Abstract
The yeast mitochondrial Oxa1 protein is a member of the conserved Oxa1/YidC/Alb3 protein family involved in the membrane insertion of proteins. Oxa1 mediates the insertion of proteins (nuclearly and mitochondrially encoded) into the inner membrane. The mitochondrially encoded substrates interact directly with Oxa1 during their synthesis as nascent chains and in a manner that is supported by the associated ribosome. We have investigated if the Oxa1 complex interacts with the mitochondrial ribosome. Evidence to support a physical association between Oxa1 and the large ribosomal subunit is presented. Our data indicate that the matrix-exposed C-terminal region of Oxa1 plays an important role supporting the ribosomal–Oxa1 interaction. Truncation of this C-terminal segment compromises the ability of Oxa1 to support insertion of substrate proteins into the inner membrane. Oxa1 can be cross-linked to Mrp20, a component of the large ribosomal subunit. Mrp20 is homologous to L23, a subunit located next to the peptide exit tunnel of the ribosome. We propose that the interaction of Oxa1 with the ribosome serves to enhance a coupling of translation and membrane insertion events.
Keywords: L23/mitochondria/Oxa1/protein insertion/ribosomes
Introduction
Oxa1 is a member of the conserved Oxa1/YidC/Alb3 protein family found throughout prokaryotes and eukaryotes (Yen et al., 2001). In prokaryotes, homologs of the Oxa1/YidC/Alb3 protein family, often termed YidC or IM60 (inner membrane 60 kDa protein), are found in both Gram-negative and Gram-positive bacteria. In eukaryotes, homologs are present in both mitochondria and chloroplasts (Yen et al., 2001). The Oxa1/YidC/Alb3 protein family represents a novel, evolutionarily conserved, membrane insertion machinery that mediates the insertion of hydrophobic proteins into their respective membranes (Luirink et al., 2001; Stuart, 2002; Kuhn et al., 2003).
Oxa1 is a component of a general protein insertion site in the inner membrane of mitochondria where it forms a homo-oligomeric complex. The Oxa1 complex mediates the insertion of both nuclear- and mitochondrially encoded proteins during their sorting to the mitochondrial inner membrane (He and Fox, 1997; Hell et al., 1997, 1998, 2001; Nargang et al., 2002; Stuart, 2002). In the yeast Saccharomyces cerevisiae, the mitochondrial genome encodes a small but important number of integral inner membrane proteins (Borst and Grivell, 1978). These proteins are essential subunits of respiratory chain complexes and include cytochrome b of the cytochrome bc1 complex, cytochrome oxidase subunits 1, 2 and 3 (Cox1, Cox2 and Cox3) and subunits of the Fo-sector of the ATP synthase, ATPase 6, 8 and 9.
The yeast OXA1-null mutant (Δoxa1) is respiratory deficient, as the assembly of the respiratory chain complexes, in particular the cytochrome oxidase, is compromised. Hence the Δoxa1-null mutants do not exhibit growth on non-fermentable carbon sources such as glycerol (Bauer et al., 1994; Bonnefoy et al., 1994). The role of Oxa1 in the insertion of proteins into the mitochondrial inner membrane was indicated from the analysis of mitochondria isolated from yeast mutants of the Oxa1 protein. In the absence of Oxa1 function, insertion of the mitochondrially encoded membrane proteins, and a subset of nuclear-encoded proteins into the inner membrane was found to be impaired (He and Fox, 1997; Hell et al., 1997, 1998, 2001). Of all the mitochondrially encoded proteins, the insertion of Cox1 and Cox2 proteins displayed the strictest dependency on Oxa1 (Hell et al., 2001). Cox1 is a polytopic protein spanning the inner membrane a total of 12 times. Cox2, also an integral membrane protein, spans the inner membrane twice in an Nout–Cout orientation. Cox2 is unique among yeast mitochondrial gene products as it is synthesized as a precursor protein (pCox2) with an additional N-terminal cleavable presequence. Attain ment of the Nout–Cout topology requires Oxa1-dependent insertion of the two transmembrane segments of Cox2 into the inner membrane, a step that is coupled to the export of hydrophilic, negatively charged N- and C-termini to the intermembrane space (Herrmann et al., 1995; He and Fox, 1997; Hell et al., 1997; Saracco and Fox, 2002). Following export across the inner membrane, the N-terminal pre-sequence is proteolytically removed by the intermembrane space-localized Imp1 peptidase (Nunnari et al., 1993). In the absence of Oxa1 function, however, pCox2 fails to undergo membrane insertion and accumulates in the mitochondrial matrix (He and Fox, 1997; Hell et al., 1997). The inner membrane proteins Pnt1, Cox18 and Mss2 recently have been reported to play a role in the insertion and translocation of the C-terminal region of Cox2 (He and Fox, 1999; Saracco et al., 2002).
Oxa1 is a 36 kDa protein located in the mitochondrial inner membrane, where it spans the inner membrane five times in a Nout–Cin orientation (Herrmann et al., 1997). The matrix-located C-terminal region of Oxa1 is ∼92 amino acid residues in length (amino acids 310–402) and is highly basic in nature (pI 11.9). Furthermore, a segment of amino acids located at the extreme C-terminal end of Oxa1 (residues 350–402) display the potential to form a coiled-coil structural motif. This feature is conserved in some but not all known mitochondrial Oxa1 homologs. The basic nature of the C-terminal region is a feature of Oxa1 that is conserved between mitochondrial Oxa1 proteins from other organisms. As previously postulated, this charged region might play an important role in the function of Oxa1 as a membrane protein insertion site and possibly in mediating a ribosomal interaction (Stuart, 2002).
Interaction of mitochondrially encoded substrate proteins with Oxa1 has been shown to occur during their synthesis as nascent chains and in a manner that is supported by the presence of the associated translating ribosome (Hell et al., 1998, 2001). We have postulated previously that the Oxa1 complex may interact directly with components of the ribosome to support or target the nascent chain–Oxa1 interaction (Hell et al., 2001; Stuart, 2002). Using a number of biochemical approaches, we provide evidence here for the association of Oxa1 with the mitochondrial large ribosomal subunit. Cross-linking data presented here indicate that a subunit of the large ribosomal subunit, Mrp20 (homolog of ribosomal subunit L23) (Fearon and Mason, 1992), is found in close proximity to Oxa1. Our current data suggest that the C-terminal region of Oxa1 supports the ribosome–Oxa1 interaction. Yeast cells harboring a C-terminal-truncated form of Oxa1 have compromised respiratory function and display a temperature-sensitive growth phenotype on non-fermentable carbon sources. Furthermore, the membrane insertion of the Cox2 and Cox1 protein is affected in the absence of the C-terminal region of Oxa1. Taken together, our data demonstrate that the basic C-terminal region of Oxa1, although not essential, plays a critical role in the function of Oxa1 as a membrane protein insertion machinery.
