Abstract
The La protein protects the 3′ ends of many nascent small RNAs from exonucleases. Here we report that La is required for efficient folding of certain pre-tRNAs. A mutation in pre-tRNAArgCCG causes yeast cells to be cold-sensitive and to require the La protein Lhp1p for efficient growth. When the mutant cells are grown at low temperature, or when Lhp1p is depleted, mature tRNAArgCCG is not efficiently aminoacylated. The mutation causes the anticodon stem of pre-tRNAArgCCG to misfold into an alternative helix in vitro. Intragenic suppressor mutations that disrupt the misfolded helix or strengthen the correct helix alleviate the requirement for Lhp1p, providing evidence that the anticodon stem misfolds in vivo. Chemical and enzymatic footprinting experiments suggest a model in which Lhp1p stabilizes the correctly folded stem. Lhp1p is also required for efficient aminoacylation of two wild-type tRNAs when yeast are grown at low temperature. These experiments reveal that pre-tRNAs can require protein assistance for efficient folding in vivo.
Keywords: aminoacylation/La autoantigen/pre-tRNA folding
Introduction
Although it is well accepted that newly synthesized proteins can require molecular chaperones to assist folding, the role of RNA-binding proteins in assisting RNA folding is less clear. As studies of RNA folding in vitro have revealed that RNA often becomes kinetically trapped in alternative helices, it has been proposed that RNA-binding proteins assist RNA folding in vivo (reviewed by Herschlag, 1995). Several RNA-binding proteins, such as Escherichia coli StpA, S12 and Hfq, as well as fragments of hnRNP A1 and HIV nucleocapsid proteins, promote formation of correctly folded RNA and/or enhance RNA:RNA pairing in vitro (Herschlag, 1995; Schroeder et al., 2002). These proteins, when overexpressed in E.coli, resolve an alternative helix that functions as a folding trap, indicating these proteins can also assist folding in vivo (Clodi et al., 1999; Schroeder et al., 2002; Moll et al., 2003). However, of these proteins, only Hfq (Zhang et al., 2002) has been demonstrated to have a normal, physiologic role in facilitating RNA folding in cells.
To date, the only well understood examples of protein-assisted RNA folding in vivo involve group I and group II introns. For these introns, specific RNA-binding proteins assist folding by stabilizing correct tertiary structures (reviewed by Schroeder et al., 2002). In addition, folding of one group I intron requires an ATP-dependent RNA helicase to disrupt misfolded structures that act as kinetic traps in the folding pathway (Mohr et al., 2002). However, for all other classes of RNA, nothing is known of the protein requirements for folding in vivo.
A protein that has often been proposed to facilitate RNA folding is the La protein. This nuclear protein binds many newly synthesized small RNAs, including pre-tRNAs, pre-5S rRNA, U6 snRNA and 7SL RNA (reviewed by Wolin and Cedervall, 2002). As part of the binding site for La is the sequence UUUOH, the majority of bound RNAs are polymerase III transcripts. In Saccharomyces cerevisiae, La also binds nascent polymerase II-transcribed small RNAs that terminate in UUUOH (Kufel et al., 2000; Xue et al., 2000). For pre-tRNAs and most La-bound RNAs, the UUUOH is removed during the maturation process. Thus, La does not bind the mature RNAs. In mammals, La has been implicated in internal initiation of mRNA translation (Wolin and Cedervall, 2002). As La binds many RNAs and has been linked to several processes, it was proposed that La binding facilitates formation of higher order structures (Svitkin et al., 1994).
Genetic and biochemical studies have revealed that a major role of La is to protect nascent RNAs from nucleases. In the yeasts S.cerevisiae and Schizosaccharo myces pombe, binding by La to pre-tRNAs prevents exonucleolytic nibbling, allowing removal of the 3′ trailers by an endonuclease (Van Horn et al., 1997; Yoo and Wolin, 1997; Intine et al., 2000). In S.cerevisiae, binding by the La protein Lhp1p stabilizes precursors to spliceosomal U snRNAs and the small nucleolar U3 RNA (Pannone et al., 1998; Kufel et al., 2000; Xue et al., 2000). In the case of pre-U4 snRNA, Lhp1p stabilizes a precursor that is preferentially bound by the snRNP protein Smd1p, suggesting that Lhp1p binding contributes to efficient snRNP assembly (Xue et al., 2000).
While there is good evidence that La protects nascent RNAs from nucleases, whether La contributes to RNA folding has remained unclear. Yeast containing a mutation that disrupts the anticodon stem of tRNASerCGA require Lhp1p for maturation of the pre-tRNA. As restoring basepairing in the stem eliminated the Lhp1p requirement, Lhp1p binding was proposed to stabilize the correctly folded pre-tRNA (Yoo and Wolin, 1997). However, as the mature tRNA did not accumulate without Lhp1p, an alternative possibility was that Lhp1p protected the mutant pre-tRNA from nucleases (Yoo and Wolin, 1997). Consistent with a role in nuclease protection, a mutant pre-tRNASerCGA with an altered variable loop requires Lhp1p for accumulation (Johansson and Bystrom, 2002). Thus, a role for La in assisting RNA folding has not been demonstrated.
Here we show that Lhp1p is required for efficient folding of certain pre-tRNAs in yeast cells. A mutation in the essential gene encoding tRNAArgCCG causes yeast to be cold-sensitive and to require LHP1 for efficient growth at 25°C. Deacylated tRNAArgCCG accumulates when Lhp1p is depleted or the cells are grown at low temperature. In vitro, the mutation causes the anticodon stem of pre-tRNAArgCCG to misfold. Chemical and enzymatic probing experiments are consistent with a model in which Lhp1p stabilizes the correctly folded stem. As intragenic suppressor mutations that weaken the incorrect helix or strengthen the correct helix alleviate the cold-sensitivity and LHP1 requirement, the pre-tRNA likely also misfolds in vivo. Moreover, Lhp1p is required for efficient aminoacylation of two wild-type tRNAs. Our experiments reveal that certain pre-tRNAs require protein assistance for efficient folding in vivo.
Results
A mutation in yeast tRNAArgCCG causes cold-sensitivity and a requirement for LHP1
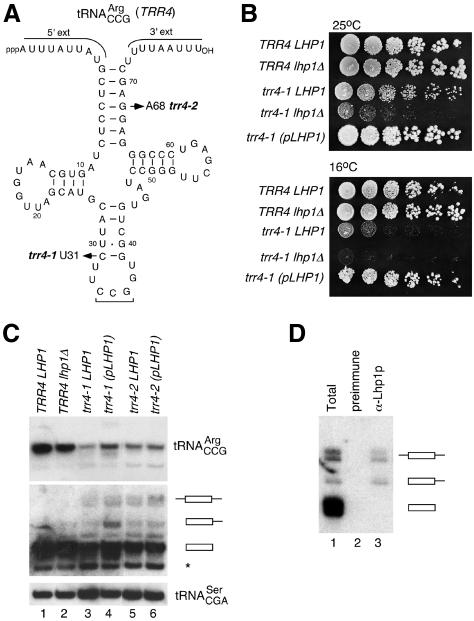
During screens for mutations that cause yeast to require LHP1 (Pannone et al., 2001; also see Materials and methods), we identified three new mutations in tRNA genes. Two mutations, trr4-1 and trr4-2, were members of one complementation group, while the third, trt2-1, defined a second group. Cloning of the genes revealed that the gene encoding tRNAArgCCG complemented the LHP1 requirement in trr4 strains while the gene encoding tRNAThrCGU complemented the trt2 strain. We named the genes TRR4 (tRNA arg4) and TRT2 (tRNAthr2). DNA sequencing revealed that trr4-1 is a C-to-U change that weakens the anticodon stem, while trr4-2 is a G-to-A change that disrupts the acceptor stem of tRNAArgCCG (Figure 1A). The trt2-1 mutation is a G-to-A at position 41 of tRNAThrCGU that disrupts the anticodon stem (data not shown). In addition to requiring LHP1 for efficient growth at 25°C, the trr4-1 strain grew poorly at 16 and 37°C, while the trr4-2 strain was inviable at 37°C. Gene disruption experiments revealed that both TRR4 and TRT2 are essential (data not shown).
