Abstract
Mucin gene 19 (MUC19) has been identified as a major gel-forming mucin in the human middle ear (ME). The objectives of this investigation were to characterize the expression and assess the regulation of MUC19 in the ME cell culture models utilized in the study of otitis media (OM). Findings demonstrate that MUC19 is expressed in both human immortalized cell culture (HMEEC) and chinchilla primary epithelial culture (CMEEC). ME exposure to inflammatory cytokines TNF-α, IL-1β, IL-6 and IL-8 up-regulate MUC19 transcription, most robustly after exposure to TNF-α. Kinetic experiments suggest a relative early response in MUC19 transcription and a down-regulation after prolonged exposure. Glycoprotein production was increased in response to the increased transcription as well. Similar to other mucin genes in the ME, MUC19 is differentially regulated after exposure to inflammatory cytokines. The large size, gel-forming properties and up-regulation in response to important inflammatory cytokines of MUC19 suggest that it has significant potential to play a role in both physiology and pathophysiology of the ME.
Keywords: MUC19, mucin, otitis media, cytokines
BACKGROUND
Infectious and inflammatory diseases of the middle ear, collectively referred to as otitis media (OM), represent one of the most common clinical entities requiring medical attention. OM is the most common diagnosis in pediatric patients who visit physicians for illness in the United States, and is responsible for approximately 5 billion dollars in health care expenditures in the US on an annual basis [1]. Increasing difficulties with antimicrobial resistance has led to an increased need for a more thorough molecular understanding of OM in attempts to limit antimicrobial usage and to develop novel and efficacious treatment options. In addition, OM is responsible for significant morbidity in children who develop chronic disease states. These include the development of chronic otitis media with effusion (OME) which is a leading cause of hearing loss in children and can lead to speech, language and developmental delays.
Mucins are high molecular weight glycoproteins (2-30MDa) that are implicated in a variety of pathologic conditions but are particularly important in respiratory epithelium such as that found in the middle ear. Variation in the quantity and character of middle ear secretions, and specifically mucin secretion, is known to be important in the pathophysiologic mechanisms of otitis media [2, 3]. Mucins are the only component of middle ear effusions responsible for its viscous properties and an over-production of these viscous mucins can prevent normal mucociliary clearance [4], which, in turn, predisposes children to OME and hearing loss. However, mucins are also known to be important in normal host defenses through participating in mucociliary clearance of pathogens, providing protective barriers to underlying epithelium and interacting with the host’s innate immune mechanisms [5]. The mounting evidence of the significance of mucin in middle ear pathophysiology and the recent discovery of MUC 19 in human middle ear epithelium (MEE) prompted the current investigation. Given this important role of mucin in the physiology of the middle ear space, investigations that provide insight into middle ear mucin function and regulation may allow meaningful new intervention strategies for otitis media by incorporating a concept of modulating mucin production by MEE.
METHODS
Human Middle Ear Epithelial Cell Culture (HMEEC)
Cells used in human middle ear cultures were HMEEC (provided by Dr. David Lim, House Ear Institute) whose primary characterization regarding transformation and growth properties have been published [6]. Stock supply of HMEEC were preserved in 106 cell/ml aliquots and stored in liquid nitrogen. The cells were thawed and grown in a 50%/50% mixture of Bronchial Epithelial Cell Basal Medium (Cambrex, East Rutherford, NJ) and Dulbecco’s Modified Eagle Medium (Invitrogen, Carlsbad, CA). Each 500 ml portion of media was supplemented with BEGM Singlequots (Cambrex) which contains: 13mg/ml bovine pituitary extract, 2 ml; 5 mg/ml insulin, 0.5 ml; 0.5mg/ml hydrocortisone, 0.5 ml; 0.1 mg/ml retinoic acid, 0.5 ml; 10 mg/ml transferrin, 0.5 ml; 6.5 mg/ml triiodothyronine, 0.5 ml; 0.5 mg/ml epinephrine, 0.5 ml; 0.5 mg/ml human epidermal growth factor, 0.5 ml; and 50 mg/ml gentamicin, 50 mg/ ml amphotericin B, 0.5 ml. Antibiotic/Antimycotic (Invitrogen) was also added to the medium. The media was changed every 3 days. When the cells reached approximately 70% confluency, they were removed from the flask using Trypsin/EDTA (Cambrex) and incubated in the flask for 5 minutes at 37°C. Once the cells were detached, Trypsin Neutralization Solution (Cambrex) was added to the flask. The cells were pelleted, washed and prepared for experimentation with excess cells refrozen.
The cells were counted by hemocytometer and plated in 12-well plates at approximately 1 × 105 cells per cm2. The cells were grown in a humidified atmosphere at 37°C containing 95% air – 5% carbon dioxide. Growth media was changed every 3rd day. After growth in this environment for approximately 6 days, immunohistochemical analysis on cells grown in a parallel plate was performed to assess middle ear epithelial growth and population. Cells were grown to near-confluency and then prepared for experimentation. Cell viability was assessed with Trypan blue staining.
Primary Chinchilla Middle Ear Epithelial Cell Cultures (CMEEC)
The techniques for establishing primary chinchilla middle ear epithelial cell cultures (CMEEC) have been previously published in our laboratory [7, 8]. Cells used in CMEEC cultures were harvested from mature (6–10 month old), 400–600g mixed breed chinchillas (R&R chinchilla, Jenera, OH). All animals were treated in accordance with the specific guidelines of the Institutional Animal Care and Use Committee at the Medical College of Wisconsin.
Briefly, after euthanasia of the chinchilla, the temporal bones were removed, and 0.09% protease Type XIV (Sigma, St. Louis, MO) in Dulbecco’s Modified Eagle’s Medium (DMEM)/F-12 (Invitrogen) with 0.5% fetal bovine serum (FBS) (Invitrogen) and antibiotic/antimycotic solution was used to fill the middle ear cavity and bulla and left to incubate overnight. The suspension within the middle ear containing the middle ear epithelial cells was then aspirated. Suspensions were pooled and centrifuged.
