Abstract
ARC92/ACID1 was identified as a novel specific target of the herpes simplex transactivator VP16 using an affinity purification procedure. Characterization of the protein revealed tight interactions with human Mediator mediated through a von Willebrand type A domain. ARC92/ACID1 further contains a novel activator-interacting domain (ACID), which it shares with at least one other human gene termed PTOV1/ACID2. The structure of ARC92/ACID1 is of ancient origin but is conserved in mammals and in selected higher eukaryotes. A subpopulation of Mediator is associated with ARC92/ACID1, which is specifically required for VP16 activation both in vitro and in mammalian cells, but is dispensable for other activators such as SP1. Despite many known targets of VP16, ARC92/ACID1 appears to impose a critical control on transcription activation by VP16 in mammalian cells.
Keywords: activator/cofactors/gene expression/RNA polymerase II/transcription regulation
Introduction
Accessory factors play a critical role in transcription of eukaryotic genes by RNA polymerase II. These transcription cofactors influence the regulatory architecture of genes (O’Riordan and Grosschedl, 2000), they modulate the structure of chromatin (Jenuwein and Allis, 2001; Rice and Allis, 2001; Roth et al., 2001; Neely and Workman, 2002) and they directly control the activity of the basal transcription apparatus (Hampsey and Reinberg, 1999). The human Mediator complexes (TRAP, ARC, CRSP and DRIP) were first isolated using activators as affinity systems (Fondell et al., 1996, 1999; Ito et al., 1999; Näar et al., 1999; Ryu and Tjian, 1999; reviewed in Malik and Roeder, 2000). They represent variants of one large evolutionarily conserved protein complex that is thought to control transcription through the recruitment of RNA polymerase II to promoters (Zaman et al., 1998; Myers and Kornberg, 2000). At least one Mediator variant is related to PC2 (positive cofactor 2), an activity that was first described in a biochemical screen for transcription cofactors (Meisterernst et al., 1991; Kretzschmar et al., 1994; Malik et al., 2000). Mechanistic studies indicated a role for the mammalian complex during pre-initiation complex formation (Johnson et al., 2002, 2003). Yeast Mediator functions during initiation as well, but seems also to be important for repeated rounds of transcription (Yudkovsky et al., 2000). Chromatin immunoprecipitations (ChIPs) in Drosophila melanogaster and Saccharo myces cerevisiae suggest that Mediator and RNA polymerase II are recruited sequentially to promoters (Park et al., 2001)
One of the widely studied models for eukaryotic activators is the prototypic acidic herpes simplex activator VP16. In eukaryotic cells, VP16 functions through the histone acetyltransferases CBP and STAGA/TFTC complexes (Brown et al., 2001; Hardy et al., 2002). In addition, the mammalian Swi/Snf and Tip60 complexes are recruited to genes following binding of this activator (Memedula and Belmont, 2003). Remodeling and histone-modifying pathways are also thought to facilitate VP16 function in other non-natural hosts, such as the yeast S.cerevisiae (Brown et al., 2001; Peterson, 2002). Within the transcription machine, VP16 targets the general factors TFIIH and TFIID (Goodrich et al., 1993; Xiao et al., 1994). Moreover, like many cellular activators, herpes simplex VP16 interacts specifically with Mediator (Ito et al., 2000; Ikeda et al., 2002). Collectively, these data suggest that the acidic activator stimulates transcription through the reversal of transcription repression in chromatin but it also affects formation and the activity of the basal transcription apparatus. It has been proposed further that proteolysis is a critical event in activation (Salghetti et al., 2001).
Despite the many possible targets, it remains unclear which protein complexes are bound by VP16 in vivo. More specifically, the critical contacts within the transcription machine remain elusive, which is reflected in the complete lack of reports of dominant-negative effects. Here we report purification, cloning and characterization of the human ARC92/ACID1 that binds herpes simplex VP16 with considerable specificity. ARC92/ACID1 is stably associated with a subpopulation of human Mediator complexes. Depletion of the ARC92/ACID1–Mediator (A-Med) complex from HeLa nuclear extracts abolishes activation by VP16, whereas SP1 and other activators remain fully active. Structure–function analysis of ARC92/ACID1 uncovered a von Willebrand factor type A (VWA) domain as the Mediator-binding module. The VP16-activating region binds an as yet uncharacterized ACID (activator interaction domain). Notably, both domains act in a strongly dominant-negative way on VP16 activation in mammalian cells. Further consistent with a role in transcription in vivo, fusion of the Mediator-binding domain to GAL4 activates transcription from synthetic reporters. ChIP experiments suggest that ARC92/ACID1 is recruited by VP16 to genes. These data uncover a novel molecular activation pathway of VP16 through A-Med complexes.
Results
Our early efforts to reconstitute transcription activation by RNA polymerase II in vitro led to the discovery of transcription coactivators, termed USA (upstream factor stimulatory activity), that supported stimulation by activators such as SP1 and USF (Meisterernst et al., 1991). The major activities in USA were identified as the positive cofactors PC4 and PC2. PC2 was later shown to be virtually identical to the CRSP-Mediator variant (Ryu and Tjian, 1999; Malik et al., 2000). Curiously, despite having many specific contacts with components of the transcription machinery, we could not reproduce the specific activation by the prototypic acidic activator VP16 efficiently in purified RNA polymerase II transcription systems using these USA-derived cofactors. On the other hand, VP16 and specifically its subregion H1 was a potent activator in crude extracts where it functions dependent on the F442P, a mutant that has been widely used to demonstrate specific activation in vivo (Ikeda et al., 2002). Thus, we concluded that our extracts contain additional critical components, which we collectively referred to as positive cofactor 6, PC6 (Meisterernst, 1999).
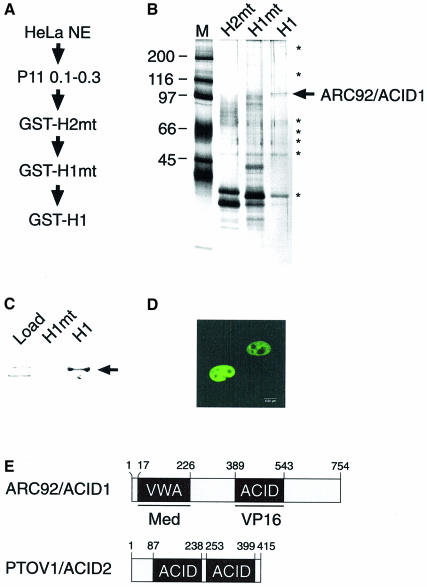
Isolation of a novel VP16-interacting protein
In an attempt to identify these missing components, we set up a biochemical screen. Fractionated HeLa nuclear extracts were subjected to affinity chromatography on two subsequent H1 and H2 mutants followed by a H1wt column (H1 and H2 are the names used for two subregions in the VP16 activation domain; Cress and Triezenberg, 1991). A single specific peptide with an apparent size of 103 kDa was purified and subsequently sequenced by N-terminal Edman degradation (Figure 1A and B). Database searches revealed a single gene that encodes a protein of ∼80 kDa with unknown function (the sequence is shown in figure 1 of the Supplementary data available at The EMBO Journal Online). Two human splice variants that differ in their C-terminal region have been described, one of which was termed p78 (Wang et al., 2002; Supplementary figure 4). Additionally, a closely related gene in Drosophila was annotated as ARC92. Furthermore, a full-length cDNA that was reported later during our investigation revealed identity to peptide sequences of a human ARC92 protein, a presumptive subunit of a previously isolated human ARC (TRAP/SMCC/DRIP-related) Mediator complex (Näar et al., 1999).