Results
Oxa1 and the large ribosomal subunit co-segregate on sucrose gradients
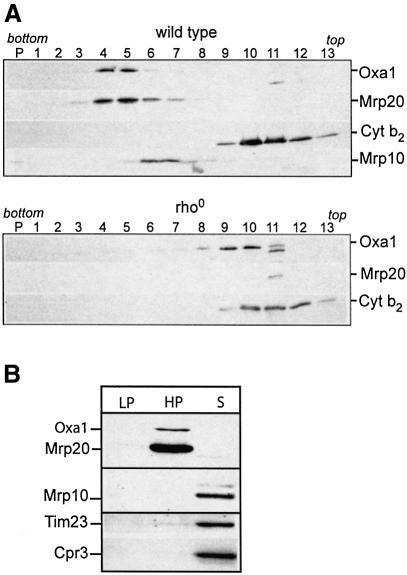
A physical interaction between Oxa1 and the mitochondrial ribosomes was analyzed using sucrose gradient centrifugation. Wild-type mitochondrial membranes were solubilized in digitonin and analyzed by sucrose gradient centrifugation (Figure 1A, upper panel). The sucrose gradient behavior of the large and small ribosomal subunits was monitored by the immunological detection of protein components of these complexes, the Mrp20 and Mrp10 proteins, respectively (Fearon and Mason, 1992; Jin et al., 1997). Western blot analysis indicated that Oxa1 from wild-type mitochondria co-fractionated in the gradient with Mrp20, towards the bottom of the gradient, consistent with the idea that the Oxa1 and ribosomal complexes may be interacting (Figure 1A, upper panel, fractions 4 and 5). Mrp10, a component of the small ribosomal subunit, did not co-fractionate with Mrp20, but was recovered in later fractions (Figure 1A, upper panel, fractions 6 and 7). The separation of Mrp20 and Mrp10 in distinct fractions indicates that the large and small ribosomal subunits did not remain together during these lysis and/or centrifugation conditions.
Fig. 1. Oxa1 solubilized from wild-type mitochondria sediments with the mitochondrial ribosomes. (A) Wild-type (upper panel) and rho0 (lower panel) mitochondria were solubilized with digitonin lysis buffer and were fractionated by a linear sucrose gradient (see Materials and methods). Fractions were analyzed by SDS–PAGE and western blotting. (B) Wild-type mitochondria were solubilized in an EDTA-containing octylglucoside lysis buffer and subjected to a clarifying spin. The resulting pellet (LP) contains non-solubilized material. The solubilized proteins were then subjected to an ultracentrifugation step, resulting in a high-speed pellet fraction (HP) and supernatant (S). Samples were analyzed by SDS–PAGE and western blotting. (A and B) Immunodecoration with antibodies specific for Oxa1, Mrp20, Mrp10, cytochrome b2 (Cyt b2), Tim23 and Cpr3 was performed, as indicated.
In further support of an Oxa1–ribosome interaction, the sedimentation behavior of Oxa1 solubilized from mitochondria lacking assembled ribosomes, i.e. rho0 mitochondria, was analyzed. Rho0 yeast strains lack mitochondrial DNA (mtDNA). As both the 15S and 21S rRNA of the mitochondrial large subunit are coded for by the mtDNA (Borst and Grivell, 1978), functional ribosomes fail to assemble in rho0 mitochondria. When solubilized from rho0 mitochondria, Oxa1 was recovered towards the top half of the gradient (Figure 1A, lower panel, fractions 9 and 10), a sedimentation behavior that was clearly different from the wild-type control sample (Figure 1A). The Mrp20 levels are reduced but detectable in the rho0 mitochondria, due to its turnover in the absence of ribosome assembly (Fearon and Mason, 1992). The recovery of Mrp20 towards the top of the sucrose gradient following solubilization of the rho0 mitochondria is also consistent with the lack of its assembly into the large ribosomal subunit (Figure 1A, lower panel). The levels of Mrp10 were so strongly reduced in rho0 mitochondria that they could not be detected in sucrose gradient fractions (data not shown). As expected, the sucrose gradient behavior of the nuclear-encoded control protein, cytochrome b2, was not changed between the wild-type and rho0 mitochondria (Figure 1A).
Thus the sucrose gradient behavior of Oxa1 is influenced by the presence or absence of the ribosomes, consistent with the idea that these two complexes may be interacting.
Oxa1 associates with the large ribosomal subunit
The observed co-sedimentation behavior of Oxa1 and Mrp20 on the sucrose gradients indicates that Oxa1 may interact with the large ribosomal subunit. We therefore analyzed the sedimentation behavior of Oxa1 following lysis of mitochondria under conditions where the large and small ribosomal subunits do not remain associated during lysis, i.e. in the absence of Mg2+, by adding EDTA (Figure 1B). When wild-type mitochondria were lysed with octylglucoside in the presence of EDTA and subjected to a clarifying spin, Oxa1 was solubilized from the membranes, as indicated by its absence from the non-solubilized pellet fraction (Figure 1B, LP). When the clarified supernatant was then subjected to an ultracentrifugation step, the solubilized Oxa1 was recovered in the high-speed pellet fraction (Figure 1B, HP). Analysis of the high-speed pellet fraction confirmed that it contained the large ribosomal subunit, as indicated by the presence of Mrp20. The small ribosomal subunit was recovered in the supernatant fraction, as indicated by the presence of Mrp10, reflecting the instability of the assembled ribosome in the absence of Mg2+. Control inner membrane protein Tim23 and a matrix soluble protein cyclophilin Cpr3 were recovered in the supernatant fraction, thus indicating the extent of the mitochondrial lysis by the octylglucoside treatment.
Taken together, we conclude that Oxa1 can be recovered in a pellet fraction together with the large ribosomal subunit, when solubilized from the mitochondrial membrane in the presence of EDTA.
Oxa1 and Mrp20, an L23 homolog, are in close proximity to each other
Mrp20 is a component of the large ribosomal subunit and homologous to the L23 ribosomal subunit (Fearon and Mason, 1992). Analysis of the crystal structure of the 50S ribosomal subunit from Haloarcula marismortui indicates that L23 is located next to the exit tunnel for nascent polypeptide chains (Ban et al., 2000; Nissen et al., 2000). Given the known interaction of nascent chains with Oxa1 and the observed co-sedimentation of Oxa1 with the large ribosomal subunit, we analyzed whether Oxa1 and Mrp20/L23 at the polypeptide exit channel are in close proximity to each other.
Using a chemical cross-linking approach with isolated mitochondria, we tested whether Mrp20 could be cross-linked to a protein whose molecular mass corresponds to Oxa1. Oxa1 was expressed in yeast either as an authentic Oxa1 (Oxa1, 36 kDa) or as a C-terminally histidine-tagged derivative (Oxa1His, 38 kDa) (see Materials and methods). Isolated mitochondria haboring the wild-type or the His-tagged Oxa1 derivative were used in the cross-linking analysis (Figure 2A). In the presence of the amino-specific cross-linking reagent, disuccinimidyl glutarate (DSG), Mrp20 (33 kDa) formed two dominant cross-linked adducts, both in the range of 66–70 kDa (Figure 2A, left panel). When cross-linking was performed in mitochondria harboring the Oxa1His derivative (12 amino acids longer than authentic Oxa1), the size of the larger adduct increased accordingly (∼72 kDa) (Figure 2A, right panel), thus indicating it to be an Mrp20–Oxa1 adduct. Furthermore, following hypotonic swelling (disrupts outer membrane, leaving inner membrane intact), the larger Mrp20 adduct was accessible to exogenously added proteinase K and gave rise to a proteolytic fragment, whose mass was ∼57 kDa (Figure 2B). The reduction in the size of the cross-linked adduct corresponds to the proteolytic cleavage of the N-terminal region of Oxa1 (∼90 residues) which is exposed to the protease in the intermembrane space (see Figure 3A for topology model of Oxa1).