Fig. 1. The trr4-1 mutation results in cold-sensitivity and a requirement for LHP1. (A) Positions of trr4-1 and trr4-2 mutations are shown on pre-tRNAArgCCG. Leader and trailer lengths were estimated from pre-tRNA sizes on denaturing gels. (B) Five-fold serial dilutions of wild-type cells (TRR4 LHP1), cells lacking LHP1 (TRR4 lhp1Δ), trr4-1 cells lacking LHP1 (trr4-1 lhp1Δ) and trr4-1 cells carrying chromosomal (trr4-1 LHP1) or plasmid LHP1 (trr4-1 pLHP1) were spotted onto YPD agar and grown for four days at 25°C (top) or six days at 16°C (bottom). (C) RNA from wild-type cells (lane 1), cells lacking LHP1 (lane 2), trr4-1 cells with chromosomal or plasmid LHP1 (lanes 3 and 4) and trr4-2 cells with chromosomal or plasmid LHP1 (lanes 5 and 6) was subjected to northern analysis to detect tRNAArgCCG (top). To detect pre-tRNAs, the blot was overexposed (middle). Asterisk, cross-hybridization with another isoacceptor. The blot was reprobed to detect tRNASerCGA (bottom). (D) A trr4-1 cell extract was incubated with pre-immune (lane 2) or anti-Lhp1p antibodies (lane 3). RNAs in the immunoprecipitate and an equivalent amount of extract (lane 1) were subjected to northern analysis to detect tRNAArgCCG.
Although we had previously identified temperature-sensitive mutations in tRNASerCGA that caused cells to require LHP1 for maturation of the pre-tRNA (Yoo and Wolin, 1997; Long et al., 2001), the trr4-1 allele was unusual in that it also resulted in cold-sensitivity. At 25°C, small trr4-1 colonies could be isolated lacking LHP1, revealing that Lhp1p was not absolutely required for viability. These strains grew very slowly (Figure 1B) and had a high rate of reversion, making them unsuitable for biochemical analyses. The trr4-1 strains containing chromosomal LHP1 grew well at 25°C, but not at the same rate as wild-type cells. However, at 16°C, these cells were retarded in growth (Figure 1B). When the sole copy of LHP1 was present on a centromeric plasmid, which raises Lhp1p levels ∼2-fold (data not shown), trr4-1 cells grew nearly as well as wild-type cells at 25 and 16°C (Figure 1B). Thus, in addition to being required for efficient growth at 25°C, LHP1 is a low-copy suppressor of the trr4-1 mutation.
To determine the effects of the mutations on tRNA levels, we grew the strains at 25°C and extracted RNA. Northern blotting to detect tRNAArgCCG and tRNAThrCGU revealed that for both trr4 strains (Figure 1C) and the trt2-1 strain (data not shown), the mutations resulted in decreased mature tRNAs (Figure 1C, lanes 2–6, top). As overexposure of the blot (middle panel) revealed pre-tRNA accumulation, processing of the mutant pre-tRNAs may be slow relative to wild-type pre-tRNAs. With plasmid LHP1, pre-tRNAArgCCG and mature tRNA levels increased ∼2-fold in the trr4-1 strain (Figure 1C, lane 4; see also Figure 3A). Thus, as described for pre- tRNAMeti (Anderson et al., 1998), overexpressed Lhp1p may stabilize trr4-1 pre-tRNAArgCCG allowing more efficient maturation. Immunoprecipitations confirmed that Lhp1p bound wild-type and mutant pre-tRNAArgCCG containing 3′ trailers, but not mature tRNA (Figure 1D and data not shown).
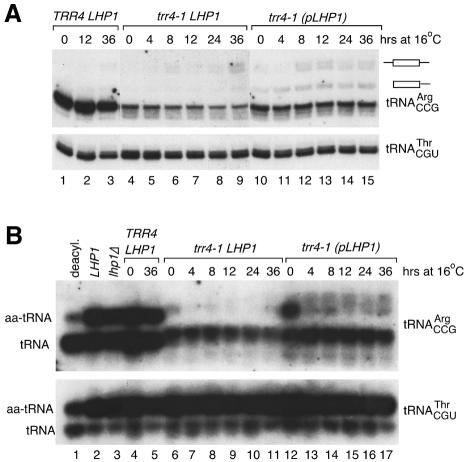
Fig. 3. Aminoacylated tRNAArgCCG declines when trr4-1 cells are grown at 16°C. (A) Wild-type (lanes 1–3) and trr4-1 cells carrying chromosomal (lanes 4–9) or plasmid LHP1 (lanes 10–16) were grown at 25°C and switched to 16°C at time 0. At indicated times, RNA was extracted and subjected to northern analysis to detect tRNAArgCCG and tRNAThrCGU. (B) At intervals, RNA was extracted under acidic conditions, fractionated in acidic acrylamide gels, and subjected to northern analysis to detect tRNAArgCCG and tRNAThrCGU.
LHP1 is required for efficient aminoacylation of tRNAArg in trr4-1 strains
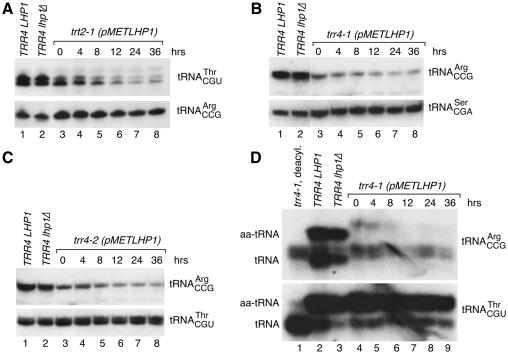
To determine why LHP1 was required, we placed LHP1 under control of the methionine-repressible MET3 promoter. When strains were grown in media lacking methionine, Lhp1p levels were similar to that of wild-type cells. Upon addition of 2 mM methionine, Lhp1p declined, becoming undetectable by 12 h. Also at 12 h, the trt2-1, trr4-1 and trr4-2 strains slowed in growth (data not shown).
RNA was extracted from the cells and subjected to northern blotting. Levels of mature tRNAThrCGU declined 3.5-fold as Lhp1p was depleted from trt2-1 cells (Figure 2A, lanes 3–8). Overexposure of the blot revealed that pre-tRNAThrCGU levels were unchanged (data not shown), consistent with a requirement for Lhp1p in tRNA maturation rather than transcription. Both findings were similar to that observed when Lhp1p was depleted from cells carrying a mutation in tRNASerCGA (Yoo and Wolin, 1997).