Fig. 1. Isolation of a novel VP16-interacting protein. (A) Purification scheme of ARC92/ACID1 through VP16 mutant (mt) and wild-type columns. Both H1 (amino acids 411–452) and H2 (amino acids 453–490) are subregions of the VP16 activation domain. (B) Silver-stained SDS– polyacrylamide gel of salt eluates from GST–H2mt, GST–H1mt and GST–H1wt columns. The arrow indicates a single specific polypeptide (ARC92/ACID1) migrating with an apparent mol. wt of 103 kDa. Unspecific bound polypeptides are marked with asterisks. (C) Specific binding of ARC92/ACID1 to GST–H1. A polyclonal antiserum directed against ARC92/ACID1 detects a single protein in H1- but not in H1F442P-purified (Mediator) complexes. (D) ARC92/ACID1 localizes to the nucleus. Transiently transfected HeLa cells were stained by indirect immunofluorescence using a ARC92/ACID1 monoclonal antibody. (E) Schematic overview of ARC92/ACID1 and PTOV1/ACID2. The locations of the VP16-binding module ACID and the Mediator-binding von Willebrand factor A domain (VWA) are indicated.
A serum raised against the N-terminal peptide of our purified factor recognized a protein with the correct size that bound specifically to the activation region H1 but not to the F442P point mutant (H1mt, Figure 1C). Expression of the protein in HeLa cells revealed a predominantly nuclear localization (Figure 1D).
Northern analysis showed ubiquitous but strongly varying levels of expression during early development and in adult tissues, respectively (Supplementary figure 5).
The analysis of the primary sequence of the protein provided strong evidence for at least two structured domains (Figure 1E), one of which resembles the A domain of the von Willebrand factor (VWA; Figure 7B, and Supplementary figure 7A and B). The factor contains a second evolutionarily conserved domain which is probably structured (supplementary figure 3, a secondary structure prediction is included in Supplementary figure 7B). We named the domain ACID as it proved to be critical for activator contacts (below, Figure 3). The search for proteins that share the ACID domain uncovered one additional transcribed gene that contains two copies of ACID, and which previously was identified as a prostate cancer overexpressed gene (PTOV-1; Santamaria et al., 2003). The genes are located next to each other on chromosome 19 (Supplementary figure 2). A third candidate in the human genome is a presumptive pseudogene (ACID3). In the light of the structural and a possible functional relationship of these proteins, we propose to add the functional annotation ACID to the corresponding proteins to be subsequently called ARC92/ACID1 and PTOV1/ACID2, respectively.
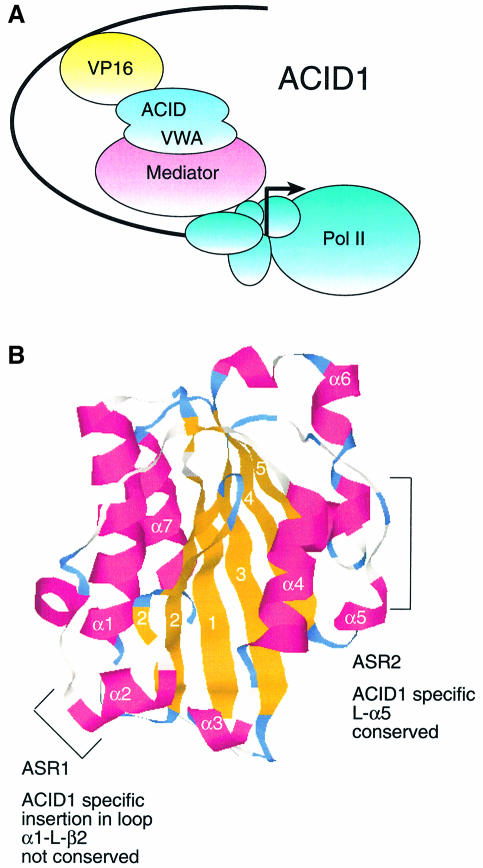
Fig. 7. Model of ARC92/ACID1 structure and function. (A) VP16 functions through ARC92/ACID1 via direct interaction with the ACID and dependent on the Mediator-binding module VWA. (B) X-ray structure of the VWA domain of integrin CD11a. Two regions specifically conserved in ARC92/ACID1 (ASR) are located on the surface.
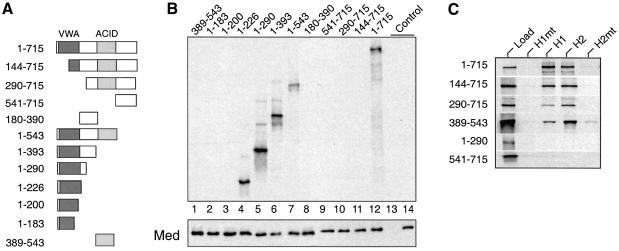
Fig. 3. VWA binds Mediator and ACID binds VP16 (A) Schematic overview of ARC92/ACID1 deletion constructs. (B) Autoradiography of an IP of 35S-labeled ARC92/ACID1 derivatives produced in reticulocytes, complemented with HeLa nuclear extracts and immunoprecipitated with the 6C9 antibody. An isotype antibody (lane 13) and nuclear extract alone (lane 14) were used as controls. Bottom panel: western blot probed with Med7 antibody to monitor levels of co-precipitated Mediator. (C) ACID binds specifically to VP16. Reticulocyte-expressed ARC92/ACID1 deletion constructs (Load), standardized via the incorporated [35S]methionine, were precipitated with GST–VP16 derivatives and bound proteins were monitored through autoradiography.