The cells were plated in 24-well plates at approximately 1 × 105 cells per cm2. The cells were grown in a humidified atmosphere at 37°C containing 95% air – 5% carbon dioxide. Immunohistochemical analysis on cells grown on separate plates was used to assess middle ear epithelial growth and population. Cells were grown to confluency and then prepared for experimentation. A total of four chinchillas were needed for generation of approximately 5 × 106 cells. Cell viability was assessed with Trypan blue staining. Cell cultures were primary cultures and did not undergo multiple passages. In order to limit animal utilization with primary cultures, corroboration of HMEEC experiments with CMEEC were only conducted for TNF-α and IL-1β given the similarity of results obtained with these 2 cytokines.
Exposure to cytokines
Cells were seeded at 1×105 cells/cm2, and grown to 70% confluency. The pre-confluent cells were exposed to different concentrations of cytokines (0, 25, 50, 100, 200ng/ml, R&D Systems, Minneapolis, MN) at various time points. After 1,2,4, and 6 hours of exposure total RNA was isolated using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. The on-column DNase I digestion was performed on each RNA sample to remove potential DNA contamination. The RNA was then reverse-transcribed using SuperScript III Frist Strand Synthesis System (Invitrogen). Random primed first strand cDNA was stored at −80°C until further use.
Reverse Transcriptase PCR
Total RNA was harvested using the RNeasy Mini Kit (Qiagen, Valencia, CA). DNase digestion was performed using RNAse-free DNAse set (Qiagen). Yield and purity were determined by spectrophotometry. Purified RNA was stored at −80°C until RT-PCR analysis. cDNA was obtained using Superscript III First Strand Synthesis System (Invitrogen, Carlsbad, CA). Each reverse transcriptase reaction used 1 µg of the previously purified RNA. A negative control that contained every component except the reverse transcriptase was included for each RNA sample. The cDNA was amplified by PCR (GeneAmp 2720, Perkin-Elmer, Waltham, MA). The 20 µl reaction mixture for PCR contained 1.0 unit Platinum Taq DNA polymerase (Invitrogen), 0.2mM each of dNTPs (Invitrogen), 0.2 µM of each primer, and 1 µl of cDNA template. The PCR reactions were run on 2% agarose gel at 100 V and the product was visualized with GelStar (Cambrex). MUC19 had been primarily identified in middle ear tissue in our laboratory previously [9]. Primer pairs (Table 1) were specifically designed for measurement of MUC19 in HMEEC and in vivo middle ear tissue [9]. Reliability of the cDNA identified in the gels and generated by the selected primer pairs was further assessed by cDNA sequencing and confirmed by comparison of the generated sequence to previously published sequence of MUC19.
Table 1.
Primer pair sequences for MUC 19 identification in human and chinchilla middle ear epithelium using RT-PCR.
| Mucin gene | Primer sequence | Size (bp) |
|---|---|---|
| human MUC19 | sense-ACCAC AAGTA TCCCA GCCAG antisense-AGCTG GTGGA AGTGA GGCTA |
94 |
| chinchilla Muc19 | sense- GTCAC GGTCC ATTGG GAGAA antisense- TAGGA GACAC AGGGG TTGCT |
274 |
Quantitative Real Time PCR
Quantitative PCR was carried out on an iCycler iQ (Biorad, Hercules, CA) using TaqMan probe chemistry (Applied Biosystems , Foster City, CA ). TaqMan primer/probes for human MUC19 (P/N # Hs00543315_m1) and HPRT (P/N #Hs99999909_m1) were obtained commercially. The primer/probes for chinchilla Muc19 and HPRT were custom designed using File Builder 3.1 software (Applied Biosystems). Samples were analyzed in triplicate and included no template and no reverse-transcriptase control for each gene. The mRNA level of MUC19 was normalized to HPRT gene. The relative fold change was then calculated using 2-ΔΔCt method [10].
Statistic analysis
Statistic analysis was performed using Sigma Stat package. Time- and dose-response in HMEEC were analyzed using two-way ANOVA to further assess the impact of interaction of both factors in MUC19 expression. Data from time response of Muc19 in CMEEC and protein assay were analyzed by t-test. Significance level was determined at p-value ≤ 0.05.
Immunohistochemistry
To determine MUC19 protein expression in human middle ear, frozen sections (8um) of middle ear mucosa obtained from patients with chronic otitis media with effusion were stained with anti-MUC19 antibody. The slide was fixed in 4% paraformaldehyde for 20 min and permeabilized with Triton-X on ice for 3 min. To block nonspecific reactions, the section was immediately incubated in blocking solution containing 5% normal goat serum and 0.3% Triton-X in PBS for 1 hr. The section was then incubated with polyclonal chicken anti-human MUC19 (a generous gift from Dr. Yin Chen, Hamner Research institute, NC, USA) at dilution 1:2000 at 4°C over night. Excess primary antibody was removed by four 15-min washing with PBS, and follow by washing with 0.1M Tris pH 8.0. The section was further incubated with Alexa Fluor 488 conjugated goat anti-chicken IgY (Invitrogen) for 90 min, followed by two 15 min washing with 0.1M Tris, pH 8.0 and then mounted. Another section staining with secondary antibody only was used as negative control.
Relative Quantitation of MUC19 by indirect ELISA
HMEEC were seeded and grown to 70% confluency in 6-well plate. The cells were exposed to TNF-α (R&D systems) at 37°C and 5% CO2. Cells were exposed to TNF-α at concentrations of 25ng/ml and 200ng/ml for time periods of 2 hours and 16 hours. Following this, ELISA was performed as described below. Although the greatest increase in MUC19 mRNA was demonstrated at 2 hours the ELISA protein measurements after exposure to 200ng/ml at 16 hours were also incorporated based on our previous work demonstrating this as the optimum time frame to detect an increase in mucin protein production from HMEEC after exposure to TNF-α [11].