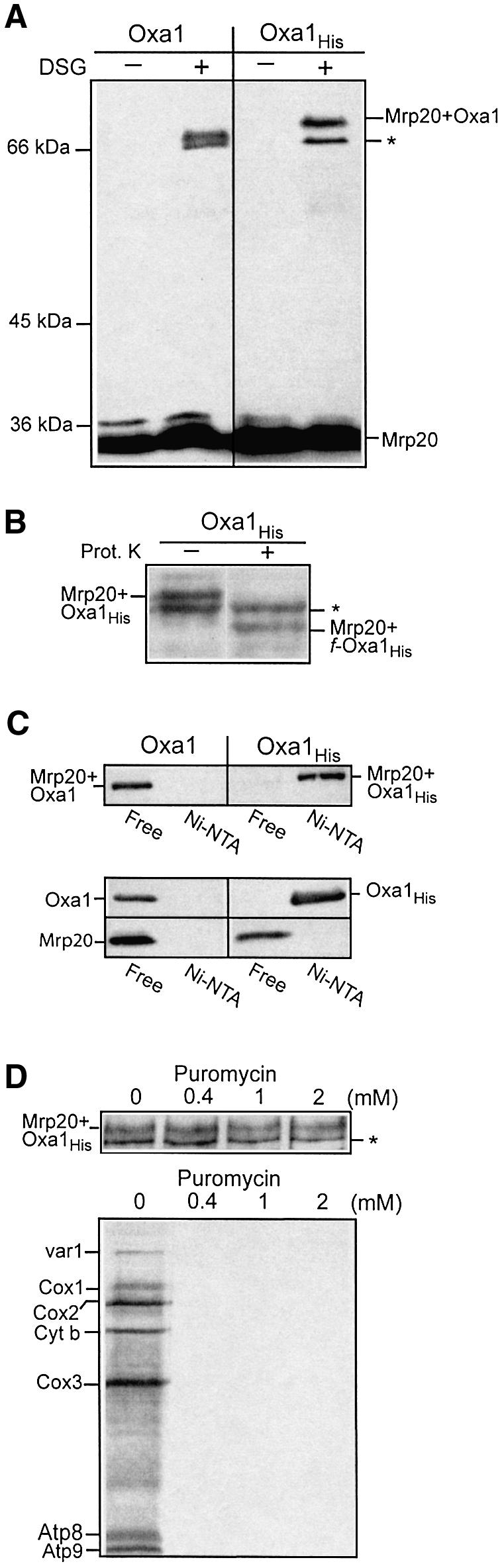
Fig. 2. Cross-linking of Oxa1 with the large ribosomal subunit Mrp20. (A) Mitochondria harboring expressed wild-type or histidine-tagged Oxa1 (Oxa1 and Oxa1His, respectively) were incubated in the presence or absence of the cross-linking reagent DSG (0.3 mM), as indicated. Following quenching, mitochondria were reisolated and analyzed by SDS–PAGE, western blotting and immunodecoration with Mrp20 monoclonal antibodies. The asterisk indicates an unknown Mrp20 adduct of ∼66 kDa. (B) Oxa1His mitochondria were hypotonically swollen, subjected to cross-linking with DSG, followed by proteinase K treatment (12.5 µg/ml), as indicated. Samples were analyzed further as in (A). (C) Mitochondria harboring expressed Oxa1 or Oxa1His derivatives treated with DSG (0.5 mM) (upper panel) or untreated (lower panel) were lysed in Triton X-100-containing buffer, and incubated with Ni-NTA beads. The Ni-NTA-bound (Ni-NTA) material and 20% of the free (non-bound) material were analyzed by SDS–PAGE and western blotting. Immunodecoration was performed with both Mrp20 and Oxa1 antiserum, as indicated. The Mrp20–Oxa1 and Mrp20–Oxa1His cross-linked adducts are shown in the upper panel; monomeric Oxa1 (Oxa1His) and Mrp20 are shown in the lower panel. (D) Mitochondria harboring the Oxa1His derivative were incubated in translation buffer with puromycin at the concentrations indicated prior to either cross-linking with DSG (upper panel) or in organello translation following the addition of [35S]methionine (15 min at 25°C) (lower panel). The cross-linked samples were analyzed further as described in (A). Abbreviations: cytochrome oxidase subunits 1, 2 and 3; Cox1, Cox2 and Cox3, respectively; cytochrome b, Cyt. b; subunits 6, 8 and 9 of the Fo-ATPase, Atp6, Atp8 and Atp9.
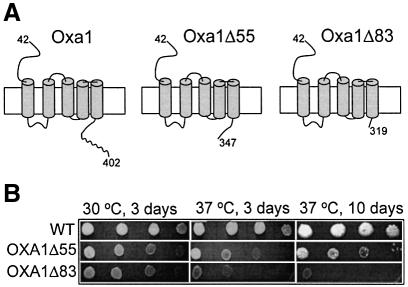
Fig. 3. Oxa1 C-terminal deletion strains display a respiratory phenotype. (A) Oxa1 (amino acids 42–402) spans the inner membrane five times. (Note the mitochondrial targeting signal is contained within residues 1–41, and undergoes proteolytic removal following import.) Oxa1Δ55 and Oxa1Δ83 bear deletions of 55 and 83 amino acid residues in the matrix-located C-terminal region of Oxa1. The putative coiled-coil structure (residues 350–400) is indicated by a curved line. (B) A dilution series of OXA1Δ55 and OXA1Δ83 strains and the corresponding isogenic wild-type (WT) was generated by serially diluting this suspension 10-fold each time. A 2 µl aliquot of each of the resulting dilutions was spotted onto YP-glycerol plates and incubated at 30 or 37°C.
The larger Mrp20 adduct was verified as an Mrp20–Oxa1 adduct in the following experiment. Cross-linking with DSG was performed in the mitochondria harboring the Oxa1His derivative, and Oxa1His and its cross-linked adducts were purified by Ni-NTA chromatography following detergent solubilization of the mitochondrial membranes (Figure 2C, upper panel). Probing with the Mrp20 antibody indicated that the Mrp20–Oxa1His adduct could be recovered on the Ni-NTA beads. The recovery of the Mrp20 adduct on the Ni-NTA beads was specific for its association with the His-tagged Oxa1 derivative. When cross-linking was performed in mitochondria harboring the wild-type Oxa1 (i.e. non-tagged version), the Mrp20–Oxa1 adduct was not recovered on the Ni-NTA beads (Figure 2C, upper panel). Control experiments show that the recovery of Oxa1His on the Ni-NTA beads is specific for the presence of the C-terminal His tag on the Oxa1 protein, as the non-tagged Oxa1 protein did not bind to the beads, but rather was recovered in the supernatant fraction (Figure 2C, lower panel). Furthermore, in the absence of cross-linking, the monomeric Mrp20 was not recovered on the Ni-NTA beads, thus indicating that the recovery of the Mrp20–Oxa1His adduct on the beads is not due to Mrp20 binding to the beads but rather is specific for the cross-linking of Mrp20 to the Oxa1His protein (Figure 2C, lower panel).