ARC92/ACID1 interacts with VP16 and human Mediator
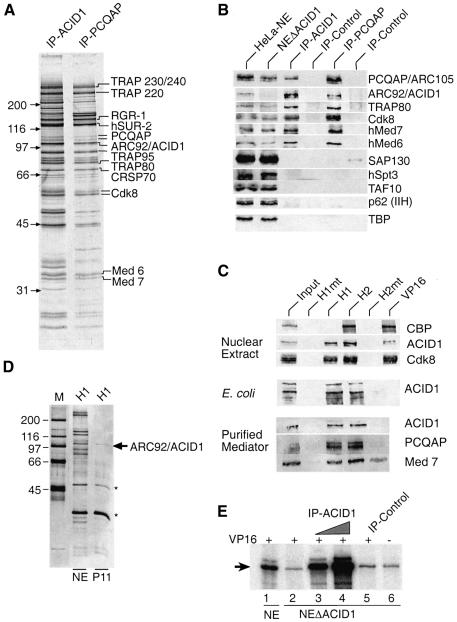
A monoclonal antibody (VC1) was generated directed against the N-terminal portion of ARC92/ACID1, and the epitope which it recognizes on ARC92/ACID1 was mapped. A bona fide Mediator complex was eluted with a peptide that comprises the corresponding epitope, if proteins were immunoprecipitated with VC1 from HeLa nuclear extracts (Figure 2A, lane 1). Evidence for this stems from the comparison with another Mediator preparation using an antibody (6C9) directed against the PCQAP/ARC105 Mediator subunit (Figure 2A, lane 2; for simplicity we use PCQAP and ACID1 in the figures). It is presently unclear whether certain differences between the two complex preparations originate from cross- reacting polypeptides or reflect as yet unknown variations in the Mediator complexes. The identity of the complexes was confirmed further in western blots which revealed co-elution of a series of defined Mediator components (Figure 2B). The western blot analysis also showed that ARC92/ACID1 is over-represented in the ARC92/ACID1-IP as compared with the PCQAP-IP (compare the ratio of ARC92/ACID1 with TRAP80). Thus, a significant percentage of ARC92/ACID1 is present in free (Mediator-unbound) form in HeLa nuclear extracts. Consistent with this, purification of proteins on GST–H1, using fractionated extracts (P11 0.3 M fraction) as input, mainly yields free ARC92/ACID1 (Figure 2D, compare with Figure 1B and Supplementary figure 6B). ARC92/ACID1-depleted extracts contained only residual ARC92/ACID1, but retained substantial amounts of several core Mediator subunits (such as TRAP80 and Med7, compare lane 1 and lane 2 of Figure 2B; Supplementary figure 6C). Thus, ARC92/ACID1 appears to be substoichiometrically represented on Mediator complexes.
Fig. 2. The interaction of ARC92/ACID1 with human Mediator is critical for activation by VP16 in vitro. (A) Silver-stained SDS–polyacrylamide gel of Mediator complexes immunopurified via ARC92/ACID1 (VC1) and PCQAP (6C9) monoclonal antibodies. The complexes were eluted with specific peptides covering the epitope of the corresponding antibodies. (B) Co-immunoprecipitation of ARC92/ACID1 with Mediator. HeLa nuclear extracts (NE), ARC92/ACID1-depleted nuclear extract (NEΔACID1), ARC92/ACID1 and PCQAP immunoprecipitates (4 U of the IP relative to input) together with antibody isotype controls (IP-control) were analyzed by western blot with the indicated Mediator and control antibodies. (C) Fractionation of HeLa extracts on GST–VP16 derivatives: ARC92/ACID1 and Mediator (here documented with Cdk8) bind specifically to H1 and H2. In contrast, CBP is specific for H2 (Ikeda et al., 2002). Below: E.coli-expressed ARC92/ACID1 and (bottom) peptide-eluted VC1-immunopurified A-Med bind specifically to H1 and H2 as analyzed in western blot with the indicated antibodies (right side). (D) Silver-stained SDS–polyacrylamide gel of fractionated HeLa extracts (P11 0.3 M KCl) purified on GST–H1. Asterisks mark non-specific proteins. (E) ARC92/ACID1–Mediator depletion of nuclear extract (NEΔACID1) impairs GAL4-VP16-activated transcription in vitro (lane 1 versus 2). The immunopurified complex (IP-ARC92/ACID1), but not an isotype antibody precipitate (IP-Control), restores transcription in depleted extract.
We next asked whether A-Med recognizes VP16 specifically. HeLa nuclear extracts were subjected to chromatography on immobilized GST–VP16 derivatives (Figure 2C, upper panel). We found that ARC92/ACID1 and Mediator (here monitored with Cdk8) bind to VP16, as well as to the independent activation regions H1 and H2, but not to the VP16 derivatives carrying point mutations (H1mt is F442P and H2mt is FFF473/475/479AAA). His-tagged ARC92/ACID1, expressed in Escherichia coli, also recognizes both H1 and H2 with a similar specificity. Importantly, a VC1-purified and peptide-eluted A-Med complex (see Figure 2A) displays comparable specificity for VP16 (Figure 2C, bottom).
A-Med is critical for VP16 activation in vitro
Immunodepletion of ARC92/ACID1 from extracts reduced GAL4-VP16-activated transcription from a GAL4-VP16-driven reporter (Figure 2E, lane 1 versus lane 2, compare with Figure 5A). Re-addition of the immunoprecipitated material strongly activated transcription above the levels seen in the extract (lanes 3 and 4). If depleted extracts were complemented with VC1- immunoprecipitated proteins, from extracts fractionated on phosphocellulose (P11) columns, they also recovered activity albeit to a significantly lower extent than non-fractionated extracts (Supplementary figure 6A and B and data not shown). This observation is generally consistent with the previously noted loss of specific activation by VP16 following fractionation of extracts (Meisterernst, 1999).
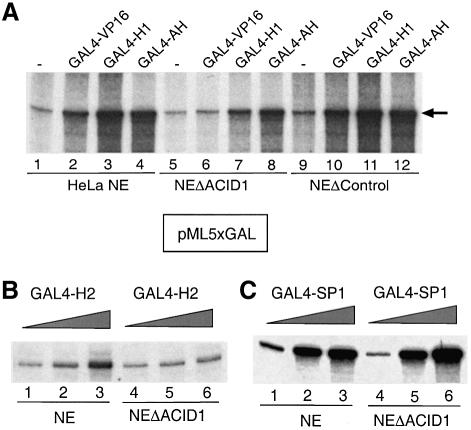
Fig. 5. ARC92/ACID1 is a gene-specific co-activator. (A) In vitro transcription experiment using the adenovirus major late core promoter carrying five upstream GAL4 sites, in the absence of activators and the presence of 100 ng of purified GAL4-VP16, GAL4-H1 and the activating helix GAL4-AH, as indicated. HeLa nuclear extracts (lanes 1–4) were compared with VC1-depleted (lanes 5–8) and isotype mock- depleted extract (lanes 9–12). (B and C) A 10, 20 and 50 ng aliquot of GAL4-H2 and 10, 20 and 100 ng of GAL4-SP1 were analyzed.
