divided and segregated to the daughter nuclei only after separation of the nucleoplasm. It is clearly essential that cytokinesis should not commence until the daughter nucleoli have completely separated. This suggests a possible rationale for linking the timing of actin ring formation to nucleolar components. The release of Nop15p from the nucleolus at this time may be functionally related to the triggering of cytokinesis, although the mechanisms involved remain to be elucidated.
Discussion
Nop15p and ribosome synthesis
We show here that Nop15p is a bona fide ribosome synthesis factor, which is required for the 5′ to 3′ exonuclease digestion that generates the 5′ end of the major, short form of the 5.8S rRNA as well as for processing of 27SB to 7S pre-rRNA. Consistent with this, and with recent proteomic analyses (Harnpicharnchai et al., 2001; Gavin et al., 2002), tagged Nop15p was associated with RNA components of the early pre-60S particles. Weaker co-precipitation was seen for the U3 snoRNA, a component of the 90S pre-ribosomes, whereas the 35S pre-rRNA was not co-precipitated detectably. We conclude that Nop15p is likely to associate with the 90S pre-ribosomes after the initial cleavage at site A0 and immediately prior to cleavage at site A2, remaining associated with the early pre-60S complexes (Fatica and Tollervey, 2002) during processing at sites A3, B1S, B1L and C2. Nop15p has a good consensus RRM and showed RNA binding activity in vitro, and is therefore likely to bind directly to the pre-rRNA. Alterations in the pre-rRNA structure induced by binding of Nop15p may be important for the recruitment of the 5′ to 3′ exonucleases Rat1p and Xrn1p (Henry et al., 1994) that generate the 5′ end of the 5.8S rRNA.
Nop15p and the cell cycle
In cells undergoing depletion of Nop15p, the growth rate was only mildly reduced until ∼16–20 h, at which time growth abruptly ceased. This was accompanied by a substantial decline in the viability of cells that were returned to permissive medium. Moreover, the abundance of the ribosomal subunits was not strongly reduced at the time at which growth ceased. This behaviour is not common for cells depleted of ribosome synthesis factors, which usually show a slow, progressive increase in doubling time, as the growth rate falls to match reduced numbers of ribosomes. A lack of cytoplasmic ribosomes is therefore unlikely to be the principal cause of the growth arrest in Nop15p-depleted cells.
Microscopic examination revealed two distinct terminal phenotypes in the Nop15p-depleted cells. Around 63% of cells were rounded and unbudded, consistent with an inability to pass the ‘Start’ cell cycle checkpoint. The remaining 37% showed an unusual, elongated morphology with duplicated and separated nuclei. A constriction was present marking the site of bud formation, but the cells had clearly been unable to complete cytokinesis. Rapid depletion of Nop15p in a ts-degron strain (Kanemaki et al., 2003) led to a similar proportion of cells apparently arrested at cytokinesis within 2 h of transfer to the non-permissive temperature. Following cell synchronization in late M-phase by nocodazole treatment and release, the defect in cytokinesis was complete, with all Nop15p-depleted cells uniformly arresting at the first cytokinesis.
In S.cerevisiae, cortical patches of polymerized F-actin support the ellipsoid outgrowth of the bud cell wall during S phase. During late mitosis, actin is partially relocated to the bud neck to form an actin/myosin contractile ring (Bi et al., 1998) that will constrict the bud neck leading to cytokinesis. Staining with rhodamine-conjugated phalloidin revealed that none of the arrested, Nop15p-depleted cells had assembled an actin ring (from >500 examined), potentially explaining their failure to complete cytokinesis (Bi et al., 1998). General defects in the actin cytoskeleton lead to defects in cell polarity and spindle formation (Palmer et al., 1992; Ayscough et al., 1997; McMillan et al., 1998; Theesfeld et al., 1999), which are predicted to lead to metaphase arrest. We did not see Nop15p cells that are arrested in metaphase, and Nop15p-depleted cells that arrested in G1 retained an apparently normal distribution of polymerized actin, indicating that the defect in actin assembly is specific for cytokinesis.
The defect in cytokinesis seen in strains lacking Nop15p has not been reported previously for any of the >100 characterized ribosome synthesis factors. More specifically, mutations in proteins that are present in the same pre-60S particles as Nop15p (Harnpicharnchai et al., 2001; Fatica and Tollervey, 2002) have not been observed to result in defects in cytokinesis. The pre-rRNA processing defect in Nop15p-depleted cells most closely resembles that seen on depletion of Nop7p/Yph1p (Adams et al., 2002; Oeffinger et al., 2002). Depletion of Yhp1p does result in a cell cycle phenotype, but this is due to defects in DNA replication (Du and Stillman, 2002) and is clearly distinct from the effects of Nop15p depletion.