Fig. 2. Depletion of LHP1 from trr4 and trt2 cells. trt2-1 (A), trr4-1 (B) and trr4-2 cells (C) containing pMETLHP1 were grown at 25°C in media lacking methionine and switched to 2 mM methionine media at time 0. At intervals (lanes 3–8), RNA was subjected to northern analysis to detect tRNAThrCGU (A, top; C, bottom), tRNAArgCCG (A, bottom; B and C, top) or tRNASerCGA (B, bottom). RNA was also analyzed from wild-type (lanes 1) and lhp1Δ cells (lanes 2). For unknown reasons, tRNAThrCGU runs as a doublet. (D) At intervals after the switch to 2 mM methionine, RNA was extracted from trr4-1 cells under acidic conditions and fractionated in acidic acrylamide gels. The northern blot was probed to detect tRNAArgCCG and tRNAThrCGU. Lane 1, deacylated trr4-1 RNA. RNA was also analyzed from wild-type cells (lane 2) and cells lacking LHP1 (lane 3). Consistent with an altered structure, charged and uncharged forms of wild-type tRNAArgCCG migrate differently than these forms of trr4-1 tRNAArgCCG. Lane 3 is underloaded.
In contrast, Lhp1p depletion from the two trr4 strains resulted in less severe decline in tRNAArgCCG levels (Figure 2B and C). This was most apparent for trr4-1 (Figure 2B, lanes 3–8), where PhosphorImager quantitation revealed that mature tRNAArgCCG declined <2-fold. To determine whether the tRNA was functional, RNA was extracted under acidic conditions to stabilize aminoacyl-tRNA linkages and fractionated in acidic acrylamide gels (Varshney et al., 1991). Northern analyses revealed that while approximately half the wild-type tRNAArgCCG was aminoacylated (Figure 2D, lane 2), only 20% of trr4-1 tRNAArgCCG was charged in the presence of Lhp1p (lane 4). Upon Lhp1p depletion, aminoacylated tRNAArgCCG declined, becoming undetectable after 12 h (lanes 4–9). Reprobing for tRNAThrCGU revealed that aminoacylation of this tRNA was unaffected (Figure 2D). Thus, Lhp1p is required in trr4-1 cells for efficient aminoacylation of tRNAArgCCG.
As Lhp1p does not bind mature tRNAArgCCG (Figure 1D), it was unlikely that Lhp1p functioned directly in aminoacylation. However, aminoacylation is sensitive to changes in tRNA structure (Giege et al., 1993), as is the export of mature tRNA from the nucleus, which uses both nuclear aminoacylation and binding of tRNA-specific export receptors as proofreading steps (Hopper and Phizicky, 2003). Thus, one possibility was that trr4-1 pre-tRNA misfolded in the absence of Lhp1p, such that the resulting mature tRNA was either incompetent for export and/or unable to undergo charging. Consistent with a structural alteration, both charged and uncharged forms of trr4-1 tRNAArgCCG migrated slower than these forms of the wild-type tRNA (Figure 2D, lanes 2–4).
Uncharged tRNAArgCCG accumulates in trr4-1 cells grown at low temperature
Since cold-sensitive mutations are associated with trapping of RNA in competing helices at low temperature (Dammel and Noller, 1993; Zavanelli et al., 1994), we examined whether deacylated tRNAArgCCG accumulated at 16°C. Wild-type and trr4-1 cells were grown at 25°C and then shifted to 16°C. RNA was analyzed by northern blotting. While tRNAArgCCG levels were initially lower in trr4-1 cells (Figure 3A, lanes 4 and 10), mature tRNA declined <2-fold during growth at 16°C (lanes 4–9 and 10–15).
To examine aminoacylation, RNA was extracted and subjected to electrophoresis under acidic conditions. At 25°C, the fraction of charged tRNAArgCCG was higher in cells carrying plasmid LHP1 than cells with chromosomal LHP1 (Figure 3B, lanes 6 and 12). Within 4 h at 16°C, charged tRNAArgCCG was undetectable in trr4-1 cells with chromosomal LHP1 (Figure 3B, lane 7). In cells with plasmid LHP1, charged tRNAArgCCG also declined (lanes 12–17), but a smear of hybridization (which may correspond to pre-tRNAs) made it hard to determine whether charged tRNA declined to undetectable levels. Nonetheless, accumulation of deacylated tRNAArgCCG at 16°C is consistent with the hypothesis that the trr4-1 mutation results in misfolding.
The trr4-1 mutation causes the anticodon stem of pre-tRNAArgCCG to misfold in vitro
To examine the structures formed by the pre-tRNAs in vitro, we synthesized the RNAs using T7 RNA polymerase. Preliminary experiments using ribonucleases revealed that both wild-type and trr4-1 pre-tRNAs were grossly misfolded when isolated under denaturing conditions (electrophoresis in urea-containing gels, phenol-extraction and ethanol precipitation). This is consistent with reports that yeast tRNAArg is one of several tRNAs that is isolated from cells in a form that is inactive for aminoacylation (Lindahl et al., 1966). Heating of the wild-type pre-tRNA in 10 mM magnesium largely alleviated the misfolding, but did not correct the trr4-1 structure (data not shown). A similar misfolding problem has been described for unmodified E.coli tRNAPhe, in that once denatured, the tRNA cannot be efficiently refolded (Uhlenbeck, 1995). To minimize the problem, in vitro-transcribed pre-tRNAs were isolated on DEAE–Sepharose columns without denaturants (see Materials and methods). As end-labeling reactions require denaturing gel electrophoresis to isolate a single labeled species, reactions were carried out on unlabeled RNAs and visualized by primer extension or northern analysis.
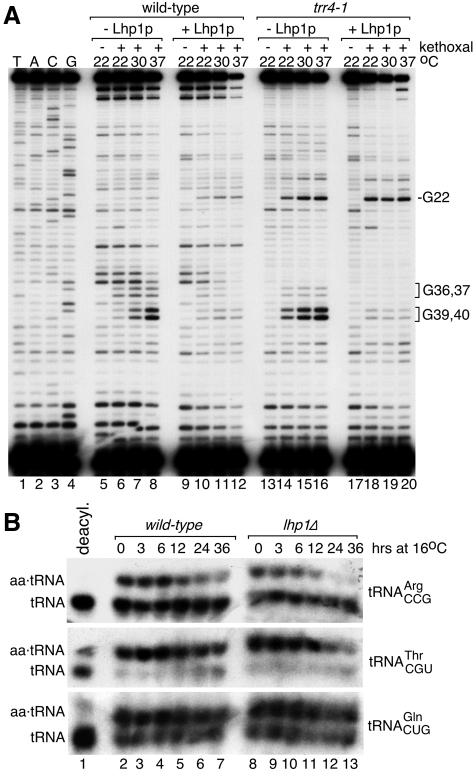
Experiments using chemical probes that modify RNA in single-stranded regions are shown in Figure 4. Sites of modification were mapped by primer extension, as reverse transcriptase stops at the nucleotide 3′ to the modification. Since pre-tRNAs are highly structured, and because some degradation is inevitable during native RNA isolation, there were many stops that were independent of chemical probes (Figure 4A and B). However, examination of the stops that depended on chemical addition revealed several differences between wild-type and trr4-1 pre-tRNAs. Most changes were in bases of the anticodon stem. Experiments using kethoxal (which reacts with unpaired Gs) revealed that G39 and G40, which form part of the weakly basepaired anticodon stem in the canonical tRNA structure, were more accessible to the chemical in trr4-1 pre-tRNA (Figure 4A, lanes 6, 7, 12 and 13). Also, G36 and G37, which are single-stranded in the wild-type structure, are less accessible in trr4-1 RNA (lanes 12 and 13; also Figure 6A). Experiments with CMCT [1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate], which modifies unpaired Us, revealed that U31 in trr4-1 pre-tRNA (which is a basepaired C in wild-type RNA) was accessible (Figure 4B, lanes 9 and 10). Lastly, G10 and G22 (at the ends of the D-stem) were more accessible in trr4-1 RNA (Figure 4A, lanes 6 and 7, 12 and 13).