VWA binds Mediator and ACID binds VP16
To identify the Mediator-binding region [35S]methionine-labeled ARC92/ACID1 deletion constructs were produced in reticulocytes (Figure 3A). Normalized amounts of expressed ARC92/ACID1 proteins were complemented with HeLa nuclear extracts as a source for the limiting human Mediator and immunoprecipitated with a highly specific monoclonal PCQAP (6C9) antibody. Incorpor ation of ARC92/ACID1 derivatives into Mediator was monitored through autoradiography. Mediator precipitated well with the N-terminal 226 amino acids and somewhat better with the 290 N-terminal amino acids. C-terminally extended proteins were also co-precipitated. Deletion of the N-terminal 200 and 143 amino acids, respectively, abolished interactions with Mediator (lanes 3 and 11). These data suggest that an intact VWA domain is required and (together with the 16 non-conserved N-terminal amino acids) is sufficient for interaction of ARC92/ACID1 with Mediator.
To monitor interaction with VP16, normalized amounts of [35S]methionine-labeled ARC92/ACID1 derivatives were loaded onto GST–VP16 derivative columns (Figure 3C). ACID (389–543) alone binds efficiently (depleting it from the reticulocyte lysate) to H2 and to H1 but not to the corresponding mutants. Other regions of ARC92/ACID1 appear to modulate binding to H1, but specific contacts of ARC92/ACID1 with H2 rely fully on ACID. From these data, we conclude that the evolutionarily conserved ACID domain binds specifically to VP16.
AC92/ACID1 is critical for VP16-activated but not for basal transcription
ARC92/ACID1 deletion constructs were cloned in eukaryotic expression vectors, normalized through analysis of the expression levels in 293T cells and tested in co-transfection experiments together with a GAL4-H1 expression vector and a synthetic GAL reporter carrying five GAL4-binding sites upstream of a core promoter (pMRG5; Xie et al., 2000). The N-terminal 290 amino acids (for simplicity called the NTD) repressed transcription activation by H1 (Figure 4A). The FLAG-tagged ACID domain (amino acids 389–543) was unstable in cells and could thus not be tested. However, constructs expressing C-terminal regions lacking the NTD (290–715 and 290–754, data not shown) or parts of VWA (144–715) but containing ACID also repressed activation by VP16. Full-length ARC92/ACID1 (1–754) usually displayed moderate negative effects, which might reflect squelching of the active A-Med complexes.
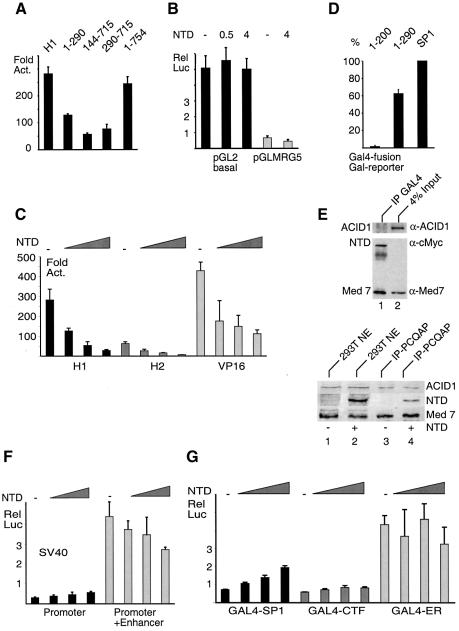
Fig. 4. Functional characterization of ARC92/ACID1 regions. (A) Normalized amounts of ARC92/ACID1 deletion constructs (corresponding to 4.0 µg of 1–290 encoding vector) were co-transfected together with 0.5 µg of GAL4-H1 expression vector and 10 µg of a GAL4-luciferase reporter (pGLMRG5) in Jurkat cells. (B) Co-transfection of 1.0 µg of pGL2 and pGLMRG5 with the indicated amount of a mouse NTD expression vector (encoding amino acids 1–290 of the mouse ARC92/ACID1 gene). (C) Co-transfection of expression vectors for VP16 derivatives (0.1 µg) with increasing concentrations of ARC92/ACID1 mouse NTD expression vector (0.05, 0.40 and 1.0 µg) and pGLMRG5 (1.0 µg) in SW13 cells. (D) Co-transfection of GAL4-NTD (amino acids 1–290 of human ARC92/ACID1 fused to GAL4 1–147) expression vector, a control lacking an intact VWA (1–200), or GAL4-SP1 expression vector (5.0 µg each) with 10.0 µg of pGLMRG5. (E) Western blot of GAL4-immunoprecipitated 293T cells transiently overexpressing GAL4-NTD (1.0 µg). NTD is detected via its Myc tag, Mediator via Med7 and ARC92/ACID1 with the VC1 antibody. Below: western blots of 293T cells transiently transfected with FLAG-ARC92/ACID1 (NTD, 1.0 µg). Input of non-transfected versus transfected cells (lanes 1 and 2) is compared with 6C9-IPs (lane 3 and 4). (F) Co-transfection of 1.0 µg of the SV40 promoter (pGL2 P) and the SV40 promoter–enhancer (pGL2 P+E, both Promega) with 0, 0.05, 0.4 and 1.0 µg of mouse NTD expression vectors in Jurkat cells. (G) Co-transfection of activator expression vectors (0.5 µg each) of GAL4-SP1, GAL4-CTF and GAL4-AF2 of the estrogen receptor (ERα) with 10 µg of pGLMRG5 and 0, 0.5, 4.0 and 10.0 µg, respectively, of mouse NTD expression vector in Jurkat cells.
Mammalian Mediator affects both activated and basal transcription by RNA polymerase II in vitro (Mittler et al., 2001; Baek et al., 2002). Next, we asked whether the presence of ARC92/ACID1 is relevant for basal transcription. Overexpression of the NTD did not reduce transcription from a luciferase reporter (pGL2, Promega), that lacks defined promoter sequences, leading to very low transcription levels from cryptic sites. The pMRG5 core promoter, comprising a TATA and an initiator sequence, was <1.5-fold repressed by NTD in the absence of GAL4 activator proteins (Figure 4B). ARC92/ACID1 (and A-Med) immunodepletion, leaving significant amounts of Mediator (measured via Med7) in the extract, moderately reduced basal transcription (<2-fold in Figure 5A; in other experiments, there was no detectable effect). Moreover, little ARC92/ACID1 was found in partially purified preparations of B-Med (basal Mediator; Mittler et al., 2001) which elutes in the P11 0.85 M KCl fraction (data not shown and Supplementary figure 6A). We conclude that ARC92/ACID1 has little influence on basal RNA polymerase II transcription.