Defects in another nucleolar protein, Net1p/Cfi1p, also result in cell cycle arrest as large-budded cells, due to a failure to activate the MEN (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). Nop15p-depleted cells lacked the characteristic features of MEN defects; a stabilized mitotic spindle and the distinctive bud projections that arise from continued spindle elongation (Jimenez et al., 1998). More directly, Nop15p-depleted cells were shown to release Cdc14p–GFP efficiently from the nucleolus following release from nocodazole arrest, demonstrating MEN activation.
Components of the septum formation machinery, including the septins and Myo1p, are localized to the future bud site early in the cell cycle (Lippincott and Li, 1998). In contrast, actin is relocated to form the actin/myosin ring only late in the cell cycle, following nucleoplasmic separation and shortly before disassembly of the mitotic spindle. Several proteins that are required for actin ring assembly have been identified (Osman et al., 2002; Tolliday et al., 2002), but the mechanism and timing of its recruitment remain poorly understood. We observed that segregation of the nucleolar markers Nop15p and Nop1p in dividing yeast cells lags behind the separation of the nucleoplasm. Dividing cells were readily detected in which the nuclei appeared well separated by DAPI staining, but were in fact bridged by nucleolar material. Late segregation of the nucleolus has been seen previously in fission yeast (Toda et al., 1981; Hirano et al., 1989), so this may be a conserved feature. In mitotic cells, Nop15p was partially delocalized from the nucleolus, with staining seen throughout the nucleoplasm. Two regulatory networks are implicated in the release of Cdc14p from the nucleolus. The FEAR network (Cdc fourteen early release) is required for initial release, while the MEN is both activated by Cdc14p and required to prevent relocation of Cdc14p back to the nucleolus (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). The roles of these networks in the triggering of nucleolar separation and/or release of Nop15p remain to be determined. The late nucleolar separation and/or the release of Nop15p would be appropriate timing markers for the initiation of actin ring formation.
The basic mechanism of cytokinesis and several factors involved in septum formation and closure are conserved from yeast to animal cells (Field et al., 1999). The closest human Nop15p homologue is NIFK, which is nucleolar in interphase but associates with condensed mitotic chromosomes (Takagi et al., 2001). Functions of Nop15p, in addition to its role in pre-rRNA processing, may therefore have been conserved in humans.
Materials and methods
Strains
Growth and handling of S.cerevisiae were by standard techniques. GAL-regulated strains were pre-grown in RGS medium, containing 2% raffinose, 2% galactose and 2% sucrose, and harvested at intervals following a shift to medium containing 2% glucose. Strains for pulse–chase analysis were pre-grown in minimal RGS medium lacking uracil and shifted to minimal glucose medium lacking uracil. Strains for immunofluorescence studies were grown in minimal glucose medium lacking leucine. The degron-Nop15p fusion strain (generously provided by K.Labib, Paterson Institute) was grown as described (Kanemaki et al., 2003).
Yeast strains used and constructed in this study are listed in table 1 of the Supplementary data available at The EMBO Journal Online. Conditional mutants under the control of the repressible GAL10 promoter were generated by a one-step PCR strategy in the strains YDL401 and BMA64 (Lafontaine and Tollervey, 1996). Transformants were selected for HIS+ prototrophy and screened by PCR. TAP-tagged strains were constructed by a one-step PCR strategy in the GAL-mediated strain GAL::nop15 (Rigaut et al., 1999). Transformants were screened by immunoblotting and PCR. TAP-tagged strains were transformed with pUN100DsRedNOP1 (kindly provided by E.Hurt, Heidelberg) to allow ready identification of the nucleolus.
Viability assay
GAL::nop15 and wild-type strains were pre-grown in RGS medium, containing 2% raffinose, 2% galactose and 2% sucrose, before being shifted to medium containing 2% glucose. A viability assay was performed by plating ∼1, 2.5 and 5 × 103 cells of samples taken at 0, 8, 16 and 24 h after the shift onto pre-warmed YGSR plates. Colonies were counted after 2 days of growth and the percentage viability was determined by comparison with the number of cells plated.