The ability of Mrp20 to become cross-linked to Oxa1 was not affected by pre-treatment with puromycin (Figure 2D, upper panel). Puromycin is a protein synthesis inhibitor and causes premature release of the nascent chain from the associated ribosome. Thus it appears that Oxa1 cross-linking to Mrp20 can occur in the absence of associated nascent chains, i.e. in a translation-independent fashion. The ability of puromycin treatment to inhibit protein translation effectively under the conditions used for the cross-linking experiment is shown (Figure 2D, lower panel).
Taking these results together, we conclude that the matrix-located Mrp20, a component of the large ribosomal subunit, can be cross-linked to the membrane-spanning Oxa1 protein, indicating a close proximity between the ribosomal and Oxa1 complexes.
Oxa1ΔC mutants display impaired ability to grow on non-fermentable carbon sources
Oxa1, an inner membrane protein, interacts with the matrix-localized ribosome. We postulated that the matrix-exposed region of Oxa1 (amino acids 310–402, Figure 3A) may be involved in supporting the ribosome–Oxa1 interaction, possibly to enhance the efficiency of membrane insertion events. We therefore created yeast mutant strains (OXA1ΔC), which bear deletions in the region of the chromosomal OXA1 gene encoding the C-terminal amino acid residues of the Oxa1 protein. Hence the expression of the truncated Oxa1 derivatives, like that of the authentic Oxa1, remains under the control of the endogenous Oxa1 promoter. Two different gene deletion strains, termed OXA1Δ55 and OXA1Δ83, were created where the resulting truncated genes encode Oxa1 derivatives bearing a C-terminal deletion of either 55 or 83 amino acids, Oxa1Δ55 (coiled-coil motif deleted) and Oxa1Δ83, respectively (Figure 3A).
When compared with wild-type cells, the two OXA1ΔC mutant strains grew slower on glycerol medium at 30°C, on plates (Figure 3B) and in liquid medium (not shown). Furthermore, the OXA1ΔC strains displayed a temperature-sensitive growth phenotype at the elevated temperature of 37°C. The most severe growth phenotype was observed with the Oxa1Δ83 yeast strain. Growth of the OXA1ΔC strains at 30°C was in contrast to the Δoxa1-null mutant (results not shown) (Bauer et al., 1994; Bonnefoy et al., 1994), thus indicating that the Oxa1ΔC derivatives do, however, exhibit some, albeit reduced, functional capacity. We conclude therefore that the C-terminal region of Oxa1, although important, is not essential for function of the yeast Oxa1. Note that the growth phenotype of the OXA1ΔC mutants reported here does not appear to be as severe as those reported for similar truncations in the accompanying manuscript by Szyrach et al. We feel the observed differences may be due to the different genetic backgrounds used to construct the Oxa1 mutants.
Association of the ribosome with Oxa1 requires the C-terminal region of Oxa1
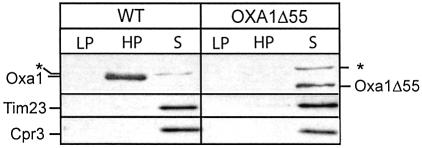
We analyzed whether the presence of the C-terminal matrix-exposed region of Oxa1 was required for the association of the ribosomes with Oxa1 following detergent lysis. Mitochondria harboring the Oxa1Δ55 derivative were solubilized with octylglucoside and the ability of Oxa1Δ55 to be recovered in the ribosomal high-speed pellet was analyzed. Following lysis in the presence of EDTA, the C-terminal truncated derivatives Oxa1Δ55 (Figure 4) and Oxa1Δ83 (results not shown) were not recovered in the ribosomal pellet fraction. Rather, in contrast to their wild-type counterpart, the Oxa1ΔC derivatives were recovered in the high-speed supernatant fraction. We conclude that the interaction of Oxa1 with the mitochondrial ribosome under these conditions is supported by the presence of the matrix-exposed C-terminal region of Oxa1.
Fig. 4. The C-terminal region of Oxa1 supports the Oxa1–ribosome interaction. Octylglucoside lysis of wild-type and Oxa1Δ55 mitochondria was performed as described in Figure 2B. Abbreviations are as in Figure 2B. The asterisk indicates a protein that cross-reacts with the antibody directed against the N-terminal region of Oxa1. Immunodecoration of control proteins Tim23 and Cpr3 was performed.
Assembly of the cytochrome oxidase complex is affected in the OXA1ΔC mutant mitochondria
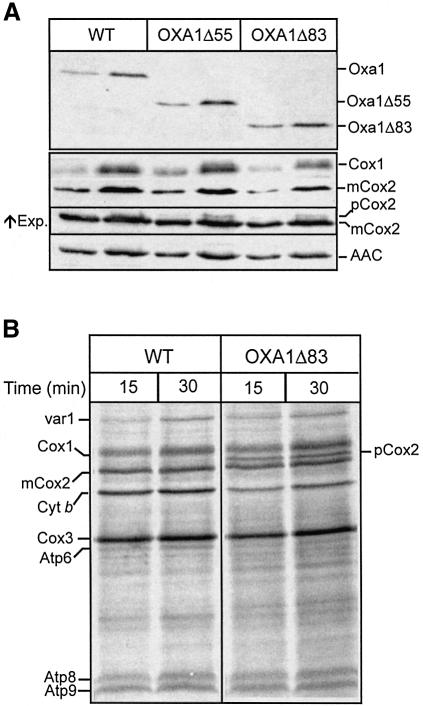
Mitochondria were isolated from the OXA1Δ55 and OXA1Δ83 yeast strains, which had been grown in lactate/galactose medium at 30°C. The presence of the C-terminally truncated Oxa1Δ55 and Oxa1Δ83 derivatives in the isolated mitochondria was detected using an N-terminal-specific Oxa1 antibody (Figure 5A). The steady-state levels of Cox1 and Cox2 in the mutant mitochondria were analyzed by western blot analysis followed by immunodecoration with subunit-specific antiserum. The levels of Cox1 in the OXA1Δ55 and OXA1Δ83 mitochondria were found to be slightly reduced from those observed in wild-type control mitochondria. The presence of Cox1 in the OXA1Δ83 mitochondria would be consistent with the observed growth of the strains on non-fermentable media. The levels of Cox2 did not appear to be strongly affected in either of the OXA1ΔC mutant mitochondria. A small amount of pCox2 species was observed, however, in the mutants, in contrast to the wild-type control mitochondria. The presence of the mature form of Cox2 in the OXA1Δ83 mitochondria is in contrast to the Δoxa1-null mutant where only pCox2 is detected (at strongly reduced levels, results not shown), and is indicative of a partial activity of the truncated Oxa1 proteins.