Efficient modulation of transcription involves the Mediator-binding domain
Overexpression of the mouse ARC92/ACID1-NTD (1–290) strongly reduced activation of pGLMRG5 by GAL4-H1 from 280- to 30-fold, by GAL4-H2 from 60- to 6-fold and by GAL4-VP16 from 430- to 110-fold (Figure 4C). The strong dominant-negative effect correlated reasonably with ∼3-fold higher expression levels of mouse compared with human NTD (data not shown). We reasoned that if the ARC92/ACID1-NTD binds Mediator in vivo, a GAL4-NTD fusion protein might activate transcription (Balciunas et al., 2003). Indeed, GAL4-NTD (comprising an integrated Myc tag), but not a mutant containing an incomplete VWA domain (GAL4-1–200), activated transcription from the pGLMRG5 reporter. The activation was reproduced several times in different cell lines. It is significant, reaching ∼60% of the capacity of a GAL4-SP1 protein at saturating activator concentrations (Figure 4D). To document the interaction of GAL4-NTD with Mediator in vivo, overexpressed GAL-NTD was immunoprecipitated with a GAL4 antibody. Precipitates were analyzed for the presence of Mediator (assayed with a Med7 antibody), GAL-NTD (assayed with a monoclonal antibody directed against the Myc tag) and endogenous ARC92/ACID1. Indeed GAL-NTD precipitates a Mediator which then contains low levels of endogenous ARC92/ACID1 (Figure 4E, top panel). This was confirmed in a slightly different experimental set-up where FLAG-tagged NTD was expressed in 293T cells leading to co-precipitation of both ARC92/ACID1 and FLAG-NTD with Mediator complexes (bottom panel). In conclusion, the activation by GAL4-NTD and the dominant-negative function of NTD correlates with the binding and the loss, respectively, of endogenous ARC92/ACID1 from Mediator.
ARC92/ACID1 establishes a gene- and activator-specific docking site on Mediator
A couple of eukaryotic genes such as the SV40 enhancer–promoter and the T-cell receptor β (TCRβ) (data not shown) promoter–enhancer pair did not respond to the ARC92/ACID1-NTD in transient reporter assays (Figure 4F). Under conditions where GAL4-H1 activation was lowered 10-fold, the estrogen receptor α (ERα), that binds Mediator via the TRAP220 subunit (Kang et al., 2002), was moderately if at all affected (Figure 4G). The proline-rich CTF activator barely responded to NTD, whereas the glutamine-rich SP1 activation region fused to GAL4, reproducibly (also seen on the SP1-controlled SV40 promoter, Figure 4F) underwent a modest, ∼2-fold increase. Preliminary analyses of other acidic activators did not show a comparable response to overexpression of NTD (L.Berti, T.Stühler and M.Meisterernst, unpublished).
The activator-specific function was studied further in vitro through the comparison of a nuclear extract with ARC92/ACID1-depleted extracts (Figure 5A). Transcrip tion from a G-less GAL4 reporter (lanes 1–4) was significantly (3- to 4-fold) reduced in depleted (lanes 5–8) but not in mock-depleted nuclear extracts (lanes 9–12), in the presence of GAL4-VP16 and GAL4-H1 but not of GAL4-AH. The synthetic GAL4-AH was included because it is a well-standardized activator that, similarly to SP1, responds to the co-activator PC2 (Meisterernst et al., 1991; Kretzschmar et al., 1994). A slight decrease of GAL4-AH activity is probably accounted for by the moderate loss of basal activity (lane 1 versus lane 5). Consistent with the in vivo analysis, activation by GAL4-H2 was reduced in depleted extracts (Figure 5B), whereas in vitro expressed and purified GAL4-SP1 (Werten et al., 1998) remained active in A-Med-depleted extracts (Figure 5C).
ARC92/ACID1 gene interactions are dynamic in vivo
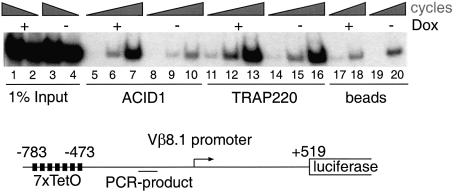
It remained to be shown that ARC92/ACID1 is found on genes in vivo. Towards this goal, a stable Epstein–Barr virus (EBV)-based Tet-VP16 reporter was established and analyzed by ChIP analysis. A HeLa cell line was used that constitutively expresses a Tet-VP16 protein, the binding of which can be rapidly induced with doxycycline. In the non-induced state, the Mediator subunit TRAP220 and low levels of ARC92/ACID1 were found on a 7xTet-Vβ 8.1 promoter. The presence of Mediator in the non-induced state may be context dependent and could relate to the fact that Vβ is under control of proximal activators, such as CREB (Halle et al., 1997). Induction of VP16 binding resulted in an ∼10-fold induction of transcription (data not shown) and led to a substantial increase of ARC92/ACID1 concentrations on the promoter (Figure 6). While the precise underlying molecular mechanism, specifically the interesting question of whether ARC92/ACID1 is recruited into a Mediator complex, remains to be elucidated, these data presently suggest that ARC92/ACID1 is recruited to genes in vivo.
Fig. 6. VP16 recruits ARC92/ACID1 to a stable episomal plasmid carrying the 7xTet-Vβ promoter. Chromatin immunoprecipitation of ARC92/ACID1 and TRAP220 was performed without and 12 h after induction with doxycycline (DOX). ARC92/ACID1 occupancy is increased 18-fold.
Discussion
In this investigation, we uncover and characterize a novel activation pathway of the prototypic acidic herpes simplex activator VP16 through ARC92/ACID1. A subpopulation of Mediator is associated with ARC92/ACID1 (termed A-Med). A-Med complexes are needed for specific activation in vitro, thereby reconstituting, at least in part, a long-missing activity previously termed PC6 (Meisterernst, 1999). Our data support a model in which VP16 critically functions through ARC92/ACID1 on human Mediator in mammalian cells (Figure 7A).