RNA extraction, northern hybridization and primer extension
RNA was extracted as described previously (Tollervey, 1987a). For high molecular weight RNA analysis, 7 µg of total RNA was separated on a 1.2% agarose gel containing formaldehyde and transferred for northern hybridization as described previously (Tollervey, 1987b). Standard 6, 8 or 12% acrylamide–8 M urea gels were used to analyse low molecular weight RNA species and primer extension reactions. Primer extensions were performed as described previously (Beltrame and Tollervey, 1992) on 5 µg of total RNA using primers 006, 007 and 033.
For pre-rRNA and rRNA analysis the following oligonucleotides were used: 002, 5′-GCTCTTTGCTCTTGCC; 003, 5′-TGTTACCTCTGGGC CC; 006, 5′-AGATTAGCCGCAGTTGG; 007, 5′-CTCCGCTTATTGA TATGC; 008, 5′-CATGGCTTAATCTTTGAGAC; 017, 5′-GCGTTGT TCATCGATGC; 020; 5′-TGAGAAGGAAATGACGCT; 033, 5′- CGC TGCTCACCAATGG; 041, 5′-CTACTCGGTCAGGCTC; and 200, 5′- UUAUGGGACUUGUU.
Immunofluorescence
For localization of Nop15p, Nop1p and tubulin, cells were fixed by incubation in 4% (v/v) formaldehyde for 30 min at 25°C, and spheroplasted. Immunofluorescence was performed as described (Grandi et al., 1993; Bergès et al., 1994). To stain nuclear DNA, DAPI was included in the mounting medium (Vectashield, Vector Laboratories). Nop15p-TAP was detected with an anti-protein A antibody coupled to fluorescein isothiocyanate (FITC). Nop1p was detected with a mouse anti-Nop1p antibody and a secondary goat anti-mouse antibody coupled to FITC (Sigma). Tubulin was detected with a rabbit anti-tubulin antibody (Sigma) and a secondary goat anti-rabbit antibody coupled to FITC (Sigma) at a 1:1000 and a 1:200 dilution, respectively. For F-actin staining, cells were fixed in 1% glutaraldehyde for 10 min at 25°C. Cells were washed twice in phosphate-buffered saline (PBS), resuspended in PBS/0.1% Triton X-100 and incubated for 10 min at 25°C. Rhodamine-conjugated phalloidin (6 U) in PBS was added and incubated for 45 min at 25°C. Cells were washed once in PBS and incubated with poly-l-lysine-coated coverslips before being mounted onto slides using moviol, containing DAPI. Cells containing pYE195-Rpl25-eGFP were grown in SGSR-URA to mid-exponential phase, transferred to SD-URA for various times and fixed in 4% (v/v) formaldehyde for 30 min and pelleted. Cells were resuspended in 100 mM KH2Ac/K2HAc/1.1 M sorbitol and mounted onto slides using moviol, containing DAPI. For synchronization in late M-phase, cells were treated with nocodazole (10 µg/ml) for 3 h, commencing 15 h after shift to glucose-containing medium. Cells were released by washing with pre-warmed medium and stained for F-actin as described above. Images were obtained with Smart Capture VP.
Immunoprecipitation of Nop15p-TAP
For immunoprecipitation of Nop15p-TAP, cells were grown in YPgal to OD600 = 2 and lysed in buffer A (150 mM KAc, 20 mM Tris-Ac pH 7.5, 5 mM MgAc) with 1 mM dithiothreitol (DTT), 0.5% Triton X-100, 2.5 mM vanadyl-ribonucleoside complexes (VRC) and 5 mM phenylmethylsulfonyl fluoride (PMSF) at 4°C using glass beads (Sigma). Immunoprecipitation with rabbit IgG–agarose beads, TEV cleavage and RNA extraction were performed as described (Rigaut et al., 1999).
Pulse–chase labelling experiments
Pulse–chase labelling of pre-rRNA was performed as previously described (Tollervey, 1987b) using 100 µCi of [5,6-3H]uracil (Amersham) for 2 min at 30°C. Total RNA was extracted with buffer AE/phenol–chloroform and ethanol precipitated (Schmitt et al., 1990). 3H-Labelled pre-rRNA and rRNA were resolved on 1.2% agarose gels for high molecular weight and 8% acrylamide–8 M urea gels for low molecular weight RNAs. RNA was transferred to Hybond-N+ Nylon membranes (Amersham), dried and exposed to X-ray film for 10 days at –80°C with an intensifying screen.