In performing ELISA, after TNF-α exposure, media was then collected for protein assay. Total protein concentration was determined using BCA protein assay kit (Pierce, Rockford, IL). To determine MUC19 level, indirect ELISA using serial dilution curves were performed as described in [12]. In brief, total protein from spent media were serially diluted in carbonate-bicarbonate buffer, pH 9.0, to coat microtiter plates at 4°C over night. Nonspecific reactions were blocked with 5% normal horse serum in PBS containing 0.05% Tween and the plates were then incubated with chicken anti-MUC19 antibody (1:4000) for 1hr. After washing, MUC19 was detected using HRP conjugated goat anti-chicken IgY at 1:10,000 (Abcam, Cambridge, MA). Color development was performed using TMB (Sigma). The range of linear response for each sample was determined by linear regression analysis. The relative amounts of MUC19 from different samples were compared using absorbance from a single concentration point within the midlinear range of the regression line.
RESULTS
Identification of MUC19
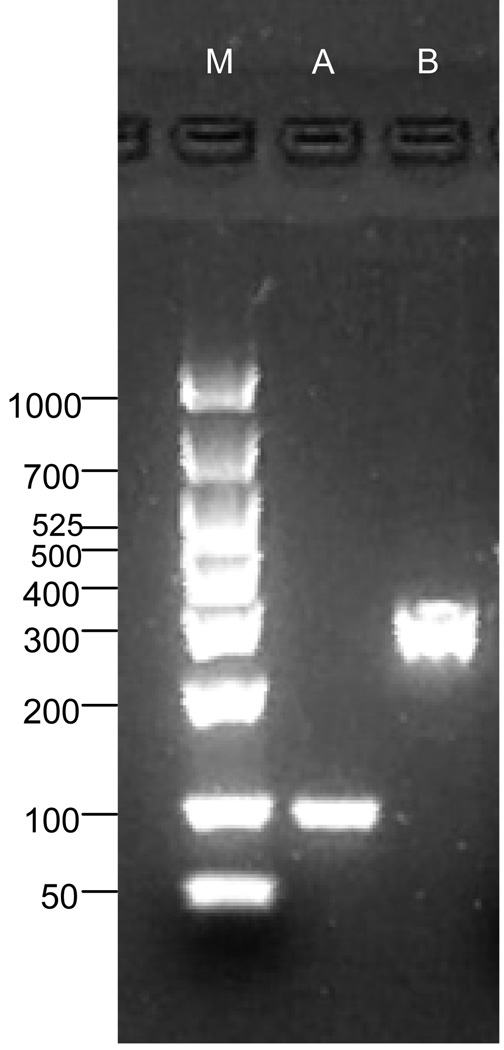
MUC19 mRNA was identified in HMEEC and demonstrated by appropriate RT-PCR bands (Figure 1, lane A) and with appropriate sequencing utilizing the primers defined in Table 1. Muc19 was also consistently identified in primary cell cultures of the chinchilla (Figure 1, lane B)
Fig. 1.
RT-PCR gel of mucin gene 19 in A) HMEEC, and B) CMEEC
qPCR with inflammatory cytokine exposure
TNF-α
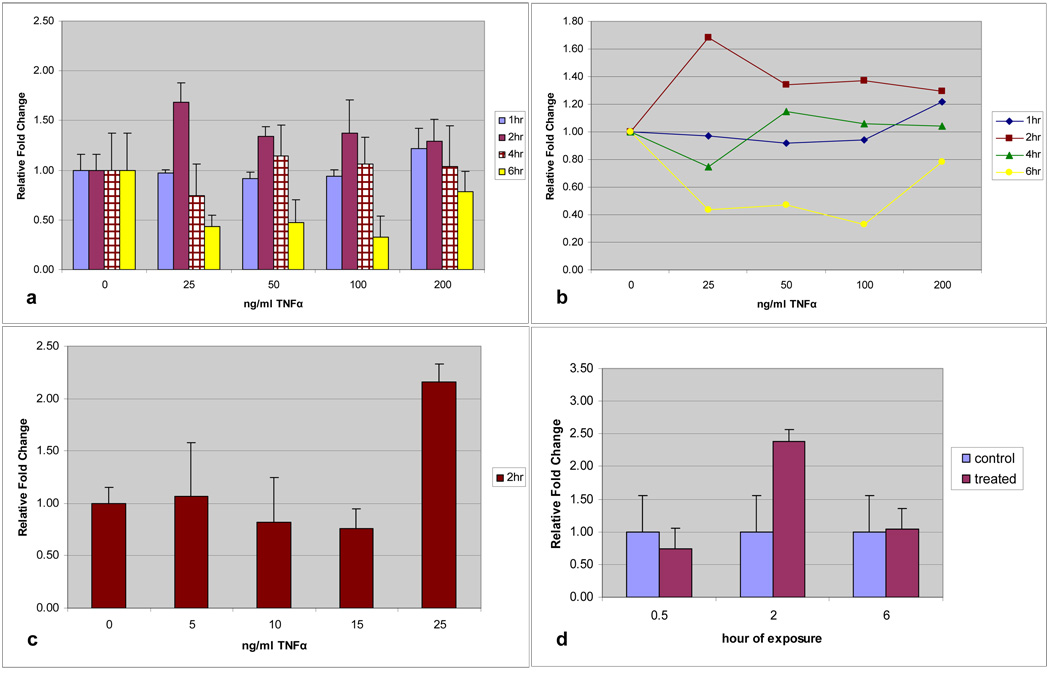
HMEEC exposed to TNF-α displayed a demonstrable increase in MUC19 expression at all concentrations, but not all time intervals (Figure 2a). Time-dose interaction plotting (Figure 2b) demonstrated the greatest effect compared with controls consistently at 2 hours of exposure and a concentration of 25ng/ml TNF-α. Utilizing time as a dependent variable the 2-hour exposure time point was significantly different than other exposure times (p=0.001). HMEEC exposed to 25ng/ml TNF-α for 2 hours demonstrated a significant (p=0.029) and greater than 2-fold increase in MUC19 expression (Figure 2c). Demonstration of the differential gene expression with this cytokine is illustrated in the down regulation of MUC19 at periods of more prolonged exposure (6-hours).