Fig. 4. trr4-1 pre-tRNA adopts an altered conformation in vitro. (A) Wild-type (lanes 5–10) and trr4-1 pre-tRNAArgCCG (lanes 11–16) (0.68 pmol each) were incubated with the indicated amounts of kethoxal in the absence or presence of 6.8 pmol Lhp1p. Modifications were detected by primer extension. Lanes 1–4, dideoxy sequencing of pre-tRNAArgCCG. Lanes are labeled according to the RNA sequence. Each extension stop is one base below the modified nucleotide. In some experiments, modification of G26 is detected in wild-type pre-tRNA. (B) Wild-type (lanes 5–7) and trr4-1 pre-tRNAArgCCG (lanes 8–10) were incubated with the indicated amounts of CMCT and subjected to primer extension. Lanes 1–4, dideoxy sequencing. (C) trr4-1 pre-tRNAArgCCG was incubated in the absence (lanes 1–4) or presence (lanes 5 and 6) of Lhp1p. Proteinase K was added to digest Lhp1p (lanes 2, 4 and 6). Following a second incubation, kethoxal was added (lanes 3–6), and modifications detected as in (A). (D) The classic structure for pre-tRNAArgCCG (left) and the proposed alternative structure (right). Sites of modification are designated by arrows (CMCT), triangles (kethoxal) and dots (DMS). Two G residues that are more accessible to kethoxal in wild-type RNA are red and two Gs that are less accessible are green. The alternative structure is predicted by MFOLD to be more stable than the classic structure by 2.3 kcal/mol. (E) 0.34 pmol of wild-type (lanes 1–6) and trr4-1 pre-tRNAArgCCG (lanes 7–12) were incubated without protein (lanes 1–7) or with 2- (lanes 2 and 8), 4- (lanes 3 and 9), 6- (lanes 4 and 10), 8- (lanes 5 and 11) or 10-fold (lanes 6 and 12) molar excess of Lhp1p. RNA and RNPs were separated in native gels and detected by northern analysis. Asterisk, a second trr4-1 pre-tRNA conformer. (F) 0.68 pmol of trr4-1 (lanes 1–3) and trr4-1 pre-tRNA lacking the 3′ 9 nt (trr4-1,Δ3′) were incubated without protein (lanes 1 and 4), or with 5- (lanes 2 and 5) or 10-fold (lanes 3 and 6) molar excess of Lhp1p. Naked RNAs and RNPs were separated in native gels and detected by northern analysis. Asterisk, a conformer of trr4-1 pre-tRNA.
Fig. 6. LHP1 is required for efficient aminoacylation of wild-type tRNAArgCCG. (A) Wild-type (lanes 5–12) and trr4-1 pre-tRNAArgCCG (lanes 13–20) were incubated at the indicated temperature in the absence or presence of Lhp1p. Following incubation, 5 µl of kethoxal was added. Modifications were detected by primer extension. Lanes 1–4, dideoxy sequencing. The primer extension stops in the wild-type tRNA (lanes 5–12) at positions 30, 34 and 35 and near the top of the gel are not kethoxal-dependent. (B) Wild-type (lanes 1–7) and cells lacking LHP1 (lanes 8–12) were grown at 25°C and switched to 16°C at time 0. At intervals, RNA was extracted under acidic conditions and subjected to northern analysis to detect tRNAArgCCG (top), tRNAThrCGU (middle) and tRNAGlnCUG (bottom panel). Lane 1, deacylated wild-type RNA.
The CMCT and kethoxal data, along with experiments with dimethylsulfate (DMS) (which modifies A and C; data not shown), indicated that trr4-1 pre-tRNAArgCCG adopts an altered conformation in vitro. The most plausible model, based on structure mapping, is shown in Figure 4D (right panel). In the model, the anticodon stem is misfolded into an alternative helix, such that the anticodon is partly basepaired. As G36 and G37 are less accessible to kethoxal in trr4-1 pre-tRNA, suggesting they are at least partly basepaired, while their proposed pairing partners U29 and U30 are accessible to CMCT, the misfolded stem may exist as two conformers, one in which the residues are basepaired, and a second in which they are unpaired (Figure 4D, right). While our model does not change positions of G10 and G22, which are more accessible in trr4-1 RNA, formation of tertiary interactions between D and T-loops in the misfolded tRNA may cause residues at the ends of the D-stem to become partly unpaired. Consistent with a largely intact D-stem, U11 (Figure 4B, lanes 9 and 10), C23 and A24 (not shown) are inaccessible to chemical probes. Although other models for the trr4-1 pre-tRNA structure are possible, the proposed misfolded stem is consistent with the finding that uncharged trr4-1 tRNA accumulates in vivo. While tRNA end maturation and CCA addition require only that the acceptor stem and TΨC stem–loop be intact (Maizels and Weiner, 1999), tRNAArg charging requires unpaired C35 and G36 in the anticodon loop (Sissler et al., 1996).
Lhp1p alters the accessibility of the mutant pre-tRNA to chemical probes
To examine effects of Lhp1p, we determined the Lhp1p concentration that shifted pre-tRNAArgCCG into an RNA–protein complex, while minimizing formation of higher order, less-specific complexes (Long et al., 2001) (Figure 4E). By comparing Lhp1p binding to full-length and 3′ truncated pre-tRNAs, we confirmed that binding was largely dependent on the 3′ trailer (Figure 4F and data not shown).
For wild-type pre-tRNA, kethoxal modifications were similar in the presence or absence of Lhp1p (Figure 4A, lanes 8–10). (As modifications are viewed by primer extension, no information is obtained about the RNA 3′ end.) Experiments with CMCT revealed that all protections were lower with Lhp1p (not shown), most likely due to side reactions that occur between CMCT and protein (Krol and Carbon, 1989).
Interestingly, Lhp1p addition to trr4-1 pre-tRNA resulted in protection of G36, G37, G39 and G40 from kethoxal modification (Figure 4A, lanes 15 and 16). These residues are all part of the misfolded stem (Figure 4D). Changes in modification were dependent on bound Lhp1p, as digestion of the RNA–protein complex with protease prior to probing restored accessibility of the mutant pre-tRNA to modification (Figure 4C, lanes 5 and 6). The changes in modification could have several explanations. Lhp1p binding could facilitate correct folding of the anticodon stem, since G39 and G40 are basepaired in the wild-type RNA (Figure 4D). However, G22 accessibility was unchanged upon Lhp1p addition, indicating that the trr4-1 structure did not fully convert to that of wild-type pre-tRNA. An alternative possibility is that Lhp1p protects G36, G37, G39 and G40 from modification. Thus, Lhp1p makes specific contacts with the misfolded stem and/or causes alterations of the trr4-1 structure in vitro.