Mobility shift assays
A PCR fragment corresponding to the YNL110c (NOP15) ORF was amplified and cloned into BamHI and EcoRI sites of the pGEX-2T vector (Pharmacia) to obtain the pGEX-NOP15 vector. Nop15p was expressed in the E.coli strain BL21, purified by using glutathione–Sepharose according to the manufacturer’s protocol and released by cleavage with thrombin (Sigma). In vitro transcription of pre-rRNA (5′-ITS1 to 3′-ITS2) was performed with PCR products amplified from an rDNA plasmid and carrying the T7 promoter region. The binding reaction was performed in 30 mM Tris–HCl pH 7.4, 150 mM KCl, 2 mM MgCl2, 0.1% Triton X-100, 20% glycerol and 1 mM DTT, in the presence of tRNA (1 µg/µl), 10 fmol of 32P-labelled pre-rRNA and 0–30 fmol of recombinant protein in a reaction volume of 15 µl. RNA was heat denatured at 65°C for 10 min, followed by slow cooling to room temperature, prior to the binding reaction. For competition experiments, a 200-fold molar excess of cold competitor RNA was added. Reactions were incubated at room temperature for 30 min and then loaded on a 6% native acrylamide/bisacrylamide (80:1)/4% glycerol gel in 0.5× TBE buffer. Prior to loading, the gel was pre-run for 1 h and then run for 5 h at 250 V in the cold room.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Anthony Leung and Angus Lamond for communicating results prior to publication, Elmar Schiebel for the Ccd14p–GFP, and Karim Labib for the ts-degron Nop15p strain. We thank Hiroyuki Ohkura and Joanna Kufel for helpful comments on the manuscript. M.O. was the recipient of a Darwin Trust Fellowship. This work was supported by the Wellcome Trust.
References
- Adams C.C., Jakovljevic,J., Roman,J., Harnpicharnchai,P. and Woolford,J.L.,Jr (2002) Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA, 8, 150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Ayscough K.R., Stryker,J., Pokala,N., Sanders,M., Crews,P. and Drubin,D.G. (1997) High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol., 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J., Grandi,P., Gadal,O., Leßmann,T., Petfalski,E., Tollervey,D., Lechner,J. and Hurt,E. (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell, 8, 517–529. [DOI] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergès T., Petfalski,E., Tollervey,D. and Hurt,E.C. (1994) Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J., 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox,P., Lew,D.J., Salmon,E.D., McMillan,J.N., Yeh,E. and Pringle,J.R. (1998) Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol., 142, 1301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi G., Saracino,F., Masnada,D. and Carbone,M.L. (2000) The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J. Cell Sci., 113, 1199–211. [DOI] [PubMed] [Google Scholar]
- Capozzo C., Sartorello,F., Dal Pero,F., D’Angelo,M., Vezzi,A., Campanaro,S. and Valle,G. (2000) Gene disruption and basic phenotypic analysis of nine novel yeast genes from chromosome XIV. Yeast, 16, 1089–1097. [DOI] [PubMed] [Google Scholar]
- De Antoni A., D’Angelo,M., Dal Pero,F., Sartorello,F., Pandolfo,D., Pallavicini,A., Lanfranchi,G. and Valle,G. (1997) The DNA sequence of cosmid 14-13b from chromosome XIV of Saccharomyces cerevisiae reveals an unusually high number of overlapping open reading frames. Yeast, 13, 261–266. [DOI] [PubMed] [Google Scholar]
- Du Y.C. and Stillman,B. (2002) Yph1p, an ORC-interacting protein. Potential links between cell proliferation control, DNA replication and ribosome biogenesis. Cell, 109, 835–848. [DOI] [PubMed] [Google Scholar]
- Fatica A. and Tollervey,D. (2002) Making ribosomes. Curr. Opin. Cell Biol., 14, 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica A., Dlakic,M. and Tollervey,D. (2002) Naf1p is a box H/ACA snoRNP assembly factor. RNA, 8, 1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Field C., Li,R. and Oegema,K. (1999) Cytokinesis in eukaryotes: a mechanistic comparison. Curr. Opin. Cell Biol., 11, 68–80. [DOI] [PubMed] [Google Scholar]
- Fujiyama S., Yanagida,M., Hayano,T., Miura,Y., Isobe,T. and Takahashi,N. (2002) Isolation and proteomic characterization of human parvulin-associating preribosomal ribonucleoprotein complexes. J. Biol. Chem., 277, 23773–23780. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Grandi P., Doyl,V. and Hurt,E.C. (1993) Purification of NSP1 reveals complex formation with ‘GLFG’ nucleporins and a novel nuclear pore protein NIC96. EMBO J., 12, 3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P. et al. (2001) Composition and functional characterization of yeast 66s ribosome assembly intermediates. Mol. Cell, 8, 505–515. [DOI] [PubMed] [Google Scholar]
- Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Konoha,G., Toda,T. and Yanagida,M. (1989) Essential roles of the RNA polymerase I largest subunit and DNA topoisomerases in the formation of fission yeast nucleolus. J. Cell Biol., 108, 243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofken T. and Schiebel,E. (2002) A role for cell polarity proteins in mitotic exit. EMBO J., 21, 4851–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J., Friedlander,G., Bergmann,S., Sarig,O., Ziv,Y. and Barkai,N. (2002) Revealing modular organization in the yeast transcriptional network. Nature Genet., 31, 370–377. [DOI] [PubMed] [Google Scholar]
- Jimenez J., Cid,V.J., Cenamor,R., Yuste,M., Molero,G., Nombela,C. and Sanchez,M. (1998) Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J. Cell Biol., 143, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa,J.L., Breitkreutz,B.J. and Tyers,M. (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science, 297, 395–400. [DOI] [PubMed] [Google Scholar]
- Kanemaki M., Sanchez-Diaz,A., Gambus,A. and Labib,K. (2003) Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature, 423, 720–4. [DOI] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and de La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (1996) One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res., 24, 3469–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J. and Li,R. (1998) Sequential assembly of myosin II, an IQGAP-like protein and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol., 140, 355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J.N., Sia,R.A.L. and Lew,D.J. (1998) A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol., 142, 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P. et al. (2001) Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell, 105, 499–509. [DOI] [PubMed] [Google Scholar]
- Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Oeffinger M., Leung,A., Lamond,A. and Tollervey,D. (2002) Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA, 8, 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M.A., Konopka,J.B. and Cerione,R.A. (2002) Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J. Cell Biol., 159, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R., Sullivan,D., Huffaker,T. and Koshland,D. (1992) Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol., 119, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Seraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W. et al. (2001) Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell, 8, 45–55. [DOI] [PubMed] [Google Scholar]
- Shou W. et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Shou,W., Dowd,G.J., Turck,C.W., Deshaies,R.J., Johnson,A.D. and Moazed,D. (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell, 97, 245–256. [DOI] [PubMed] [Google Scholar]
- Sueishi M., Takagi,M. and Yoneda,Y. (2000) The forkhead-associated domain of Ki-67 antigen interacts with the novel kinesin-like protein Hklp2. J. Biol. Chem., 275, 28888–92. [DOI] [PubMed] [Google Scholar]
- Takagi M., Sueishi,M., Saiwaki,T., Kametaka,A. and Yoneda,Y. (2001) A novel nucleolar protein, NIFK, interacts with the forkhead associated domain of Ki-67 antigen in mitosis. J. Biol. Chem., 276, 25386–25391. [DOI] [PubMed] [Google Scholar]
- Theesfeld C.L., Irazoqui,J.E., Bloom,K. and Lew,D.J. (1999) The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol., 146, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Yamamoto,M. and Yanagida,M. (1981) Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe: video fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J. Cell Sci., 52, 271–287. [DOI] [PubMed] [Google Scholar]
- Tollervey D. (1987a) High level of complexity of small nuclear RNAs from fungi and plants. J. Mol. Biol., 196, 355–361. [DOI] [PubMed] [Google Scholar]
- Tollervey D. (1987b) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J., 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen,H., Carmo-Fonseca,M. and Hurt,E.C. (1991) The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J., 10, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday N., VerPlank,L. and Li,R. (2002) Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol., 12, 1864–1870. [DOI] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang,E.S. and Amon,A. (1999) Cfl1p prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature, 398, 818–823. [DOI] [PubMed] [Google Scholar]
- Warner J.R. (2001) Nascent ribosomes. Cell, 107, 133–136. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu,Z., Fu,X. and Liang,C. (2002) Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell, 109, 849–860. [DOI] [PubMed] [Google Scholar]
- Zimmerman Z.A. and Kellogg,D.R. (2001) The Sda1 protein is required for passage through start. Mol. Biol. Cell, 12, 201–219. [DOI] [PMC free article] [PubMed] [Google Scholar]