Abstract
Y-chromosome and mitochondrial DNA markers can reveal phylogenetic patterns by allowing tracking of male and female lineages, respectively. We used sequence data from a recently discovered Y-linked marker and a mitochondrial marker to examine phylogeographic structure in the widespread and economically important rainbow trout (Oncorhynchus mykiss). Two distinct geographic groupings that generally correspond to coastal and inland subspecies were evident within the Y marker network while the mtDNA haplotype network showed little geographic structure. Our results suggest that male-specific behavior has prevented widespread admixture of Y haplotypes and that gene flow between the coastal and inland subspecies has largely occurred through females. This new Y marker may also aid conservation efforts by genetically identifying inland populations that have not hybridized with widely stocked coastal-derived hatchery fish.
Keywords: phylogeography, sex-biased gene flow, sex-linked marker, subspecies, Y chromosome
1. Introduction
The comparison of intraspecific variability between the maternally-inherited mitochondrial genome and paternally-inherited Y chromosome has provided insights into sex-biased life history patterns for many organisms (Boissinot and Boursot 1997, Eriksson et al. 2006, Pidancier et al. 2006). The mitochondrial genome has been used extensively to reconstruct phylogenies and examine post-glacial dispersal and phylogeographic patterns (Avise 1986, Danzmann et al. 1998, Bernatchez 2001). Use of Y haplotypes in comparable studies has been less extensive and limited to humans (e.g., Underhill and Kivisild 2007) and other mammals (e.g., Tosi et al. 2003, Sundqvist et al. 2006). This limitation is because the X and Y chromosomes in fishes and amphibians are similar in genetic content and their Y-specific regions are relatively small and challenging to isolate (Schartl 2004, Kondo 2006, Smith and Voss 2009). Although only about 10% of all fish species show morphologically apparent sex chromosomes, a high proportion appear to show a simple genetic mechanism with a single chromosome having a major influence on sex determination (Devlin and Nagahama 2002). Thus, the isolation of additional Y-specific sequences in fishes and other non-mammalian vertebrates will likely enable the evolution of Y-specific markers to be studied in many of these animals.
The rainbow trout is an economically important sport and food fish with a native distribution on both sides of the North Pacific Ocean. The subspecies and classification of rainbow trout have been reviewed by Behnke (2002). The two most widespread subspecies are the coastal rainbow (Oncorhynchus mykiss irideus) along the Pacific Coast of North America and the inland redband rainbow (O. m. gairdneri) in areas east of the Cascade Mountains in the USA and the Coast Range in British Columbia, Canada. Interest in sport fishing and the ease of hatchery rearing has resulted in a world-wide distribution of the coastal rainbow trout subspecies (Halverson 2010). Within the United States, stocking practices have distributed the coastal subspecies widely throughout the range of the inland subspecies. The coastal and inland lineages are recognized by differences in coloration and numbers of pyloric caecae, scales along the lateral line, vertebrae and gill rakers (Behnke 1992). Differences in microsatellite and allozyme frequencies have often been used to study introgression and hybridization between these subspecies (Utter 2001, Knudsen et al. 2002, Small et al. 2007).
The salmonids are famously known for their anadromy and life history of spawning in fresh water, followed by hatch and juvenile residence in fresh water prior to parr-smolt transformation and oceanic migration. After several years of oceanic maturation, the anadromous form completes this life-cycle by precise homing to their natal stream to spawn and, among Pacific salmon, die (Dittman and Quinn 1996; Quinn 2005). Homing has lead to reproductive isolation within spawning populations, and selection for adaptations to specific environments. The trouts including O. mykiss are able to optionally remain in fresh water throughout their entire life cycle and survive spawning. Rainbow trout have an ocean-going form (anadromous steelhead) when given access to the sea. Steelhead do not show consistent genetic distinctions from non-migratory resident rainbow trout (McMillan et al. 2007, Narum et al. 2004). Non-anadromous “resident” males, which can mature and spawn without going to sea are common in some populations. Even though the resident males are significantly smaller than the sea-going migrants of the same population, mature resident males contribute successfully during spawning, effectively creating higher homing fidelity (Hendry et al. 2004). In years of low steelhead spawning return, resident precocial parr can sometimes reproductively contribute more than the anadromous form (Seamons et al. 2004).
Previous evaluations of salmonid sex-biased dispersal in migrating and resident populations have yielded mixed results. Male-biased dispersal has been predicted to be a response to reduced mate competition where females are limiting, and this speculation is supported in mark-recapture evaluations of brook trout populations, Salvelinus fontinalis (Hutchings and Gerber 2002). Evaluations of sex-biased dispersal in rainbow trout (Olsen et al. 2006), and Atlantic salmon do not detect sex-biased dispersal (Consuegra and Garcia de Leaniz 2007). Conversely, other evaluations of lake-dwelling brook trout sex-biased dispersal have detected a spatial component in which male-mediated gene flow is restricted, while the females are more widely dispersing (Fraser et al. 2004). Female-biased dispersal has also been reported in evaluations of stream-dwelling Dolly Varden (Salvelinus malma) populations (Koizumi et al. 2006).
The recent discovery of a polymorphic Y-linked marker in rainbow trout (Brunelli et al. 2008) has allowed us to study the geographic distribution of this new genetic marker among rainbow trout populations. This marker was initially identified through homology to a sequence isolated in the closely related Chinook salmon (Oncorhynchus tshwytscha) from an AFLP band present in males but not in females (Brunelli and Thorgaard 2004). Because the male-specific region of the sex chromosome does not recombine it provides an accurate legacy of paternal lineages.