Intragenic suppressors that stabilize the correct helix or weaken the competing helix relieve the LHP1 requirement
To obtain evidence that the alternative conformation forms in vivo, we selected intragenic suppressors of the trr4-1 cold-sensitivity. We expected to identify mutations that stabilized the correct helix or weakened the competing helix. A library of trr4-1 genes containing random second mutations in trr4-1 was constructed. The DNA was integrated into strains in which the only copy of TRR4 was on a URA3-containing plasmid. Transformants were tested for growth on media containing 5-fluoro-orotic acid (FOA) at 25°C. As FOA selects for cells that have lost the URA3 plasmid, cells that grew received a mutated trr4-1 allele that functions at 25°C. Strains were tested for growth at 16°C. One strain, which changed G40 to U, was identified. This change destabilizes the central C–G basepair of the competing helix, but should have only a mild effect on the correct stem, as it disrupts a non-Watson–Crick basepair adjacent to unpaired bases (Figure 5A). Cells carrying the mutation (trr4-1,U40) grew better than trr4-1 cells at 16°C (Figure 5B, bottom panel). Mating to a strain lacking LHP1, followed by tetrad dissection, revealed that cells carrying trr4-1,U40 did not require LHP1 for efficient growth at 25°C (Figure 5B). As mutations stabilizing the correct stem were not obtained (most likely because our screen did not reach saturation), we constructed such a strain by mutating G39-to-A (Figure 5A). This strain (trr4-1,A39) grew well at 16°C and did not require LHP1 for efficient growth (Figure 5B).
Fig. 5. Intragenic suppressor mutations favor formation of the correct helix. (A) Standard and proposed competing structures of pre-tRNAArgCCG are shown along with positions of the trr4-1 (U31) mutation and A39 and U40 suppressors. (B) Top: 5-fold serial dilutions of wild-type, trr4-1 LHP1 and trr4-1 LHP1 strains carrying suppressors A39 (trr4-1,A39 LHP1) and U40 (trr4-1,U40 LHP1) were spotted on YPD agar and grown at 25°C for four days (left) or 16°C for six days (right). Bottom: growth of wild-type and trr4-1 strains lacking LHP1 at 25°C was compared with trr4-1 strains carrying mutations A39 (trr4-1,A39 lhp1Δ) and U40 (trr4-1,U40 lhp1Δ). (C) RNA from wild-type (lanes 1 and 5), trr4-1 (lanes 2 and 6), trr4-1,U40 (lanes 3 and 7) and trr4-1,A39 strains (lanes 4 and 8) containing LHP1 was subjected to northern analysis to detect tRNAArgCCG (top) and tRNAThrCGU (bottom). The band below mature tRNAArgCCG in trr4-1,U40 strains may represent tRNA lacking CCA. (D) RNA was extracted under acidic conditions and fractionated in acidic acrylamide gels. The blot was probed to detect tRNAArgCCG and tRNAThrCGU. Charged species are indicated by dots. For trr4-1,U40, two species represent uncharged tRNA (lanes 4 and 8), one of which may lack CCA. Lane 1, deacylated TRR4 LHP1 RNA.
To determine the mechanism of suppression, we examined tRNAArgCCG levels and charging. At 25 and 16°C, tRNAArgCCG levels in strains carrying suppressor mutations (trr4-1,U40 and trr4-1,A39) were similar to the trr4-1 strain (Figure 5C, lanes 2–4 and 6–8). However, the fraction of charged tRNAArgCCG was higher in the trr4-1,A39 strain (51% for trr4-1,A39 versus 20% for trr4-1) (Figure 5D, lanes 3 and 5). For the trr4-1,U40 strain, the presence of shorter species (Figure 5C, lanes 3 and 7; Figure 5D, lanes 4 and 8), which may be due to inefficient CCA addition, made it difficult to quantitate the charged tRNA. However, after 4 h at 16°C, charged tRNAArgCCG remained detectable in the suppressors, but was undetectable in the trr4-1 strain (lanes 7–9). Thus, both suppressors function by preventing tRNAArgCCG misfolding.
As mutations predicted to stabilize the correct helix (trr4-1, A39) or weaken the competing helix (trr4-1, U40) alleviate the cold-sensitivity and requirement for LHP1, we conclude that the trr4-1 anticodon stem likely misfolds in vivo.
LHP1 is required for efficient aminoacylation of two wild-type tRNAs
Since the anticodon stem of wild-type pre-tRNAArgCCG is weakly basepaired, the wild-type RNA may also misfold. We used kethoxal modification to examine pre-tRNA structure as a function of temperature in vitro (Figure 6A). For both wild-type and trr4-1 pre-tRNAArgCCG, raising the temperature to 30 or 37°C resulted in enhanced accessibility of G39 and G40 to kethoxal (lanes 7, 8, 15 and 16). As decreased accessibility at G36 and G37 was not detected, enhanced modification of G39 and G40 probably reflects increased opening of the weakly basepaired wild-type and trr4-1 anticodon stems at 37°C, rather than misfolding. Lhp1p addition largely eliminated the changes in modification at G39 and G40 (lanes 10–12 and 18–20). Thus, Lhp1p prevents opening of the anticodon stem and/or protects the unpaired residues from modification.
We examined whether efficient aminoacylation of the wild-type tRNA depended on LHP1. As low temperature should trap misfolded forms (Zavanelli and Ares, 1991; Dammel and Noller, 1993), we examined aminoacylation at 16°C. Cells were grown first at 25°C, and then shifted to 16°C. In wild-type cells, there was a small (∼2-fold) decrease in charged tRNAArgCCG during growth at 16°C (Figure 6B, lanes 2–7, top). This decrease was exacerbated in cells lacking Lhp1p, as charged tRNAArgCCG declined >5-fold by 24 h at 16°C (lanes 8–12). Reprobing for tRNAThrCGU and tRNAGlnCUG revealed that tRNAThrCGu aminoacylation also declined slightly in cells lacking LHP1 during growth at 16°C, while tRNAGlnCUG charging was unaffected. Thus, LHP1 is required for efficient aminoacylation of at least two wild-type tRNAs at low temperature.
We examined tRNAArgCCG aminoacylation during growth at higher temperatures. While the fraction of tRNAArgCCG that was charged increased at 30 and 37°C, no differences were detected between wild-type and lhp1Δ strains (data not shown). Thus, if the anticodon stem opening that we detect at elevated temperatures in vitro (Figure 6A) occurs in vivo, it is not detrimental to aminoacylation. Nonetheless, the finding that Lhp1p increases aminoacylation of two wild-type tRNAs at 16°C suggests that binding by Lhp1p to these pre-tRNAs facilitates correct folding.
Lhp1p may contact the acceptor stem and anticodon loop of pre-tRNAArgCCG
A question raised by our studies was how a protein that binds the 3′ trailer could influence folding of the anticodon stem. However, while 3′ uridylates are a major determinant for La binding, La must also recognize other structural features (Wolin and Cedervall, 2002). To examine sites of contact between Lhp1p and pre-tRNAArgCCG, we performed enzymatic footprinting using 5′ end-labeled RNA. As only wild-type pre-tRNAArgCCG could be largely refolded following denaturing gel purification, the analysis was confined to the wild-type RNA.
We performed footprinting using ribonucleases T1, T2 and V1. T1 cuts after guanosines, preferentially in single-stranded regions, T2 cuts in single-stranded regions and V1 prefers helical regions (Krol and Carbon, 1989). Cleavage of the naked RNA revealed strong V1 cuts in the acceptor and anticodon stems, but weak to undetectable cuts in the T and D stems, consistent with previous tRNA probing (Lowman and Draper, 1986). T1 and T2 yielded strong cuts in the anticodon loop and weaker cleavage of the D and T loops (Figure 7A, lanes 5, 7 and 9). We also detected weak T1 and T2 cleavage in the T-stem, suggesting that a fraction of the RNA is single-stranded in this region. As these cleavages were more prominent in pre-tRNA that was not refolded, they may represent a fraction of the wild-type RNA that remains incorrectly folded.