ARC92/ACID1 is restricted to selected higher eukaryotes
Mediator is highly conserved from yeast to man (Boube et al., 2000; Borggrefe et al., 2002). Nonetheless, we failed to detect a gene with significant homology to ARC92/ACID1 in yeast. The gene is present in Drosophila, but neither ARC92/ACID1 nor the related PTOV1/ACID2 was found in the genome of Caenorhabditis elegans. In line with a recent hypothesis of Levine and Tjian (2003), that the evolution of eukaryotes is generally accompanied by an increase in complexity of transcription control, the integration of an additional adaptor into a core Mediator complex may serve specific functions, i.e. in the development of mammals. This is also supported indirectly by the tissue-specific ARC92/ACID1 mRNA distribution. Mouse knock-out experiments will be necessary to substantiate this hypothesis. Curiously, however, Ciona intestinalis, more commonly known as sea squirt, contains a highly conserved gene that comprises both the VWA and the ACID domain of ARC92/ACID1 (Supplementary figure 7B). The recent sequencing of the genome of C.intestinalis serves as a model for some 550 million-year-old chordates, which are the precursors of vertebrates (Dehal et al., 2002). Thus, the Mediator-binding domain and ACID is of ancient origin. The fact that herpes simplex VP16 uses ARC92/ACID1 may be considered fortuitous. However, the virus is indeed restricted to vertebrates. It will be of interest to elucidate whether activation through ARC92/ACID1 is a coincidence or, via squelching of A-Med complexes, helps the virus to manipulate cellular functions (i.e. immune response, proliferation, etc.) and to support the viral life cycle.
The VWA domain comprises typical features of a protein–protein interaction module
The presence of the VWA domain is predicted by standard BLAST search and is suggested further by secondary structure predictions (Supplementay figure 7A). The A region of von Willebrand factor establishes contacts with collagen, while the corresponding domain in the integrins CD11a/b mediates CD18 ligand interactions. The predicted secondary structure of the ARC92/ACID1 VWA (indicated below the alignment of Supplementary figure 7A) is indeed related to the true secondary structure of CD11a as evident from the 1.8 Å resolution X-ray structure (indicated on top of the alignment of Supplementary figure 7A). VWA domains have typical features of a protein–protein interaction module. They consist of a rigid scaffold composed of helices grouped around a core that is formed from β-sheets. These are connected through loops that are less conserved in the structure and, therefore, could be used for individual protein–protein interactions. Two ARC92/ACID1-specific regions (ASRs) surrounding an α-helix 5 in CD11a (amino acids 145–167) and a short insertion (amino acids 51–63) are probably located on the surface and are non-structured. Specifically, the α5 region shows little homology within the other VWA proteins but is highly conserved through evolution in ARC92/ACID1 (Supplementary figure 7B). Hence, it must be involved in a critical function of ARC92/ACID1. This could be the interaction with Mediator or another function. Presently, the general features of the domain provide a plausible model for its involvement in interactions with Mediator.
Mechanistic implications and perspective
There is precedence for distinct gene-specific functions of Mediator subunits. The deletion of Mediator subunit Sur2 (Stevens et al., 2002) and TRAP220 (Ito et al., 2000) affected specific mitogen-activated protein (MAP) kinase and thyroid hormone receptor activation pathways with no obvious consequences on activation by VP16. In mammals, the relevance of Mediator for VP16 activation has largely been suggested through in vitro studies. Our data imply that ARC92/ACID1 introduces one additional critical level of control for the activation by the prototypic activator VP16 in mammalian cells that may be absent in lower eukaryotes. To our knowledge, this is the first report where a domain that is not a direct target of VP16, i.e. the VWA domain, causes a significant dominant effect in mammalian cells. One conclusion is that other targets of VP16, such as STAGA/TFTC, CBP, TFIID and TFIIH, that could theoretically bypass a need for Mediator, might contribute moderately to activation. Alternatively, VP16 could act sequentially through STAGA/TFTC, p300, Mediator and GTFs, leading to chromatin opening and activation of RNA polymerase II (Malik and Roeder, 2000; Lemon et al., 2001; Soutoglou and Talianidis, 2002). It remains to be elucidated whether the activation process also involves dynamic changes in the conformation (Taatjes et al., 2002) and/or the composition of the Mediator complex. The latter may be superficially concluded from the observation that ARC92/ACID1-TRAP220 ratios change following induction of transcription by VP16. Clearly, proof for this requires further investigations.
Notably, Mediator has been proposed to interact with VP16 through the TRAP80 subunit (Ito et al., 1999). We have confirmed the interaction using TRAP80 expressed in reticulocytes which bound specifically to the H2 domain of VP16 (T.Stühler and M.Meisterernst, unpublished). Hence, ARC92/ACID1 could be specifically important for binding to and the activation by H1. However, H2 and the intact VP16 activation domain also rely on ARC92/ACID1–Mediator interactions. Altogether, our data do not exclude the possibility that VP16 interacts with Mediator through both ARC92/ACID1 and TRAP80. Another interesting possibility, for which we have preliminary evidence, is that free ARC92/ACID1 acts as a co-activator on selected cellular genes (L.Berti, L.Santolin and M.Meisterernst, unpublished observation).
SP1 appears to function slightly better in the absence of ARC92/ACID1 and A-Med, respectively. The molecular reason for the stimulation is presently unclear but could relate to the observation that SP1, as well as GAL4-AH, which performed similarly in vitro, is specifically dependent on the PC2/CRSP form of human Mediator in vitro (Kretzschmar et al., 1994; Ryu and Tjian, 1999; Malik et al., 2000). Our preliminary data do not support the notion that other acidic activators such as p53 operate as efficiently through ARC92/ACID1 as VP16 (unpublished results). Yet, we envisage that certain defined activators, in a manner comparable with VP16, use this pathway. It will be of considerable interest to understand the interplay of ARC92/ACID1 with cellular genes and with the related factor PTOV1/ACID2.
Materials and methods
Gene isolation
The locus was identified on two genomic cosmids (AC006942 and AC018766). Mouse and human full-length cDNAs of ARC92/ACID1 and a neighboring gene PTOV1/ACID2 were obtained by screening cDNA libraries and via expressed sequence tag (EST) clones obtained from public resources.
Plasmids
hARC92/ACID1 deletion expression vectors were constructed by cloning of PCR products in the multiple cloning site of a previously modified pCI-neo Vector (Promega), which contains a FLAG tag cloned between the NheI and XhoI sites, followed by an in-frame NLS and the ARC92/ACID1 sequences. Alternatively, the GAL4 DNA-binding domain (amino acids 1–147) was fused to a Myc epitope positioned between the NheI and the EcoRI sites. ARC92/ACID1 derivatives correspond to the human factor unless indicated otherwise. All clones were verified by sequencing. The GAL4 reporter pGLMRG5 is derived from pMRG5 (Xie et al., 2000), but contains a luciferase gene instead of a G-free cassette.
In vitro transcription
Transcription from a linearized G-free cassette containing (5xGAL4)-Ad2ML promoter (100 ng) was conducted in HeLa S3 nuclear extracts (20–50 µg of total protein) complemented with 10–100 ng of purified His-tagged GAL4 and GAL4-VP16 derivatives under standard conditions (Ikeda et al., 2002). Depleted extracts were prepared using 6C9 antibody essentially as described (Mittler et al., 2001).