Fig. 2.
a) Time and dose response of MUC19 expression in HMEEC following TNF-α exposure. Greatest up-regulation at 2 hours and 25ng/ml. b) Time-dose interaction plot for MUC19 expression following TNF-α exposure. c) TNF-α dose up-regulation of MUC19 at level below 25ng/ml. Only significant increase at 25ng/ml (p=0.029). d) Time response of MUC19 expression in CMEEC following TNF-α exposure at 100ng/ml with significant increase at 2 hours (p=0.005).
Similarly, up-regulation for Muc19 was demonstrated in CMEEC. The greatest response was demonstrated at 100ng/ml and kinetic experiments demonstrated that, similar to HMEEC, the greatest effect on MUC19 transcription compared with controls occurred at 2 hours of exposure. CMEEC exposed to 100ng/ml TNF-α for 2 hours demonstrated a significant increase (p=0.005) of greater than 2-fold MUC19 gene expression (Figure 2d).
IL-1β, IL-6 and IL-8
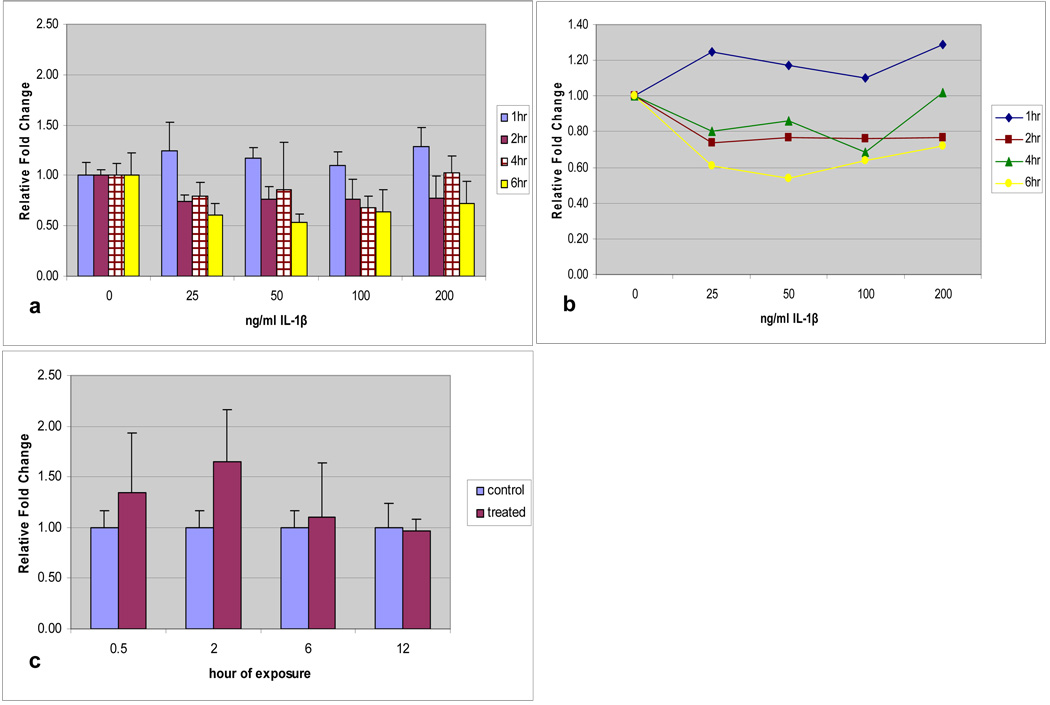
HMEEC exposed to IL-1β also displayed a demonstrable increase in MUC19 expression at all concentrations, but not all time intervals (Figure 3a). Utilizing a time-dose interaction plot (Figure 3b), time was a significant factor with exposure at 1-hour significantly different than other exposure times (p<0.001). Again, prolonged exposure demonstrated an eventual down-regulation of MUC19.
Fig. 3.
a) Time and dose response of MUC19 expression in HMEEC following IL-1β exposure. Greatest up-regulation at 1 hour. b) Time-dose interaction plot for MUC19 expression following IL-1β exposure. c) Time response of Muc19 expression in CMEEC following IL-1β exposure at 100ng/ml with significant increase at 2 hours (p=0.024).
Utilizing a dose-response curve, CMEEC demonstrated the greatest up-regulation at 100ng/ml. Utilizing this dose, to reduce total numbers of animals required for primary cell culture experimentation, up-regulation of Muc19 was greatest at 2 hours of exposure to IL-1β (p=0.024) (Figure 3c). This is similar to the kinetics of Muc19 response to TNF-α in CMEEC which also demonstrated the greatest up-regulation at 2 hours.