Fig. 7. Lhp1p may contact the acceptor stem and anticodon loop of pre-tRNAArgCCG. (A) 5′ labeled pre-tRNAArgCCG was incubated without (lanes 3, 5, 7 and 9) or with (lanes 4, 6, 8 and 10) Lhp1p, followed by cleavage with T1 (lanes 5 and 6), T2 (lanes 7 and 8) or V1 (lanes 9 and 10) ribonucleases. In the experiment, ∼85% of the labeled RNA was bound by Lhp1p. Lanes 1 and 2, T1 ribonuclease and alkaline hydrolysis ladders. Asterisks, sites of weak protection by Lhp1p. (B) Phosphorimager quantitation of V1 cleavage of the acceptor stem (top) and the anticodon stem (bottom). (C) Quantitation of T2 cleavage of the anticodon loop (top) and D loop (bottom). (D) Pre-tRNAArgCCG was modeled on the structure of tRNAArgICG (Delagoutte et al., 2000) using SPOCK. Bases protected from cleavage by Lhp1p are shown in red.
Lhp1p protected the 3′ strand of the acceptor stem from V1 RNase digestion (Figure 7A, lanes 9 and 10; Figure 7B, top). The V1 cleavages and Lhp1p protection extended into the trailer, consistent with the proposal that the trailer and leader basepair (Lee et al., 1997). In contrast, V1 cleavages in the anticodon stem were unchanged with Lhp1p (Figure 7B, bottom). However, in multiple experiments, Lhp1p addition resulted in slight protection of parts of the anticodon loop (nucleotides U34–G36) from T2 ribonuclease (Figure 7A, lanes 7 and 8, asterisks). PhosphorImager quantitation confirmed that these nucleotides were less accessible in the presence of Lhp1p (Figure 7C, top). T2 cleavages in the D-loop were unchanged with Lhp1p (Figure 7C, bottom), revealing that the very weak protection of the anticodon loop was not due to underloading in the lane. While changes in nuclease accessibiity can be due to either protein binding or structural alterations, protection of the 3′ acceptor stem and trailer is consistent with the known binding of Lhp1p to 3′ ends (Wolin and Cedervall, 2002). Moreover, the slight protection of the anticodon loop from nuclease suggests that bound Lhp1p may also contact the anticodon loop. Regions of pre-tRNAArgCCG protected from nuclease digestion by Lhp1p are shown in red in Figure 7D.
Discussion
Our experiments have revealed that binding by Lhp1p to both wild-type and a mutant pre-tRNAArgCCG is important for formation of the correctly folded mature tRNAs. In the absence of Lhp1p, the pre-tRNAs undergo end maturation, but the mature tRNAs are inefficiently aminoacylated. Biochemical and genetic experiments support a role for Lhp1p in stabilizing pre-tRNA structure. To our knowledge, this is the first evidence that pre-tRNAs can require proteins to assist folding into their correct structures in vivo.
Previous studies have documented that certain tRNAs have a propensity to misfold. Several tRNAs are isolated from cells in two conformations, only one of which is a substrate for aminoacylation (Gartland and Sueoka, 1966; Lindahl et al., 1966; Herschlag, 1995). Studies of point mutations in human mitochondrial tRNAIle have revealed that wild-type tRNAIle has a fragile T stem that is susceptible to structural rearrangements (Kelley et al., 2001). The tRNAArgCCG may be particularly susceptible to misfolding, as it is the only S.cerevisiae tRNA with a mismatch in the anticodon stem. The trr4-1 mutation further weakens the already fragile stem, increasing the fraction of RNA that misfolds. As tRNAThrCGU also requires Lhp1p for efficient aminoacylation at 16°C, a fraction of this tRNA may similarly misfold.
Our data are most consistent with a model in which Lhp1p stabilizes the correctly folded anticodon stem. First, as treatment of Lhp1p–pre-tRNA complexes with protease prior to kethoxal addition restores the modification pattern of the misfolded anticodon stem to that of naked RNA (Figure 4C), Lhp1p probably acts by stoichiometric binding rather than through a catalytic mechanism. Second, the weak protection of the wild-type anticodon loop from ribonuclease, coupled with the finding that Lhp1p eliminates the increased kethoxal accessibility of the trr4-1 and wild-type anticodon stems at 30 and 37°C (Figure 6A) suggests that bound Lhp1p stabilizes the anti-codon stem–loop. In our model, the mutant pre-tRNAArgCCG is kinetically trapped in the alternative helix at 16°C. Raising the temperature lowers the kinetic barrier, allowing more pre-tRNA to fold into the correct helix. Binding by Lhp1p stabilizes the correct anticodon stem, preventing misfolding.
As Lhp1p is removed upon cleavage of the trailer (Yoo and Wolin, 1997), the correct mature tRNAArgCCG structure must be maintained by other mechanisms. One possibility is that once the structure is formed, tertiary and/or base stacking interactions stabilize the correctly folded tRNA. As pre-tRNAs bound by La lack certain modifications (Rinke and Steitz, 1982), acquisition of these modified nucleotides may contribute to stability. Also, subsequent proteins, such as modifying enzymes, the aminoacyl-tRNA synthetase, tRNA export factors and elongation factor 1A may stabilize the functional conformation (Johansson and Bystrom, 2002). We note that pre-tRNAArgCCG regions likely contacted by Lhp1p (the acceptor stem and anticodon stem–loop), are contacted in mature tRNA by arginyl-tRNA synthetase (Delagoutte et al., 2000). Interestingly, we identified a mutation in yeast arginyl-tRNA synthetase that causes cells to require Lhp1p (unpublished data). Characterization of the mutation may reveal if the synthetase redundantly stabilizes folding of tRNAArgCCG.
Although our data suggest that Lhp1p facilitates folding by stabilizing the anticodon stem–loop, other proteins could also be involved. To date, we have been unable to fully convert the misfolded trr4-1 pre-tRNA structure to that of the wild-type RNA by adding Lhp1p. One possibility is that the misfolded structure, once formed, is thermodynamically stable in vitro. Alternatively, other proteins may assist in resolving the misfolded pre-tRNA in vivo. One candidate would be a member of the DEAD box family of RNA helicases. Human La associates with DDX-15, the orthologue of yeast Prp43, an RNA- dependent ATPase that functions in pre-mRNA splicing (Fouraux et al., 2002). Such a mechanism would resemble group I intron folding in Neurospora crassa, where CYT-18, which stabilizes correct tertiary structure, functions together with the CYT-19 RNA helicase (Mohr et al., 2002). Thus far, our attempts to use cell extracts to examine roles of other proteins have been complicated by the fact that cleavage of the 3′ trailer occurs in extracts, removing Lhp1p from the pre-tRNA.
While base modifications enhance tRNA structural stability, it is unlikely that Lhp1p acts by facilitating a modification that affects basepairing. HPLC analyses of tRNA from strains lacking Lhp1p failed to detect differences in modified nucleosides (Johansson and Bystrom, 2002), making it unlikely that Lhp1p is required for an abundant modification. Also, most modified nucleosides in cytosolic tRNAs do not influence basepairing. An exception is the dimethylguanosine at position 26 in many tRNAs, which prevents G26 from pairing with C (Steinberg and Cedergren, 1995). This modification, which is carried out by TRM1, would weaken the proposed alternative helix. However, tRNAArgCCG lacks identity elements for TRM1 dimethylation (G–C basepairs at positions G10–C25 and C11–G24 and a variable loop of at least five nucleotides) (Edqvist et al., 1994). Also, deletion of TRM1 in the trr4-1 strain did not affect the cold-sensitivity or LHP1 requirement (unpublished data), revealing that a failure to modify G26 does not contribute to either phenotype.