Fig. 5. Analysis of OXA1Δ55 and OXA1Δ83 mitochondria. (A) Mitochondria (20 and 50 µg of protein) were subjected to SDS–PAGE and analyzed by western blotting for the presence of the Oxa1 (using an N-terminal-specific antibody), Cox1 and Cox2 proteins. An increased exposure of the Cox2 blot is shown (indicated by ↑Exp.), to document the accumulation of pCox2 species in the mutant mitochondria. The blots were also immunodecorated with an ADP/ATP carrier protein antibody (AAC). (B) Mitochondria were pre-incubated at 37°C for 10 min and in organello translation was then monitored at 25°C for the times indicated, following the addition of [35S]methionine. Abbreviations used are as in Figure 2D.
We further analyzed the nature of the defect in the Oxa1ΔC mutants and present here the further characterization of the Oxa1Δ83 mitochondria only, as this strain exhibited the strongest growth phenotype. The levels of newly synthesized Cox1, Cox2 and other mitochondrially encoded proteins were next determined by performing in organello translation in the presence of [35S]methionine using isolated wild-type and OXA1Δ83 mitochondria (Figure 5B). The levels of newly synthesized mtDNA gene products in the OXA1Δ83 mitochondria were similar to those obtained in the wild-type control. A slight reduction in the levels of cytochrome b relative to the wild-type control was observed. An accumulation of pCox2 was observed in the OXA1Δ83 mitochondria, in contrast to the wild-type control, thus indicating a possible translocation defect of the pCox2 in the OXA1Δ83 mitochondria. Furthermore, the OXA1Δ83 mitochondria displayed the capacity to synthesize wild-type levels of Cox1 (Figure 5B). Similar observations were made following analysis of in organello translation in the OXA1Δ55 mitochondria (results not shown).
In summary, truncation of the C-terminal region of Oxa1 does not significantly affect the ability of the mitochondria to synthesize Cox1 and Cox2 subunits of the cytochrome oxidase complex. Rather, the observed accumulation of the pCox2 species would indicate that the Oxa1ΔC proteins display an impaired capacity to support insertion of these proteins into the inner membrane, thus hindering their efficient assembly into a functional and stable cytochrome oxidase complex, in particular at the elevated temperature of 37°C.
The insertion activity of Oxa1 is impaired in the OXA1Δ83 mitochondria
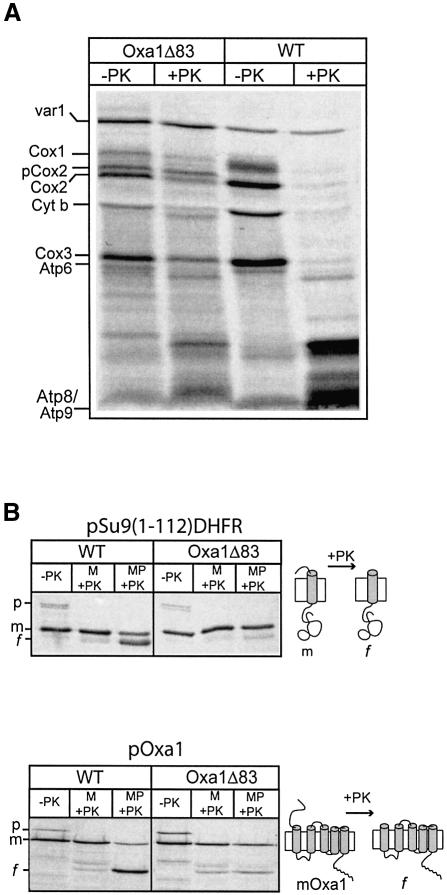
The membrane insertion of newly synthesized Cox1 and Cox2 proteins was directly investigated following translation in mitoplasts generated by hypotonic swelling of mitochondria isolated from the OXA1Δ83 mutant strains. Mitochondrial protein synthesis in the presence of [35S]methionine was performed in these mitoplasts at 25°C (Figure 6A). The insertion of the newly synthesized Cox1 and Cox2 proteins into the inner membrane was then assessed by accessibility of their intermembrane space-localized segments to exogenously added proteinase K, as previously described (Hell et al., 2001). In wild-type mitoplasts, the newly synthesized Cox1 and Cox2 proteins were degraded by the added protease, confirming the accessibility of the intermembrane space-exposed regions of the proteins to the protease (Figure 6A), as had been reported previously (Hell et al., 2001). The radiolabeled precursor species of Cox2 (pCox2) synthesized in the OXA1Δ83 mitochondria was localized in the mitochondrial matrix, where it was inaccessible to the added protease (Figure 6A). A similar inhibition of export of pCox2 was observed in the OXA1Δ55 mitoplasts (data not shown). The membrane insertion of the newly synthesized Cox1 protein was also affected in the OXA1Δ83 mitoplasts, where ∼50% of the Cox1 species was inaccessible to the added protease (Figure 6A). A partial inhibition of Cox1 insertion (∼30%) was also observed following translation in OXA1Δ55 mitoplasts (data not shown).
Fig. 6. Oxa1Δ83 exhibits compromised insertion activity for both mitochondrially and nuclear-encoded substrates. (A) Mitoplasts prepared from OXA1Δ83 and wild-type (WT) mitochondria were incubated in translation buffer supplemented with 2 mM NADH for 5 min at 37°C, prior to the addition of [35S]methionine. Following translation at 25°C for 20 min, samples were divided and were either mock treated or treated with proteinase K, as indicated. Abbreviations: see Figure 2D. (B) Radiolabeled pSu9(1–112)-DHFR (upper panel) and pOxa1 (lower panel) were imported into isolated wild-type or OXA1Δ83 mitochondria for 15 min at 25°C in the presence of 2 mM NADH. Following import, samples were divided and either mock treated or subjected to proteinase K (PK) treatment under swelling (mitoplasts, MP) or non-swelling (mitochondria, M) conditions, as indicated. p, precursor; m, mature form; f, specific fragment of imported protein generated by the degradation of the intermembrane space (IMS)-exposed segment of mSu9(1–112)-DHFR or Oxa1 by proteinase K under the swelling conditions. The swelling efficiency of both types of mitochondria (as judged by release of cytochrome c peroxidase) was similar and in the range of 90–95% efficient (results not shown).
We also analyzed the ability of the Oxa1Δ55 and Oxa1Δ83 proteins to mediate the insertion of post-translational substrates of Oxa1, i.e. those sorted via Oxa1 following their initial import into mitochondria. Radiolabeled pSu9(1–112)-DHFR, a previously characterized Oxa1-dependent substrate (Rojo et al., 1995; Hell et al., 1998), was imported into isolated OXA1Δ83 and wild-type control mitochondria (Figure 6B). Insertion of this protein into the inner membrane from the matrix to obtain an Nout–Cin orientation was analyzed by proteinase K accessibility studies, as previously published (Rojo et al., 1995). Export of the N-terminal tail of this protein was inhibited in the OXA1Δ83 mitochondria, and the non-exported protein was observed to accumulate in the matrix (Figure 6B). A similar inhibition of the export of pSu9(1–112)-DHFR was observed following import into the OXA1Δ55 mitochondria (data not shown).