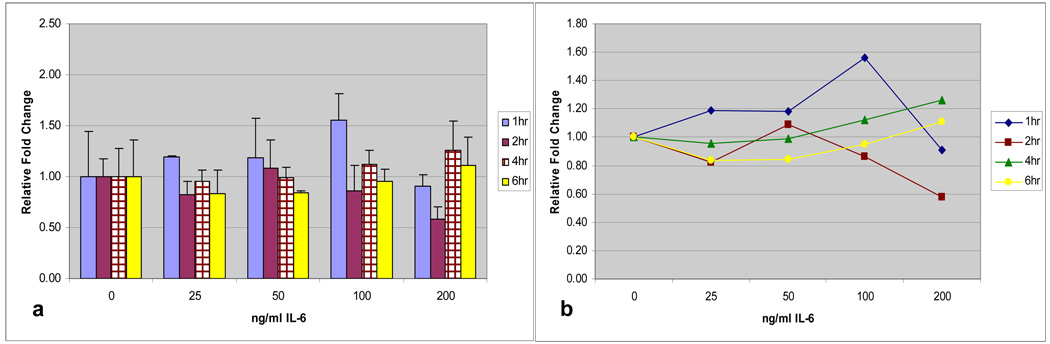
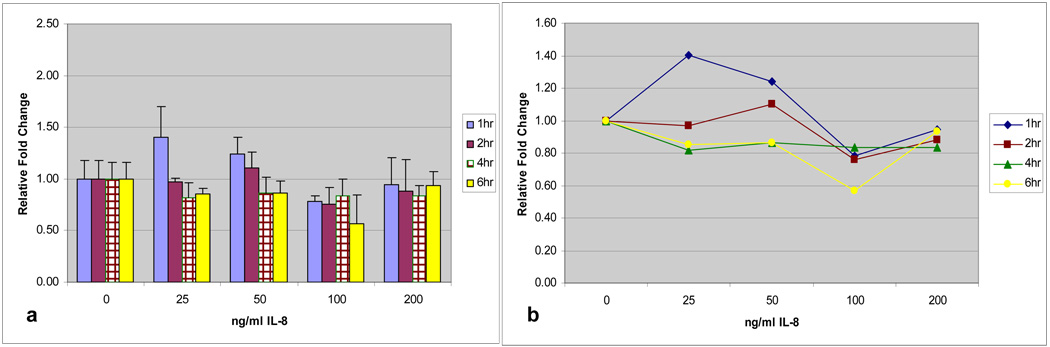
The patterns of regulation demonstrated for TNF-α and IL-1β were similar for IL-6 and IL-8 (Figures 4a and 5a). Early exposure at 1 and 2-hours demonstrated the greatest levels of MUC19 up-regulation compared with controls and time-dose interaction plots (Figures 4b and 5b) showed that time was a significant factor with exposure at 1-hour being significantly different than other exposure times (p=0.007 for IL-6 and p=0.002 for IL-8). Prolonged exposure, especially at 6 hours, continued to demonstrate a down-regulation in MUC19 expression. Additionally, for IL-8, dose at 25ng/ml for 1-hour demonstrated a significantly increased up-regulation compared with controls (p=0.017).
Fig. 4.
a) Time and dose response of MUC 19 expression in HMEEC following IL-6 exposure. Greatest up-regulation is at 1 hour. b) Time-dose interaction plot for MUC19 expression following IL-6 exposure.
Fig. 5.
a) Time and dose response of MUC 19 expression in HMEEC following IL-8 exposure. Greatest up-regulation at 1-hour. b) Time-dose interaction plot for MUC19 expression following IL-8 exposure.
MUC19 glycoprotein expression
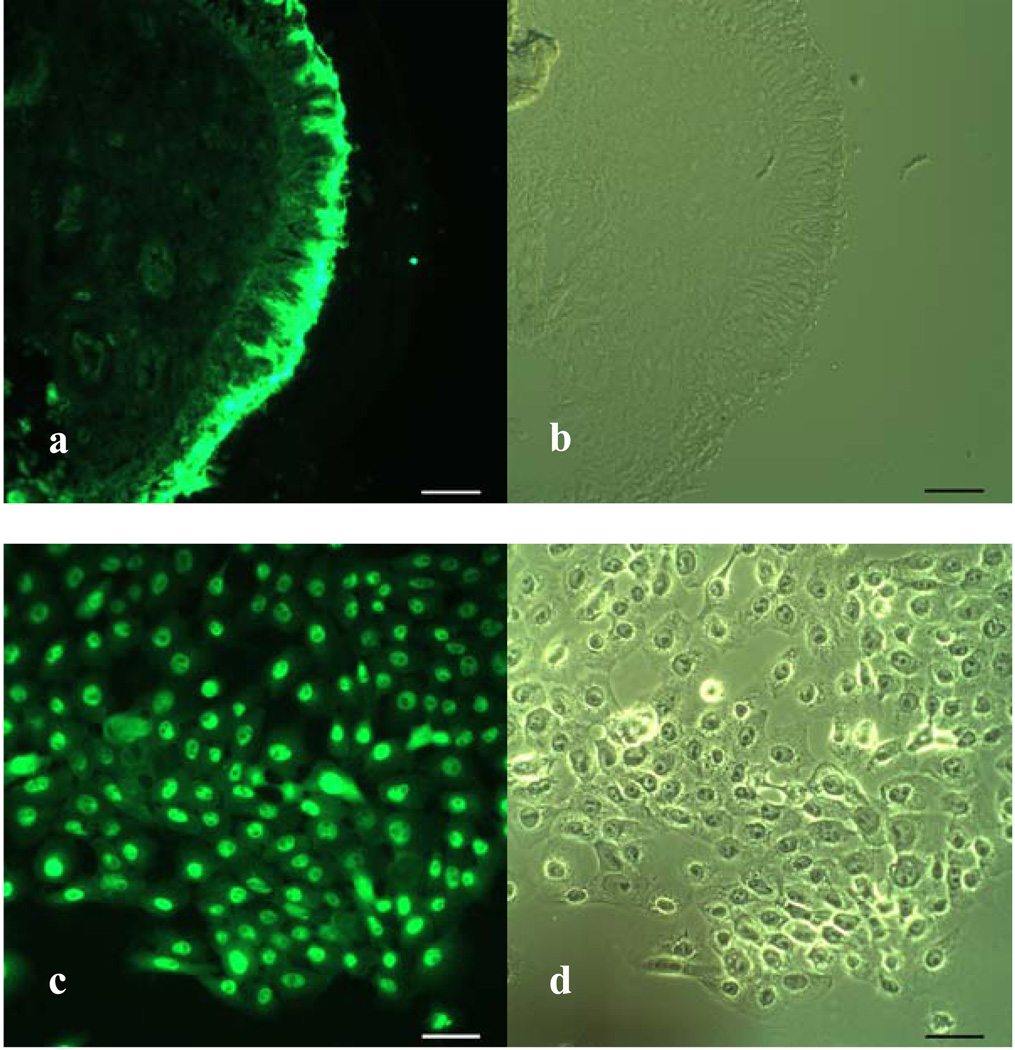
Immunofluorescence staining with a specific antibody revealed that MUC19 protein was very strongly and specifically expressed in the middle ear mucosa (Figure 6a). In human tissue, the protein demonstrated bright staining at the apical layer of the epithelium. In HMEEC, the MUC19 was identified in each of the epithelial cells in culture. Given that the cell culture model does not contain significant depth of tissue, two dimensional imaging was performed (Figure 6b).