Our finding that binding by Lhp1p to certain pre-tRNAs facilitates folding suggests that La may assist folding of other nascent small RNAs. In many RNA polymerase III transcripts, such as pre-tRNAs, 5S rRNA and signal recognition particle (SRP) RNA, 3′ sequences basepair with internal or 5′ sequences to form a long stem. As Lhp1p contacts the pre-tRNAArgCCG acceptor stem, La may contact the analogous stems in other nascent RNAs, stabilizing the helix. Moreover, our identification of an essential pre-tRNA that misfolds in vivo may be useful for genetic identification of other proteins that resolve misfolded RNAs or stabilize correctly folded RNA structures.
Materials and methods
Yeast media and strains
Yeast media were according to Sherman (1991). Wild-type and lhp1::LEU2 strains were CY1 (MATα ura3 lys2 ade2 trp1 his3 leu2 LHP1) and CY2 (MATα ura3 lys2 ade2 trp1 his3 leu2 lhp1::LEU2). Other strains were SK100 (MATα trr4-1 LHP1 ura3 lys2 ade2 trp1 his3 leu2), SK110 (MATα trr4-1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 + pATL), SK130 (MATα trr4-1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 + pMETLHP1), SK201 (MATa trr4-2 LHP1 ura3 lys2 ade2 trp1 his3 leu2), SK210 (MATa trr4-2 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 + pATL), SK230 (MATα trr4-2 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 + pMETLHP1), SK310 (MATa trt2-1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 + pATL) and SK330 (MATa trt2-1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 + pMETLHP1). Suppressors were isolated using SK160 (ura3 lys2 ade2 his3 leu2 trr4::HIS3 + TRR4 in pRS316).
Synthetic lethal screen
trr4-1 and trt2-1 alleles were identified previously (Pannone et al., 2001). The trr4-2 allele was isolated largely as described (Pannone et al., 1998). CY2 carrying pATL (LHP1, TRP1, ADE2; Pannone et al., 1998) was mutagenized with ultraviolet light to 15% survival. After plating on media containing limiting adenine, colonies were screened for the inability to lose pATL and form red sectors. Colonies that did not sector were transformed with pSLL28 (Yoo and Wolin, 1997) and tested for the ability to lose pATL. Strains that lost pATL were tested on media containing 1 µg/ml FOA to confirm they required pSLL28.
To clone TRR4 and TRT2, a genomic library in YCp50 was introduced into SK110 and SK310 and transformants screened for loss of pATL. Subcloning revealed that TRR4 with 81 bp of 5′ and 543 bp of 3′ flanking sequence eliminated the LHP1 requirement in SK110. For SK310, TRT2 with 286 bp of 5′ and 46 bp of 3′ sequence eliminated the requirement. Mutations were identified by sequencing genomic DNA.
Lhp1p depletion and coldshift experiments
To deplete Lhp1p, strains SK130, SK230 and SK330 carrying pMETLHP1 (Pannone et al., 1998) were grown in synthetic complete (SC) minus methionine at 25°C to OD600 = 0.5, diluted into SC + 2 mM methionine and grown for 36 h. Cells were kept below OD600 = 0.5. For the coldshift, CY1, SK100 and SK110 were grown at 25°C in YPD to OD600 = 0.4 and shifted to 16°C. Cells were kept below OD600 = 0.5.
Immunoprecipitations and northerns
Immunoprecipitations were as described (Xue et al., 2000). Total RNA was extracted using hot phenol/SDS and subjected to northern blotting (Pannone et al., 1998). For aminoacylation analyses, cells were lysed in pH 4.5 phenol as described (Varshney et al., 1991). Deacylation was at pH 9.0. RNAs were fractionated in 6.5% polyacrylamide, 8 M urea, 0.1 M NaOAc pH 5.0 gels at 4°C. Probes: tRNAArgCCG: 5′-ACTCGAACCCGGATCACAGCCACCGGAAGAATGCATGCTAACCATT-3′, tRNAThrCGU: 5′-AATTGAACCCACGATCCCCGCATTACGAGTGCGATGCCTTACCACT-3′, tRNAGlnCUG: 5′-ATTCGAACTGGGGTTGTTCGGATCAGAACCGAAAGTGATAACCACT-3′, tRNASerCGA: 5′-AGCCCAAGAGATTTCGAGTCTCTCG-3′.
RNA purification, gel shifts and structure mapping
To put pre-tRNAArgCCG behind the T7 promoter, 5′-CGGCGAATTCTAATACGACTCACTATAGTTTATTATGCTCCTCTAGTGC-3′ and 5′-GGCCGGATCCTTTAAATTAAAAGCTCCTCCCGGG-3′ were used to amplify TRR4 or trr4-1 DNA. The DNA was inserted into EcoRI/BamHI sites of pSP64 (Promega). After DraI cleavage and T7 transcription, reactions were diluted 10-fold with 10 mM MOPS pH 7.0, 100 mM NaCl, 2 mM MgCl2 and bound to DEAE–Sepharose (100 µl) equilibrated in this buffer. Unincorporated nucleotides were removed with 250 mM NaCl in this buffer. RNAs were eluted with 500 mM NaCl. Samples were dialyzed against 10 mM HEPES pH 7.9, 2 mM MgCl2, frozen on dry ice and stored at –80°C.
For gel shifts, 0.34 pmol of wild-type and trr4-1 pre-tRNAArgCCG in HMK buffer (Black and Pinto, 1989) was incubated with Lhp1p (0.68–6.8 pmol) and 0.04 mg/ml E.coli tRNA in 10 µl for 10 min at 22°C. After 10 min on ice, samples were loaded on 5% acrylamide/5% glycerol gels (Long et al., 2001) and run at 15 V/cm for 3 h.
Chemical modifications using DMS, kethoxal and CMCT were as described (Black and Pinto, 1989). Pre-tRNAArgCCG (0.68 pmol) and 0.04 mg/ml E.coli tRNA were incubated for 15 min with or without 6.8 pmol of Lhp1p in modification buffer. Kethoxal stock (37 µl kethoxal/ml), CMCT (140 mg/ml) or DMS (5% in dioxane) was added and incubated 15 min. RNAs were phenol-extracted, precipitated, and modifications visualized by extending a primer complementary to nucleotides 55 to 89 of the pre-tRNA. For proteinase K digestion, 1 µl of 10 mg/ml proteinase K was added and incubated for 15 min at 22°C prior to kethoxal addition.
For enzymatic footprinting, labeled RNA was heated to 50°C in 50 mM Tris pH 8.0, 10 mM MgCl2 and cooled to 22°C. 0.34 pmol pre-tRNA was incubated with 3.4 pmol Lhp1p and 0.02 mg/ml E.coli tRNA in 20 mM Tris pH 8, 1 mM EDTA, 10 mM MgCl2, 5 mM DTT, 100 mM NaCl, 10% glycerol, 0.01% NP-40 in 10 µl at 22°C for 20 min, then placed on ice for 30 min. Next, 5 U of RNase T2, 0.02 U RNase V1 or 0.05 U RNase T1 was added and incubated on ice for 10 min. Reactions were stopped with 300 µl 0.2 mg/ml proteinase K, 0.03 mg/ml E.coli tRNA, 50 mM Tris pH 7.5, 50 mM NaCl, 5 mM EDTA, 0.5% SDS and incubated 30 min at 55°C. Following phenol extraction and ethanol precipitation, samples were fractionated in 15% polyacrylamide, 8 M urea gels.