Oxa1 is a nuclear-encoded protein, which is sorted to an Nout–Cin orientation in the inner membrane following complete import into the mitochondrial matrix. Sorting of newly imported Oxa1 from the matrix into the inner membrane is supported by the assembled Oxa1 complex (Hell et al., 1998). The import and sorting of newly synthesized radiolabeled Oxa1 was also analyzed in the OXA1Δ83 mitochondria. The insertion of Oxa1 into the inner membrane from the matrix was judged by the accessibility of the N-terminal hydrophilic tail to proteinase K in hypotonically swollen mitochondria following import. The insertion efficiency of radiolabeled Oxa1 was reduced significantly in the OXA1Δ83 mitochondria (Figure 6B). A reduction in the export efficiency of radiolabeled Oxa1 was also observed in the OXA1Δ55 mitochondria (results not shown).
Thus it appears that the mitochondria isolated from the OXA1ΔC mutants display a reduced capacity to facilitate the co- and in particular the post-translational membrane insertion of substrate proteins. These data indicate that the function of the C-terminal region of Oxa1 may not be limited to interacting with the translating ribosome and mediating the insertion of nascent chain substrates.
Discussion
Oxa1 is a component of a general insertion site in the mitochondrial inner membrane, which facilitates the integration of both mitochondrially and nuclear-encoded substrate proteins (Stuart, 2002). The yeast Oxa1 is an integral inner membrane protein and contains a C-terminal segment of ∼92 amino acid residues that protrudes into the mitochondrial matrix. Oxa1 directly interacts with its substrate proteins while supporting their insertion into the membrane. The interaction of Oxa1 with its mitochondrially encoded substrate proteins occurs early during their synthesis as nascent chains and in a manner that is supported by the presence of the associated ribosome. A close coupling of the ribosome undergoing translation and the Oxa1 complex facilitating insertion was proposed previously (Hell et al., 2001; Stuart, 2002). Evidence for a physical association between Oxa1 and the ribosome, however, was lacking to date.
The data presented here indicate that the Oxa1 complex may be found in physical association with the ribosomal complex. Moreover, our results are consistent with an interaction of Oxa1 with the large ribosomal subunit. First, Oxa1 was observed to co-purify with the large ribosomal subunit in the presence of EDTA, conditions which do not maintain the association of the large and small ribosomal subunits, a Mg2+-dependent process. Secondly, cross-linking between Oxa1 and the large ribosomal subunit Mrp20 was obtained. The observation that Oxa1 associates with the large ribosomal subunit is consistent with our previous findings demonstrating that nascent polypeptides associate with Oxa1 during their synthesis (Hell et al., 1998, 2001). Furthermore, our data indicate that cross-linking of Mrp20 to Oxa1 can occur in a nascent chain-independent fashion. This observation would suggest that the association of the Oxa1 complex with the translational machinery is not mediated through the nascent chains, i.e. the substrates of Oxa1.
Mrp20 is homologous to the L23 ribosomal subunit, a component of the large ribosomal subunit (Fearon and Mason, 1992). The crystal structure of the 50S subunit from H.marismortui shows that L23 is located next to the exit tunnel for nascent polypeptides (Ban et al., 2000; Nissen et al., 2000). Thus it would appear that Oxa1, the site of membrane insertion, is maintained in close proximity to the region of the ribosomes where nascent chains emerge from the ribosomal large subunit. A close coupling of Oxa1 and the ribosome in this manner may enhance the co-translational insertion steps and minimize the possibility of hydrophobic substrates being synthesized and aggregating in the matrix. The close association of Mrp20/L23 with Oxa1 may have other functional significance. In bacteria, the ribosomal L23 subunit has been shown to interact with the chaperone trigger factor and components of the signal recognition particle (SRP) (Kramer et al., 2002; Gu et al., 2003; Ullers et al., 2003). In eukaryotic cells, the cytoplasmic homolog of L23, L23a, has been shown to physically interact with SRP also during signal peptide recognition (Pool et al., 2002). In addition, L23a was identified as a region of the ribosome that participates in forming a connection between the large subunit of the ribosome and the Sec61 channel of the endoplasmic reticulum (Morgan et al., 2002). Hence the L23 (L23a) subunit has been postulated to play a central role in directly linking protein synthesis to targeting and membrane insertion events in both prokaryotic and eukaryotic cytosolic translational systems (Kramer et al., 2002; Ullers et al., 2003). Due to the position of the Mrp20/L23 protein in the ribosome and the available data indicating the role of L23 proteins in membrane protein targeting and insertion events, it is possible that Mrp20 may also play an important role in coordinating mitochondrial protein translation and Oxa1-dependent protein insertion events. Although our cross-linking studies of Oxa1 with Mrp20 indicate a close physical relationship between these two proteins, it is unclear presently whether these two proteins are directly interacting with each other.
Analysis of the Oxa1Δ55 and Oxa1Δ83 proteins has indicated that elements contained within the C-terminal region of Oxa1 appear to support the ribosome–Oxa1 interaction. Furthermore, these Oxa1ΔC derivatives display an impaired ability to support the insertion of its substrates synthesized on the mitochondrial ribosomes. The membrane insertion of newly synthesized Cox2 and Cox1, both mitochondrially encoded substrates, was affected in the OXA1ΔC mitochondria. Mitochondrial respiration thus was compromised, as evidenced by the reduced ability of the OXA1ΔC yeast strain to grow on non-fermentable carbon sources, in particular at the elevated temperature of 37°C. The impaired ability of Oxa1ΔC proteins to support the insertion of mitochondrially encoded substrates might be accounted for by a reduction in the efficiency of ribosome binding to Oxa1 or stability of the ribosome–Oxa1 complex. Interestingly, the Oxa1-faciliated insertion of the post-translational substrates, pSu9(1–112)-DHFR and Oxa1 itself, was also inhibited in the OXA1ΔC mitochondria. Thus it appears that the presence of the C-terminal region of Oxa1 may be required for efficient insertion of both co- and post-translationally inserted substrate proteins. It is possible that the reduced efficiency of the Oxa1-mediated insertion of these post-translational substrates may be due to a reduced membrane potential in the OXA1ΔC mitochondria. The mitochondria used in our analysis were, however, isolated from cultures grown at 30°C, conditions where the respiratory defect of the OXAΔC mutants is weak and the levels of the COX complex in the isolated mitochondria are only slightly reduced. Furthermore, a similar export defect of post-translational substrates was observed in the OXA1Δ55 mitochondria, where the respiratory phenotype was weaker than that observed for the OXA1Δ83 mitochondria. Finally, the in vitro import of radiolabeled precursor proteins (a membrane potential-dependent step) into the OXA1ΔC mitochondria was not affected. Although we do not wish to rule out a possible effect of a membrane potential, we consider it also possible that the C-terminal region of Oxa1 may directly influence the ability of Oxa1 to recognize and/or insert post-translational substrates.