Fig. 6.
MUC19 protein in human middle ear mucosa (a,b) and in human middle ear epithelial culture (c,d). Fluorescent staining (a,c) and bright-field microscopic image (b,d). Scale bars; 40µm.
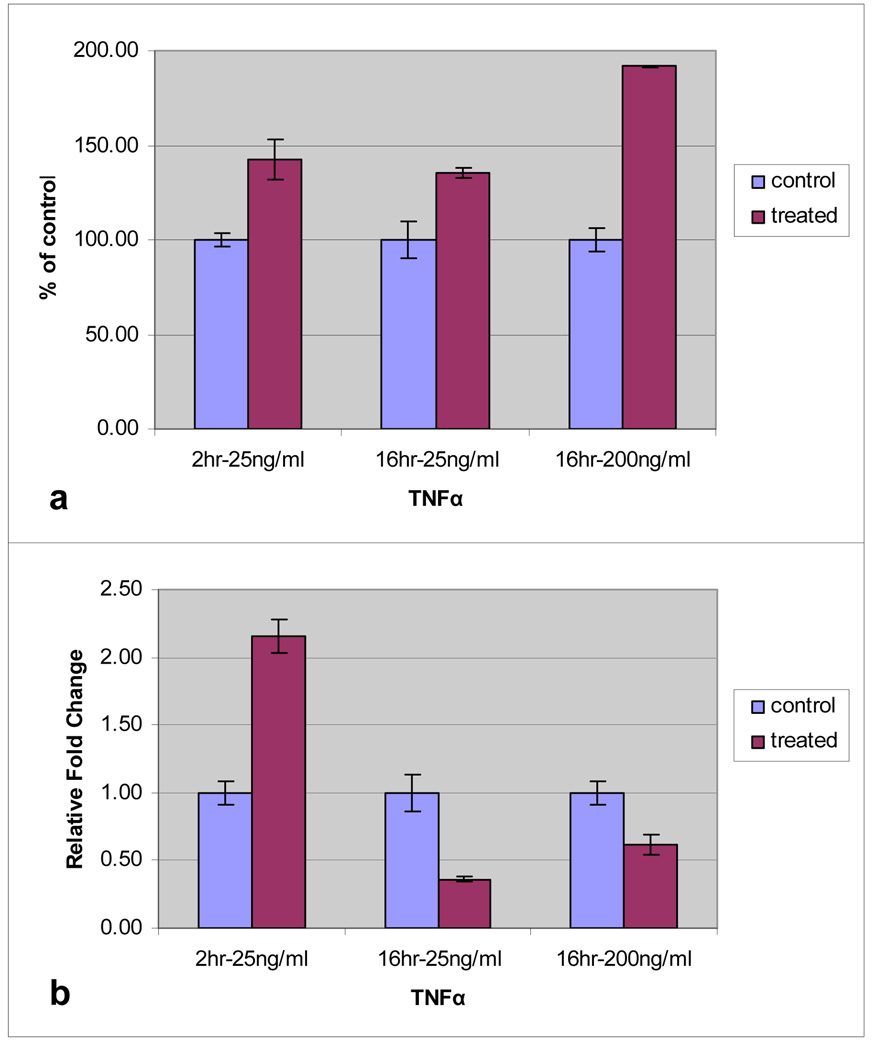
Indirect ELISA measuring secreted MUC19 from HMEEC exposed to 25ng/ml TNF-α for 2 hours demonstrated a difference in level of protein product from that of the control cells at the two-hour time point, p=0.019 (Figure 7a). However, in experiments utilizing data from previous work [11] highlighted that the maximum mucin protein production occurred after a longer exposure to inflammatory cytokines. Exposure of HMEEC to 16 hours of TNF-α at concentrations of 25ng/ml and 200ng/ml, demonstrated a significant elevation in the level of secreted MUC19 (p= 0.022 for 25ng/ml; p≤0.001 for 200ng/ml). In these experiments it was confirmed that the transcript level was decreased after this 16 hour exposure (p=0.009 for 25ng/ml; p=0.028 for 200ng/ml) (Figure 7b)
Fig. 7.
Level of MUC19 expression in HMEEC following TNFα exposure. Cells were exposed to TNFα at 25ng/ml (for 2hr and 16hr) and 200ng/ml (for 16hr). Culture media was assayed for MUC19 protein using indirect ELISA (a). Level of MUC19 mRNA was determined by quantitative PCR (b).
DISCUSSION
The characterization of mucoglycoproteins, or mucins, has expanded to include 20 unique human mucin genes [9]. However, recent data would suggest that previously described mucin genes 11 and 12 actually represent a single glycoprotein; bringing the number of unique human mucin genes back to 19. (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov). Mucins are expressed in many human tissues, and impact many organ systems including the gastrointestinal tract, genitourinary tract and the respiratory tract. In the upper respiratory tract mucins are important in the pathophysiology of OM, sinusitis, laryngeal pathology and head and neck cancer [13,14]. Given the importance of mucins in OM pathology and the prevalence of this disease, our laboratory has recently examined the profile of mucin gene expression in both in vivo human middle ear tissue and an in vitro human middle ear cell line [9]. These experiments demonstrated that a majority of human mucins are expressed in the middle ear, including membrane bound mucins 1 and 4 and secretory, gel-forming mucins 2, 5AC and 5B.
Although each of the mucins produced in a given tissue type undoubtedly has an important function in the normal physiology of that epithelium and has the potential to contribute to pathophysiology, the precise mechanisms and interactions for the varied mucins has not been well-characterized. Each mucin gene product has somewhat different characteristics and an increase or decrease in production of any one mucin creates changes in the composition and function of the mucous blanket overlying the epithelium. These changes affect viscosity, mucociliary clearance, epithelial repair mechanisms and bacterial pathogenesis in the middle ear. As the mucus blanket overlying an epithelium functions as a unit composed of the various mucin gene products, characterizing the entire spectrum of mucin gene products and their response to given stimuli in the middle ear is crucial in understanding the function of the produced mucin during periods of middle ear inflammation as occurs in OM.