Transfection
Transient transfection of HEK 293T and SW13 cells (plasmid amounts refer to 6 cm dishes) with calcium phosphate was carried out using standard protocols. Jurkat T cells (plasmid amounts refer to 1.2 × 107 cells) were transfected by electroporation as described (Halle et al., 1997). Total amounts of DNA were adjusted by supplementation with the respective empty expression vectors. Luciferase units were normalized to protein concentrations. Generally, the means of 3–5 experiments and the standard errors are shown. GAL-ERα (AF2) transactivation was measured in the presence of 20 nM estradiol.
Protein purification
A 200 ml aliquot of a HeLa nuclear extract was loaded on phosphocellulose (P11) and step eluted with 0.3 M KCl in buffer BC (Meisterernst et al., 1991). The P11 0.3 M KCl fractions (containing ∼1000 mg of protein) were adjusted with buffer HD0 [20 mM HEPES NaOH pH 7.6, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT), 0.1% NP-40, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 µg/ml benzamidine] to 100 mM salt and incubated with VP16 derivatives fused to GST and immobilized on glutathione–Sepharose at concentrations of ∼1 mg/ml. Incubation for 4 h with 1.5 ml of GST–H2mt (FFF473/475/479AAA) was followed by incubation of the supernatant with 1.2 ml of GST–H1mt (F442P) column (1 mg/ml) for 16 h at 4°C. Upon gentle removal of beads, the supernatant was loaded on 1.3 ml of a GST–H1 (wild type) column, incubated for 4 h, washed extensively with HD100 (500 mM NaCl; 1× 5 ml, 2× 10 ml), further washed with 2 ml of HD250 (250 mM NaCl) and eluted with 3 ml of HD500 (500 mM NaCl). The HD500 eluate was analyzed by SDS–PAGE, subjected to preparative chromatography, eluted from the gel and analyzed by N-terminal Edman degradation. A peptide corresponding to the 10 N-terminal amino acids of ARC92/ACID1 was determined. GST pull-down experiments were performed essentially as described (Ikeda et al., 2002) except that in certain cases 35S-labeled reticulocyte expressed proteins substituted for nuclear extracts. For immunopurification, 4 ml of HeLa nuclear extracts were adjusted to 150 mM KCl, pre-cleared with 400 µl of protein G–beads, 400 µl of an isotype antibody column, followed by binding to specific antibody columns (400 µl) 6C9 and VC1, respectively. Antibodies were cross-linked to protein G–beads at a concentration of 1 mg/ml. Beads were washed extensively with buffer C (Mittler et al., 2001) at 150 and 1000 mM KCl (20 and 30 column volumns, respectively) and eluted with the peptides containing the corresponding epitope (sequences are available upon request). Two column volumes were added at a final concentration of 100 mg/ml peptide in buffer C.
Antibodies, immunoprecipitation, immunofluorescence and ChIP
Polyclonal antisera against TRAP220 were purchased from Santa Cruz Inc. A monoclonal antibody against the NTD (amino acids 1–290) of murine ARC92/ACID1, generated in rats using a GST fusion protein, recognizes both the human and the mouse protein. Polyclonal sera against amino acids 1–14 of human ARC92/ACID1 were generated in rabbits (Eurogentec, Belgium). For nuclear localization studies, both ARC92/ACID1 and FLAG antibodies were used in a standard procedure. For immunoprecipitation of overexpressed proteins, nuclear extracts were prepared from HEK 293T cells as described (Berti et al., 2001). To map the Mediator-interacting region, ARC92/ACID1 deletion constructs were expressed and radioactively labeled in vitro using [35S]methionine (TNT Coupled Reticulocyte Lysate System kit, Promega). 6C9 was immobilized on protein G–Sepharose at a concentration of 1 mg/ml. A 10 µg aliquot of beads was incubated at 4°C overnight, with normalized amounts of expressed proteins and together with 800 µg of total protein of HeLa nuclear extract. Beads subsequently were washed at 0.15 and 0.45 M KCl in buffer C and analyzed by SDS–PAGE, followed by either autoradiography or western blotting. As a control for the specificity of the method, 35S-labeled ARC92/ACID1 (1–715) was analyzed with an isotype antibody. ChIP has been performed essentially as described (Orlando et al., 1997) with modifications reported elsewhere (S.Jünemann and M.Meisterernst, in preparation) using the antibodies indicated in the figure legends.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to the former members of the laboratory, specifically Keiko Ikeda and Klaus Kaiser, for their contribution to the characterization of PC6, Rob Chapman for critical reading of the manuscript, Henk Stunnenberg for expression plasmids, Làszlò Tora for GAL-ERα-(AF2) plasmid and antibodies, Gema Merchan-Olea for technical assistance, and Gertraud Stelzer for support. This work was supported by grants from the DFG (transregio5), the TMR research training program of the EC (HPRN-CT-2002-00261) and the BMBF proteomics platform technology program (031U101F and 0313030A) to M.M.