What features does the C-terminal segment of Oxa1 have which renders it important for the insertion activity of the yeast Oxa1 protein? The alignment of sequences from known mitochondrial Oxa1 homologs has indicated that the sequence similarities are mainly limited to the regions of the proteins encompassed by the five transmembrane segments (Yen et al., 2001). In contrast, the N- and C-terminal regions appear to display little primary sequence similarities between homologs (Yen et al., 2001). Despite this apparent lack of primary sequence similarity, the basic nature of the C-terminal matrix-exposed segment is a feature which is conserved throughout the known mitochondrial Oxa1 homologs (pI range between 9.7 and 11.9). In addition, a stretch of amino acid residues (∼350–402 for yeast Oxa1) contained within this C-terminal region display the potential to form a coiled-coil structure. Although present in a number of mitochondrial homologs from other organisms (e.g. mouse, human and Neurospora crassa), the coiled-coil region may not represent a universal feature of all mitochondrial Oxa1 homologs. The current database sequence information indicates that Oxa1 homologs from Schizosaccharomyces pombe (Oxa1-2) and Caenorhabditis elegans do not contain this motif, and a second S.pombe homolog (Oxa1-1) and the Drosophila melanogaster homologs display only weak potentials to form one. Interestingly, it was reported recently that deletion of the C-terminal putative coiled-coil region from the N.crassa Oxa1 homolog did not appear to have any affect on growth (Nargang et al., 2002). We show here that the deletion of the coiled-coil region of yeast Oxa1 (Oxa1Δ55), however, did result in a growth phenotype, in particular at 37°C, and an impaired insertion activity of Oxa1. The more extensive deletion of the C-terminal region, Oxa1Δ83 (where 83 out of 92 matrix-exposed residues were removed), did result in a stronger growth phenotype. Taking this observation together with the fact that the potential to form a coiled-coil is not conserved in all mitochondrial Oxa1 proteins, it is possible, therefore, that the phenotype of the OXA1ΔC strains reported here is caused primarily by removal of important positively charged residues contained within the C-terminal region.
Does the C-terminal region of Oxa1 constitute an essential ribosome-binding site? Both our cross-linking analysis and detergent lysis–sedimentation experiments support a physical interaction between the Oxa1 complex and the large ribosomal subunit. The C-terminal region of Oxa1 supports this ribosome interaction, at least under the detergent lysis conditions used here. Furthermore, we have not yet observed cross-links between the Oxa1ΔC and Mrp20 proteins (results not shown). However, our data indicate that the C-terminal region of Oxa1 plays an important, but non-essential role in the insertion of mitochondrially encoded proteins. It is possible that other matrix-exposed regions of Oxa1, or components associated with Oxa1, may cooperate with the C-terminal region of Oxa1 to bind and/or stabilize the ribosome. Furthermore, it is conceivable that membrane-bound translational activators (Fiori et al., 2003; Naithani et al., 2003) may also cooperate and direct the targeting of ribosomes to the Oxa1 membrane insertion site. Aside from this, it is currently unknown whether Oxa1 itself directly interacts with the large ribosomal subunit or if other component(s), interacting either with Oxa1 in the membrane or with the ribosome/nascent chain in the matrix, directly mediate this interaction.
In addition, our data indicate that the post-translational sorting of well-characterized Oxa1-dependent substrates was strongly impaired in both the OXA1Δ83 and OXA1Δ55 mitochondria. Thus our data would argue that the function of the C-terminal region of Oxa1 is not limited to the coordination of ribosome binding and nascent chain insertion. It is possible that the C-terminal region of Oxa1 may recognize and bind a component involved in targeting of substrates, which would be common to both the co- and post-translational targeting pathways. Although there is no evidence to date for the presence of an SRP in yeast mitochondria, it is possible that a component may exist which acts in an SRP-like fashion, targeting both ribosome-associated nascent chains and fully synthesized proteins to the Oxa1 insertion site. In chloroplasts, SRP has been shown to operate in both post- and co-translational targeting pathways to the thylakoid membrane (Moore et al., 2000; Groves et al., 2001; Woolhead et al., 2001). It has been proposed that chloroplast SRP may act as a chaperone to maintain post-translational substrates in a competent form for subsequent membrane insertion (Eichacker and Henry, 2001). We currently are investigating whether the interaction between Oxa1 and the ribosome is a direct one.
Materials and methods
Yeast strains
Yeast strains used in this study were wild-type W303-1A (Mat a, leu2, trp1, ura3, his3, ade2), rho0 W303-1A (Mat a, leu2, trp1, ura3, his3, ade2) and the oxa1-null mutant, Δoxa1 (W303-1A, leu2, trp1, ura3, ade2, OXA1::HIS3) (Hell et al., 1998). Construction of the strains bearing truncations in the C-terminal region of Oxa1 protein (OXA1ΔC) was performed by the introduction of the kanamycin resistance gene (KANr) into the OXA1 locus of wild-type cells, resulting in the introduction of a premature translational stop codon and a partial deletion of the OXA1 open reading frame in the 3′ end, essentially as described by Wach et al. (1994). Correct homologous recombination of the KANr gene at the OXA1 gene locus was verified by PCR analysis (results not shown).
Cross-linking of Mrp20 and affinity purification of Oxa1His
Mitochondria (240 µg of total protein) were suspended in 600 µl of SH buffer (0.6 M sorbitol, 20 mM HEPES-KOH pH 7.2) and cross-linked by DSG (0.33 or 0.5 mM, as indicated) for 30 min on ice. Excess DSG was quenched by adding 50 µl of glycine (1 M). Cross-linking analysis in the presence of puromycin was performed in translation buffer (Hell et al., 2001) (amino acids were omitted, however, as they quench the amino-specific cross-linking reagent).
For the Ni-NTA purification of Oxa1His and cross-linked adducts, mitochondria were solubilized in 200 µl of TNT buffer (1% Triton X-100, 300 mM NaCl, 60 mM Tris–HCl pH 7.4) for 5 min on ice. After a clarifying spin (20 860 g, 15 min at 4°C), the supernatants were incubated for 1 h at 4°C with the Ni-NTA beads (equilibrated in the TNT buffer containing 20 mM imidazole). The beads were washed three times with TNT-imidazole buffer and bound proteins were eluted with SDS sample buffer containing 5% (v/v) β-mercaptoethanol and 0.5 M imidazole.
Digitonin solubilization of mitochondria and sucrose gradient centrifugation
Mitochondria (300 µg of protein) were solubilized with 600 µl of lysis buffer [0.25% (w/v) digitonin, 20 mM HEPES-KOH pH 7.2, 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] for 30 min on ice. Following a clarifying spin (21 000 g, 15 min at 4°C), the supernatant was loaded on top of a linear 20–40% linear sucrose gradient (11 ml) (in 20 mM HEPES-KOH pH 7.2, 0.1% digitonin, 0.5 mM PMSF). Following centrifugation (26 000 r.p.m., Beckman: SW41 Ti rotor, Optima L90K ultracentrifuge, 16 h at 4°C), fractions (0.9 ml) were harvested from the bottom of the gradient. Samples were trichloroacetic acid precipitated and subjected to SDS–PAGE and western blot analysis.
Octylglucoside solubilization of Oxa1
Mitochondrial membranes (100 µg of protein) (wild-type, OXA1Δ55 or OXA1Δ83, as indicated) were solubilized with octylglucoside [(1.5% (w/v)] for 30 min on ice in lysis buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris–HCl pH 7.4). Lysed mitochondria initially were subjected to a clarifying spin (30 000 g, 15 min). The supernatant was then subjected to an ultracentrifugation step (227 000 g, 30 min, 4°C). The fractions were analyzed by SDS–PAGE and western blotting.