Identification of intragenic suppressors
To construct a mutant trr4-1 library, two oligonucleotides, degenerate in the coding sequence (99.1% correct, 0.9% incorrect nucleotides), were synthesized. After annealing and extending, DNA was ligated into BamHI/EcoRI sites of pRS304 (Sikorski and Hieter, 1989). After transformation, DNA was pooled from 5 × 105 colonies, cut with BspMI and integrated into TRP1 of SK160. Integrants were tested on FOA for the ability to lose the URA3 plasmid. After testing strains for growth at 16°C, trr4-1 DNA was sequenced. To confirm that trr4-1,U40 and trr4-1,A39 rescued the cold-sensitivity, these alleles, along with TRR4 and trr4-1 alleles, were synthesized with PCR and integrated into the trr4::HIS3 locus of SDK160. To examine if trr4-1,U40 and trr4-1,A39 strains required LHP1, strains were mated to CY2.
Acknowledgments
Acknowledgements
We thank O.Uhlenbeck, L.Szewszak, S.Aigner, D.Xue and B.Pannone for advice, T.Cedervall for plasmids, J.Pata for tRNA modeling, and A.Pyle, M.Solomon, E.Ullu and C.Waldsich for comments on the manuscript. The work was supported by N.I.H. grant R01-GM48410. S.L.W. is an Associate Investigator of the Howard Hughes Medical Institute.
References
- Anderson J., Phan,L., Cuesta,R., Carlson,B.A., Pak,M., Asano,K., Bjork,G.R., Tamame,M. and Hinnebusch,A.G. (1998) The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev., 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. and Pinto,A.L. (1989) U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol. Cell. Biol., 9, 3350–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clodi E., Semrad,K. and Schroeder,R. (1999) Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J., 18, 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammel C.S. and Noller,H.F. (1993) A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev., 7, 660–670. [DOI] [PubMed] [Google Scholar]
- Delagoutte B., Moras,D. and Cavarelli,J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J., 19, 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist J., Blomqvist,K. and Straby,K.B. (1994) Structural elements in yeast tRNAs required for homologous modification of guanosine-26 into dimethylguanosine-26 by the yeast Trm1 tRNA-modifying enzyme. Biochemistry, 33, 9546–9551. [DOI] [PubMed] [Google Scholar]
- Fouraux M.A., Kolkman,M.J., Van der Heijden,A., De Jong,A.S., Van Venrooij,W.J. and Pruijn,G.J. (2002) The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA, 8, 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartland W.J. and Sueoka,N. (1966) Two interconvertible forms of tryptophanyl sRNA in E.coli. Proc. Natl Acad. Sci. USA, 55, 948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege R., Puglisi,J.D. and Florentz,C. (1993) tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol., 45, 129–206. [DOI] [PubMed] [Google Scholar]
- Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- Hopper A.K. and Phizicky,E.M. (2003) tRNA transfers to the limelight. Genes Dev., 17, 162–180. [DOI] [PubMed] [Google Scholar]
- Intine R.V., Sakulich,A.L., Koduru,S.B., Huang,Y., Pierstorff,E., Goodier,J.L., Phan,L. and Maraia,R.J. (2000) Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol. Cell, 6, 339–348. [DOI] [PubMed] [Google Scholar]
- Johansson M.J. and Bystrom,A.S. (2002) Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA, 8, 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley S.O., Steinberg,S.V. and Schimmel,P. (2001) Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J. Biol. Chem., 276, 10607–10611. [DOI] [PubMed] [Google Scholar]
- Krol A. and Carbon,P. (1989) A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol., 180, 212–227. [DOI] [PubMed] [Google Scholar]
- Kufel J., Allmang,C., Chanfreau,G., Petfalski,E., Lafontaine,D.L. and Tollervey,D. (2000) Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol., 20, 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kindelberger,D.W., Lee,J.Y., McClennen,S., Chamberlain,J. and Engelke,D.R. (1997) Nuclear pre-tRNA terminal structure and RNase P recognition. RNA, 3, 175–185. [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Adams,A. and Fresco,J.R. (1966) Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc. Natl Acad. Sci. USA, 55, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K.S., Cedervall,T., Walch-Solimena,C., Noe,D.A., Huddleston,M.J., Annan,R.S. and Wolin,S.L. (2001) Phosphorylation of the Saccharomyces cerevisiae La protein does not appear to be required for its functions in tRNA maturation and nascent RNA stabilization. RNA, 7, 1589–1602. [PMC free article] [PubMed] [Google Scholar]
- Lowman H.B. and Draper,D.E. (1986) On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem., 261, 5396–5403. [PubMed] [Google Scholar]
- Maizels N. and Weiner,A.M. (1999) The genomic tag hypothesis: what molecular fossils tell us about the evolution of tRNA. In Gesteland,R., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 79–111. [Google Scholar]
- Mohr S., Stryker,J.M. and Lambowitz,A.M. (2002) A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell, 109, 769–779. [DOI] [PubMed] [Google Scholar]
- Moll I., Leitsch,D., Steinhauser,T. and Blasi,U. (2003) RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep., 4, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B.K., Xue,D. and Wolin,S.L. (1998) A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J., 17, 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B.K., Kim,S.D., Noe,D.A. and Wolin,S.L. (2001) Multiple functional interactions between components of the Lsm2–Lsm8 complex, U6 snRNA and the yeast La protein. Genetics, 158, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J. and Steitz,J.A. (1982) Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell, 29, 149–159. [DOI] [PubMed] [Google Scholar]
- Schroeder R., Grossberger,R., Pichler,A. and Waldsich,C. (2002) RNA folding in vivo. Curr. Opin. Struct. Biol., 12, 296–300. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissler M., Giege,R. and Florentz,C. (1996) Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J., 15, 5069–5076. [PMC free article] [PubMed] [Google Scholar]
- Steinberg S. and Cedergren,R. (1995) A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. RNA, 1, 886–891. [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause,A. and Sonenberg,N. (1994) La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol., 68, 7001–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O.C. (1995) Keeping RNA happy. RNA, 1, 4–6. [PMC free article] [PubMed] [Google Scholar]
- Van Horn D.J., Yoo,C.J., Xue,D., Shi,H. and Wolin,S.L. (1997) The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA, 3, 1434–1443. [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee,C.P. and RajBhandary,U.L. (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem., 266, 24712–24718. [PubMed] [Google Scholar]
- Wolin S.L. and Cedervall,T. (2002) The La protein. Annu. Rev. Biochem., 71, 375–402. [DOI] [PubMed] [Google Scholar]
- Xue D., Rubinson,D.A., Pannone,B.K., Yoo,C.J. and Wolin,S.L. (2000) U snRNP assembly in yeast involves the La protein. EMBO J., 19, 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.J. and Wolin,S.L. (1997) The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]
- Zavanelli M.I. and Ares,M.,Jr (1991) Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 structural element. Genes Dev., 5, 2521–2533. [DOI] [PubMed] [Google Scholar]
- Zavanelli M.I., Britton,J.S., Igel,A.H. and Ares,M.,Jr (1994) Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol. Cell. Biol., 14, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman,K.M., Ortega,J., Steven,A.C. and Storz,G. (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell, 9, 11–22. [DOI] [PubMed] [Google Scholar]