The secreted, or gel-forming mucins are of particular interest in middle ear pathogenesis as they are the primary mucins determining the viscoelastic properties of middle ear fluid. As such, these particular mucins are associated with airway epithelial pathology due to their ability to compromise normal mucociliary clearance. This is especially important in chronic disease states such as chronic bronchitis, asthma and chronic otitis media. In addition to the gel-forming mucins, MUC2, MUC5AC and MUC5B, recently, an additional mucin gene, MUC19 has been identified [15]. Sequence information demonstrates that the protein product from this gene is a gel-forming mucoglycoprotein that is the largest of mucin proteins identified to date. As such this mucin has the potential to: significantly contribute to the viscosity of the mucous blanket in epithelial tissues where it is expressed, have an important role in the normal protective functions of mucous in these epithelial tissues and also contribute to pathologic conditions where mucous secretions attain a viscosity that prevents normal secretion clearance [15]. This mucin gene was first identified in human middle ear epithelium in our laboratory [9] and in this investigation it was also demonstrated that this mucin gene is conserved in its expression in the most important animal model of OM, the chinchilla (Figure 1). This expression and identification in the chinchilla is significant to allow for further functional studies of this mucin in this well-established animal model, particularly in the realm of bacterial pathogenesis, and these experiments are currently ongoing.
The investigations presented here focused on MUC19 response to inflammatory mediators. This cascade is complex with numerous distinct proteins having been identified as important regulators in infectious processes. Clinical reports have outlined the importance of TNF-α, IL-1β, IL-6, and IL-8 in otitis media [16]. Elevated levels of IL-1β are present in effusions of younger children and in purulent otitis, increased levels of TNF-α are present in purulent effusions, older children, children who require multiple tympanostomy tubes and children with chronic effusions. IL-6 levels are increased in younger children with acute otitis media and this cytokine has been described as an important regulator during both the early and later stages of otitis media with effusion. IL-8 is an important mediator in chronic otitis media with effusion formation and increased levels of IL-8 in middle ear effusions have correlated with increased recurrence rates for acute otitis media [17]. Our laboratory has demonstrated that these inflammatory cytokines regulate mucin secretion in MEE and produce a differential regulation of mucin gene expression in MEE, specifically with up-regulation of MUC2, in a dose and time-dependent fashion with TNF-α and IL-1β exposure [7].
The experiments in this manuscript represent the first data set examining the potential of MUC19 to be regulated by these important inflammatory cytokines in OM. Understanding that MUC19 expression is increased in response to each of these cytokines, and most significantly by TNF-α, (Figures 2–5) gives insight into how it is incorporated into the overall inflammatory response that takes place in the pathophysiology of OM. These results indicate that this gene can be up-regulated during periods of inflammation in the ME. Given its large size, up-regulation of MUC19 during periods of inflammation in the ME may have important protective roles for the ME. However, over-production or chronically increased production may ultimately lead to decreased mucociliary clearance in the ME due to increased ME fluid viscosity. It is interesting to note that throughout this experimental data set that MUC19 gene expression is up-regulated very early in the period of exposure to inflammatory cytokines, in the one to two hour time period (Figures 2–5). This is then followed by a period of down-regulation of MUC19 mRNA after longer-term exposure to TNF-α. This might suggest an “understanding” of the MEE of the need for early production of this large mucoglycoprotein to provide protection of the underlying mucosa during periods of injury or inflammation but the importance of avoiding long-term up-regulation to avoid difficulties with mucociliary clearance or increased viscosity. Finally, the experiments presented demonstrate ongoing similarities between the culture methods utilized in both the HMEEC and CMEEC models.
Importantly, the experiments detailed in this manuscript also elucidate some of the differences in dose-response and kinetics of mucin gene expression in different model systems utilized (Figures 2–5). These findings will be important for future investigations investigating the function or response of MUC19 in studies of respiratory epithelium.
The protein expression data generated in these investigations also provides important information for future experiments as well as developing an understanding of the complex interaction of the multiple mucin gene products in the middle ear. Our laboratory has previously demonstrated that there is a significant increase in overall production of mucin in human middle ear epithelium exposed to inflammatory cytokines [11]. These experiments, however, did not elucidate which specific mucin proteins expressed in the MEE were responsible for the overall up-regulation of mucin production. This current work demonstrates that the protein MUC19 is expressed in both human in vivo and in vitro ME tissue and in chinchilla MEE (Figure 2). Tissue biopsies of human MEE examined by immunohistochemistry demonstrate a robust expression of MUC19 at the apical area of the epithelium. This apical expression would be expected given the secretory nature of this gel-forming mucin. Similarly, HMEEC, demonstrates uniform staining with MUC19 antibody (Figure 6).
ELISA assessment of MUC19 glycoprotein production in HMMEC demonstrated significant increased production in multiple conditions (Figure 7). These experiments demonstrate a response to the early up-regulation of MUC19 transcription with significant increase in glycoprotein production. The demonstration that these inflammatory events lead to both transcriptional and translational changes in MUC19 in HMEEC suggests the need for further investigations into the specific functionality of MUC19 in the ME. These results also further corroborate earlier work that the HMEEC model is effective as an experimental model in the study of mucin regulation in the ear and provides the ability to study both transcriptional and translational events.
Although the precise biologic functions of MUC19 have not been completely investigated it has been demonstrated that MUC19 is expressed in conjunctival goblet cells and therefore may also be regulated by mechanisms controlling goblet cell function [18]. Goblet cells certainly have been identified as important in respiratory epithelium, including MEE. The primary cell culture model utilized for these experiments precludes goblet cell experimentation, however, this is an area that warrants further study in the ME and is ongoing in our laboratory. Interestingly, in the eye, another important gel-forming mucin produced by goblet cells, MUC5AC, was expressed in the conjunctiva and lacrimal gland along with MUC19. However, in the cornea, MUC19 was expressed in the absence of MUC5AC. These observations further highlight the specific functionality that each mucin product likely exhibits and the need for specific regulatory understanding of each of these mucins to more completely decipher the meaning of molecular changes occurring within the full spectrum of MEE mucin genes during periods of ME inflammation such as OM. These avenues of investigation, along with of more thorough study of pathogen interactions with mucin products are areas of ongoing interest and investigation.