Expression of the Oxa1 and Oxa1His derivatives
Overexpression of Oxa1 in wild-type yeast cells was achieved using a yeast-integrating vector Yip351-GAL10-OXA1, where the OXA1 gene had been cloned downstream of the galactose-inducible GAL10 promoter. For the expression of Oxa1His, a DNA fragment encoding Oxa1His (an Oxa1 derivative bearing a His12 tag at the C-terminus) was used. The recombinant plasmids were integrated into the yeast genome of the oxa1-null mutant at the LEU2 locus following linearization with BstEII.
Miscellaneous
Mitochondria were isolated from cultures grown at 30°C in YP-0.5% lactate, 2% galactose medium. Isolation of mitochondria, in organello labeling with [35S]methionine, in vitro protein import of radiolabeled precursors and protease accessibility studies were performed as described in Hell et al. (2001).
Acknowledgments
Acknowledgements
We thank Dr Thomas Langer (Universität Köln) for the Yip351-GAL10-OXA1 plasmid and the N-terminal Oxa1 antibody, Dr Thomas L.Mason (University of Massachusetts, Amherst) for the Mrp20 monoclonal antibodies, Dr Alex Tzagoloff (University of Columbia, NY) for the rho0 yeast strain and Mrp10 and COX complex antibodies, and Drs Gail Waring and Pinfen Yang for their stimulating discussions. This research was supported by NSF grants MCB 0077961 and DBI 0100667 to R.A.S. M.M. and M.S. were supported by an NSF-REU award.
References
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Bauer M., Behrens,M., Esser,K., Michaelis,G. and Pratje,E. (1994) PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet., 245, 272–278. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Chalvet,F., Hamel,P., Slonimski,P.P. and Dujardin,G. (1994) OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol., 239, 201–212. [DOI] [PubMed] [Google Scholar]
- Borst P. and Grivell,L.A. (1978) The mitochondrial genome of yeast. Cell, 15, 705–723. [DOI] [PubMed] [Google Scholar]
- Eichacker L.A. and Henry,R. (2001) Function of a chloroplast SRP in thylakoid protein export. Biochim. Biophys Acta, 1541, 120–134. [DOI] [PubMed] [Google Scholar]
- Fearon K. and Mason,T.L. (1992) Structure and function of Mrp20 and Mrp49, the nuclear genes for two ribosomal proteins of the 54S subunit of the yeast mitochondrial ribosome. J. Biol. Chem., 267, 5162–5170. [PubMed] [Google Scholar]
- Fiori A., Mason,T.L. and Fox,T.D. (2003) Evidence that synthesis of the Saccharomyces cerevisiae mitochondrially encoded ribosomal protein Var1p may be membrane localized. Eukaryot. Cell, 2, 651–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves M.R., Mant,A., Kuhn,A., Koch,J., Dubel,S., Robinson,C. and Sinning,I. (2001) Functional characterization of recombinant chloroplast signal recognition particle. J. Biol. Chem. 276, 27778–27786. [DOI] [PubMed] [Google Scholar]
- Gu S.Q., Peske,F., Wieden,H.J., Rodnina,M.V. and Wintermeyer,W. (2003) The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA, 9, 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. and Fox,T.D. (1997) Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N- and C-termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell., 8, 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. and Fox,T.D. (1999) Mutations affecting a yeast mitochondrial inner membrane protein, Pnt1p, block export of a mitochondrially synthesized fusion protein from the matrix. Mol. Cell. Biol., 19, 6598–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Herrmann,J.M., Pratje,E., Neupert,W. and Stuart,R.A. (1997) Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett., 418, 367–370. [DOI] [PubMed] [Google Scholar]
- Hell K., Herrmann,J.M., Pratje,E., Neupert,W. and Stuart,R.A. (1998) Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl Acad. Sci. USA, 95, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Neupert,W. and Stuart,R.A. (2001) Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J., 20, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J.M., Koll,H., Cook,R.A., Neupert,W. and Stuart,R.A. (1995) Topogenesis of cytochrome oxidase subunit II: mechanisms of protein export from the mitochondrial matrix. J. Biol. Chem., 270, 27079–27086. [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Neupert,W. and Stuart,R.A. (1997) Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear encoded Oxa1p. EMBO J., 16, 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Myers,A.M. and Tzagoloff,A. (1997) Cloning and characterization of MRP10, a yeast gene coding for a mitochondrial ribosomal protein. Curr. Genet., 31, 228–234. [DOI] [PubMed] [Google Scholar]
- Kramer G., Rauch,T., Rist,W., Vorderwulbecke,S., Patzelt,H., Schulze-Specking,A., Ban,N., Deuerling,E. and Bukau,B. (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature, 419, 171–174. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Stuart,R.A, Henry,R. and Dalbey,R.E. (2003) The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol., 10, 510–516. [DOI] [PubMed] [Google Scholar]
- Luirink J., Samuelsson,T. and de Gier,J.W. (2001) YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett., 501, 1–5. [DOI] [PubMed] [Google Scholar]
- Morgan DG., Menetret,J.F., Neuhof,A., Rapoport,T.A. and Akey,C.W. (2002) Structure of the mammalian ribosome–channel complex at 17 Å resolution. J. Mol. Biol., 324, 871–886. [DOI] [PubMed] [Google Scholar]
- Moore M., Harrison,S., Peterson,E.C. and Henry,R. (2000) Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem., 275, 1529–1532. [DOI] [PubMed] [Google Scholar]
- Naithani S., Saracco,S.A., Butler,C.A. and Fox,T.D. (2003) Interactions among COX1, COX2 and COX3 mRNA specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell., 14, 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargang F.E., Preuss,M., Neupert,W. and Herrmann,J.M. (2002) The Oxa1 protein forms a homo-oligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J. Biol. Chem., 277, 12846–12853. [DOI] [PubMed] [Google Scholar]
- Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Nature, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Fox,T.D. and Walter,P. (1993) A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science, 262, 1997–2003. [DOI] [PubMed] [Google Scholar]
- Pool M.R., Stumm,J., Fulga,T.A., Sinning,I. and Dobberstein,B. (2002) Distinct modes of signal recognition particle interaction with the ribosome. Science, 297, 1345–1348. [DOI] [PubMed] [Google Scholar]
- Rojo E.E., Stuart,R.A. and Neupert,W. (1995) Conservative sorting of Fo-ATPase subunit 9: export from matrix requires ΔpH across inner membrane and matrix ATP. EMBO J., 14, 3445–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco S.A. and Fox,T.D. (2002) Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell, 13, 1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart R.A (2002) Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochim. Biophys Acta, 1592, 79–87. [DOI] [PubMed] [Google Scholar]
- Ullers R.S., Houben,E.N. Raine,A., Ten Hagen-Jongman,C.M., Ehrenberg,M., Brunner,J. Oudega,B., Harms,N. and Luirink,J. (2003) Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol., 161, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Poehlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Woolhead C.A., Thompson,S.J., Moore,M., Tissier,C., Mant,A., Rodger,A., Henry,R. and Robinson,C. (2001) Distinct Albino3-dependent and -independent pathways for thylakoid membrane protein insertion. J. Biol. Chem., 276, 40841–40846. [DOI] [PubMed] [Google Scholar]
- Yen M.-R., Harley,K.T., Tseng,Y.-H. and Saier,M.H. (2001) Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol. Lett., 204, 223–231. [DOI] [PubMed] [Google Scholar]