References
- Baek H.J., Malik,S., Qin,J. and Roeder,R.G. (2002) Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol., 22, 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Hallberg,M., Bjorklund,S. and Ronne,H. (2003) Functional interactions within yeast mediator and evidence of differential subunit modifications. J. Biol. Chem., 278, 3831–3939. [DOI] [PubMed] [Google Scholar]
- Berti L. et al. (2001) Isolation and characterization of a novel gene from the DiGeorge chromosomal region that encodes for a mediator subunit. Genomics, 74, 320–332. [DOI] [PubMed] [Google Scholar]
- Borggrefe T., Davis,R., Erdjument-Bromage,H., Tempst,P. and Kornberg,R.D. (2002) A complex of the Srb8, -9, -10 and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem., 277, 44202–44207. [DOI] [PubMed] [Google Scholar]
- Boube M., Faucher,C., Joulia,L., Cribbs,D.L. and Bourbon,H.M. (2000) Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes Dev., 14, 2906–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Howe,L., Sousa,K., Alley,S.C., Carrozza,M.J., Tan,S. and Workman,J.L. (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science, 292, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Cress W.D. and Triezenberg,S.J. (1991) Critical structural elements of the VP16 transcriptional activation domain. Science, 251, 87–90. [DOI] [PubMed] [Google Scholar]
- Dehal P. et al. (2002) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science, 298, 2157–2167. [DOI] [PubMed] [Google Scholar]
- Fondell J.D., Ge,H. and Roeder,R.G. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA, 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J.D., Guermah,M., Malik,S. and Roeder,R.G. (1999) Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl Acad. Sci. USA, 96, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.A., Hoey,T., Thut,C.J., Admon,A. and Tjian,R. (1993) Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell, 75, 519–530. [DOI] [PubMed] [Google Scholar]
- Halle J.P., Haus-Seuffert,P., Woltering,C., Stelzer,G. and Meisterernst,M. (1997) A conserved tissue-specific structure at a human T-cell receptor β-chain core promoter. Mol. Cell. Biol., 17, 4220–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. and Reinberg,D.(1999) RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev., 9, 132–139. [DOI] [PubMed] [Google Scholar]
- Hardy S., Brand,M., Mittler,G., Yanagisawa,J., Kato,S., Meisterernst,M. and Tora,L. (2002) TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J. Biol. Chem., 277, 32875–32882. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Stuehler,T. and Meisterernst,M. (2002) The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells, 7, 49–58. [DOI] [PubMed] [Google Scholar]
- Ito M. et al. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. [DOI] [PubMed] [Google Scholar]
- Ito M., Yuan,C.X., Okano,H.J., Darnell,R.B. and Roeder,R.G. (2000) Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell, 5, 683–693. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Johnson K.M. and Carey,M. (2003) Assembly of a Mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr. Biol., 13, 772–777. [DOI] [PubMed] [Google Scholar]
- Johnson K.M., Wang,J., Smallwood,A., Arayata,C. and Carey,M. (2002) TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev., 16, 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.K., Guermah,M., Yuan,C.X. and Roeder,R.G. (2002) The TRAP/Mediator coactivator complex interacts directly with estrogen receptors α and β through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl Acad. Sci. USA, 99, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M., Stelzer,G., Roeder,R.G. and Meisterernst,M. (1994) RNA polymerase II cofactor PC2 facilitates activation of transcription by GAL4-AH in vitro. Mol. Cell. Biol., 14, 3927–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B., Inouye,C., King,D.S. and Tjian,R. (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature, 414, 924–928. [DOI] [PubMed] [Google Scholar]
- Levine M. and Tjian,R. (2003) Transcription regulation and animal diversity. Nature, 424, 147–151. [DOI] [PubMed] [Google Scholar]
- Malik S. and Roeder,R.G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. [DOI] [PubMed] [Google Scholar]
- Malik S., Gu,W., Wu,W., Qin,J. and Roeder,R.G. (2000) The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell, 5, 753–760. [DOI] [PubMed] [Google Scholar]
- Meisterernst M. (1999) Structure and function of proteins that modulate RNA polymerase II transcription. In Chambon,P., Fukasawa,T., Coath,C. and Kornberg,R.D. (eds), Transcription Regulation in Eukaryotes. Human Frontier Science Program, Strasbourg, Vol. VII, pp. 131–133. [Google Scholar]
- Meisterernst M., Roy,A.L., Lieu,H.M. and Roeder,R.G. (1991) Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell, 66, 981–993. [DOI] [PubMed] [Google Scholar]
- Memedula S. and Belmont,A.S. (2003) Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol., 13, 241–246. [DOI] [PubMed] [Google Scholar]
- Mittler G., Kremmer,E., Timmers,H.T. and Meisterernst,M. (2001) Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep., 2, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L.C. and Kornberg,R.D. (2000) Mediator of transcriptional regulation. Annu. Rev. Biochem., 69, 729–749. [DOI] [PubMed] [Google Scholar]
- Näar A.M., Beaurang,P.A., Zhou,S., Abraham,S., Solomon,W. and Tjian,R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature, 398, 828–832. [DOI] [PubMed] [Google Scholar]
- Neely K.E. and Workman,J.L. (2002) The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys Acta, 1603, 19–29. [DOI] [PubMed] [Google Scholar]
- O’Riordan M. and Grosschedl,R. (2000) Transcriptional regulation of early B-lymphocyte differentiation. Immunol. Rev., 175, 94–103. [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Park J.M., Werner,J., Kim,J.M., Lis,J.T. and Kim,Y.J. (2001) Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell, 8, 9–19. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. (2002) Chromatin remodeling: nucleosomes bulging at the seams. Curr. Biol., 12, R245–R247. [DOI] [PubMed] [Google Scholar]
- Rice J.C. and Allis,C.D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol., 13, 263–273. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Ryu S. and Tjian,R. (1999) Purification of transcription cofactor complex CRSP. Proc. Natl Acad. Sci. USA, 96, 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S.E., Caudy,A.A., Chenoweth,J.G. and Tansey,W.P. (2001) Regulation of transcriptional activation domain function by ubiquitin. Science, 293, 1651–1653. [DOI] [PubMed] [Google Scholar]
- Santamaria A., Fernandez,P.L., Farre,X., Benedit,P., Reventos,J., Morote,J., Paciucci,R. and Thomson,T.M. (2003) PTOV-1, a novel protein overexpressed in prostate cancer, shuttles between the cytoplasm and the nucleus and promotes entry into the S phase of the cell division cycle. Am. J. Pathol., 162, 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E. and Talianidis,I. (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science, 295, 1901–1904. [DOI] [PubMed] [Google Scholar]
- Stevens J.L., Cantin,G.T., Wang,G., Shevchenko,A. and Berk,A.J. (2002) Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science, 296, 755–758. [DOI] [PubMed] [Google Scholar]
- Taatjes D.J., Näar,A.M. andel,F.,3rd, Nogales,E. and Tjian,R. (2002) Structure, function and activator-induced conformations of the CRSP coactivator. Science, 295, 1058–1062. [DOI] [PubMed] [Google Scholar]
- Wang C., McCarty,I.M., Balazs,L., Li,Y. and Steiner,M.S. (2002) A prostate-derived cDNA that is mapped to human chromosome 19 encodes a novel protein. Biochem. Biophys. Res. Commun., 296, 281–287. [DOI] [PubMed] [Google Scholar]
- Werten S., Stelzer,G., Goppelt,A., Langen,F.M., Gros,P., Timmers,H.T., Van der Vliet,P.C. and Meisterernst,M. (1998) Interaction of PC4 with melted DNA inhibits transcription. EMBO J., 17, 5103–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H. et al. (1994) Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol., 14, 7013–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Collart,M., Lemaire,M., Stelzer,G. and Meisterernst,M. (2000) A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J., 19, 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N., Ranish,J.A. and Hahn,S. (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature, 408, 225–229. [DOI] [PubMed] [Google Scholar]
- Zaman Z., Ansari,A.Z., Gaudreau,L., Nevado,J. and Ptashne,M. (1998) Gene transcription by recruitment. Cold Spring Harbor Symp. Quant. Biol., 63, 167–171. [DOI] [PubMed] [Google Scholar]