Acknowledgments
Supported by NIH grant NIDCD: DC007903 (JEK), Also supported in part through funding provided by the Department of Otolaryngology and Communication Sciences, Medical College of Wisconsin
Footnotes
Portions of this manuscript presented at the Association for research in Otolaryngology Annual Midwinter Meeting, AZ, February 17, 2008.
References
- 1.Rosenfeld RM, Casselbrant ML, Hannley MT. Implications of the AHRQ evidence report on acute otitis media. Otolaryngol. Head Neck Surg. 2001;125:440–448. doi: 10.1067/mhn.2001.119326. [DOI] [PubMed] [Google Scholar]
- 2.Brown DT, Litt M, Potsic WP. A study of mucus glycoproteins in secretory otitis media. Arch. Otolaryngol. 1985;111:688–695. doi: 10.1001/archotol.1985.00800120082011. [DOI] [PubMed] [Google Scholar]
- 3.Carrie S, Hutton DA, Birchall JP, Green GG, Pearson JP. Otitis media with effusion: components which contribute to the viscous properties. Acta. Otolaryngol. 1992;112:504–511. doi: 10.3109/00016489209137432. [DOI] [PubMed] [Google Scholar]
- 4.FitzGerald JE, Green GG, Stafford FW, Birchall JP, Pearson JP. Characterization of human middle ear mucus glycoprotein in chronic secretory otitis media (CSOM) Clin. Chim. Acta. 1987;169:281–297. doi: 10.1016/0009-8981(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 5.Zeiher BG, Hornick DB. Pathogenesis of respiratory infections and host defenses. Curr. Opin. Pulm. Med. 1996;2(3):166–173. doi: 10.1097/00063198-199605000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Jono H, Shuto T, Xu H, Kai H, Lim DJ, Gum JR, Jr, Kim YS, Yamaoka S, Feng XH, Li JD. Transforming Growth Factor-β-Smad Signaling Pathway Cooperates with NF-B to Mediate Nontypeable Haemophilus influenzae-induced MUC2 Mucin Transcription. J. Biol. Chem. 2002;277:45547–45557. doi: 10.1074/jbc.M206883200. [DOI] [PubMed] [Google Scholar]
- 7.Kerschner JE, Meyer TK, Burrows A. Chinchilla middle ear epithelial mucin gene expression in response to inflammatory cytokines. Arch. Otolaryngol. Head Neck Surg. 2004;130:1163–1167. doi: 10.1001/archotol.130.10.1163. [DOI] [PubMed] [Google Scholar]
- 8.Kerschner JE, Meyer TK, Burrows A, Ehrlich G, Post JC. Mucin gene cDNA sequence characterization in chinchilla middle ear epithelium. Int. J. Pediatr. Otorhinolaryngol. 2006;70(8):1449–1456. doi: 10.1016/j.ijporl.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Kerschner JE. Laryngoscope. 9. Vol. 117. 2007. Mucin gene expression in human middle ear epithelium. American Laryngological, Rhinological and Otological Society Thesis; pp. 1666–1676. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Samuel EA, Burrows A, Kerschner JE. Cytokine regulation of mucin secretion in a human middle ear epithelial model. Cytokine. 2008;41(1):38–43. doi: 10.1016/j.cyto.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argueso P, Gipson IK. Quantitative analysis of mucins in mucosal secretions using indirect enzyme-linked immunosorbent assay. In: Brockhausen I, editor. Glycobiology Protocols: Methods in Molecular Biology. v.347. New Jersey: Humana Press Inc; 2006. [DOI] [PubMed] [Google Scholar]
- 13.Baek SK, Woo JS, Kwon SY, Lee SH, Chae YS, Yung KY. Prognostic significance of the MUC1 and MUC4 expressions in thyroid papillary carcinoma. Laryngoscope. 2007 May;117(5):911–916. doi: 10.1097/MLG.0b013e31803d1720. [DOI] [PubMed] [Google Scholar]
- 14.Samuels TL, Handler E, Syring ML, Blumin JH, Kershner JE, Johnston N. Mucin gene expression in human laryngeal epithelium: effect of laryngopharyngeal reflux. Arch. Otolaryngol. Head Neck Surg. doi: 10.1177/000348940811700911. In Press. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Zhao YH, Kalaslavadi TB, Hamati E, Nehrke K, Le AD, Ann DK, Wu R. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am. J. Respir. Cell Mol. Biol. 2004;30(2):155–165. doi: 10.1165/rcmb.2003-0103OC. [DOI] [PubMed] [Google Scholar]
- 16.Sato K, Nonomura N, Kawana M, Nakano Y. Course of IL-1b, IL-6, IL-8 and TNF-a in the middle ear fluid of the guinea pig otitis media model induced by nonviable Haemophilus influenza. Ann. Otol. Rhinol. Laryngol. 1999;108:559–563. doi: 10.1177/000348949910800606. [DOI] [PubMed] [Google Scholar]
- 17.Chonmaitree T, Patel JA, Garofalo R, et al. Role of leukotrienne B4 and interleukin-8 in acute bacterial and viral otitis media. Ann. Otol. Rhinol. Laryngol. 1996;105:968–974. doi: 10.1177/000348949610501207. [DOI] [PubMed] [Google Scholar]
- 18.Yu DF, Chen Y, Han JM, Zhang H, Chen XP, Zou WJ, Liang LY, Xu CC, Liu ZG. MUC19 expression in human ocular surface and lacrimal gland and its alteration in Sjögren syndrome patients. Exp. Eye. Res. 2008 Feb;86(2):403–411. doi: 10.1016/j.exer.2007.11.013. [DOI] [PubMed] [Google Scholar]