We present the first Y-haplotype phylogeographic evaluation of a fish species. We have found a surprisingly deep divergence between the Y chromosome sequences of inland and coastal rainbow trout populations without a corresponding difference between their mitochondrial DNAs. The difference in patterns between Y markers and mtDNA may be related to differences in dispersal and evolutionary history between the sexes. The two main Y haplotype lineages correspond geographically to the two main subspecies. We also examined mtDNA haplotypes from the same individuals and found that the mtDNA haplotypes do not show the same subspecies associations. Our results suggest that male-specific behaviors may have contributed to the maintenance of subspecific Y haplotypes and that introgression between the subspecies is mediated by females.
2. Materials and methods
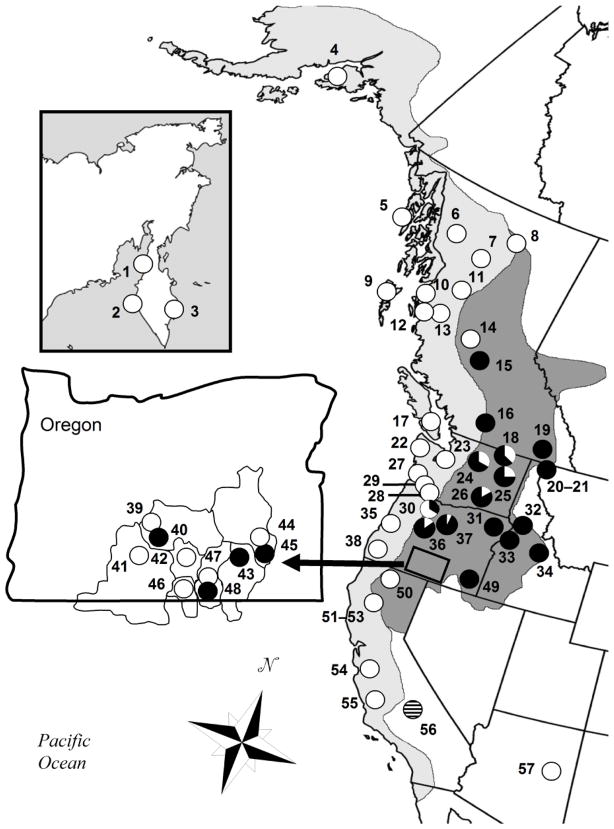
We sampled 333 male rainbow trout (Oncorhynchus mykiss) representing four subspecies (coastal [O. m. irideus], inland [O. m. gairdneri], Kamchatka [O. m. mykiss] and California golden [O. m. aquabonita]) from 57 localities in western North America and Russia (Figure 1; S1). Samples from a closely related species, the Apache trout (O. apache), were also included for comparisons. All samples were sequenced for Y-linked and mitochondrial markers (Table 1).
Fig. 1.
Range and sampled localities of rainbow trout (Oncorhynchus mykiss) within western North America. Light grey indicates approximate range of coastal subspecies (O. m. irideus) and dark grey indicates inland subspecies (O. m. gairdneri). Insets show sampled localities on the Kamchatka Peninsula in Russia, and within the internally draining basins of Oregon. Pie charts represent proportion of individuals sampled with inland (black) or coastal (white) Y haplotypes. Numbers correspond to localities in Table 1. Locality 56 represents Golden trout which has an intermediate Y haplotype (Figure 2).
Table 1.
Locality, sample size, haplotypes recovered for samples. Populations sampled are described in Supplementary Table 1.
| Number | Location | Y-marker Haplotype (Freq.) | mtDNA MYS Haplotype (Freq.) |
|---|---|---|---|
| 1 | Voyampolka R., Russia | C2(2) | 1K(2) |
| 2 | Sedanka R., Russia | C2(2) | 1K(2) |
| 3 | Zhupanova R., Russia | C1(2); C2(1) | 1K(2); N4(1) |
| 4 | Swanson R., AK | C1(9) | 1K(4);12D(4); N8(1) |
| 5 | Sashin Crk., AK | C1(4) | 12D(3); 21(1) |
| 6 | Ealue Lake, BC | C2(4) | 1K(4) |
| 7 | Moosevale Crk, BC | C1(1) | 12D(1) |
| 8 | Turnagain R., BC | C1 (9) | 12D(7); 12E(2) |
| 9 | Yakoun R., BC | C1(7); C2(1) | 1K(2); 12D(5); CUT(1) |
| 10 | Copper R., BC | C1(3); C2(4) | 1K(2); 12D(5) |
| 11 | Canyon Crk, BC | C2(1) | 12D(1) |
| 12 | Zymoetz R., BC | C2(1) | N6(1) |
| 13 | Morice R., BC | C2(1) | 12D(1) |
| 14 | Blackwater R., BC | C3(8) | 1K(7); N6(1) |
| 15 | Tzenzaicut Lake, BC | I2(9) | 1K(7); N7(2) |
| 16 | Pennask Lake, BC | I2(1); I3(8) | 1H(1); N3(8) |
| 17 | Cowichan R., BC | C1(3); C3(1) | 1K(2); 12D(1); N9(1) |
| 18 | W. Fork Trout Crk, WA | C1(3); I2(5) | 1K(8) |
| 19 | Kootenay Lake, BC | I2(2) | 1E(1); 1K(1) |
| 20 | Basin Crk, MT | I2(7); I4(2) | 1K(3); 9(6) |
| 21 | Fisher River, MT | I2(4); I5(2) | 1K(4); 3B(1); N5(1) |
| 22 | Hoh R., WA | C1(5) | 1K(4); 12D(1) |
| 23 | White R., WA | C1(2) | 12D(2) |
| 24 | Wells Hatchery, WA | C1(2); I1(1) | 1K(2); 3B(1) |
| 25 | N. Fork Little Deep Crk., WA | C1(2); I3(6) | 1K(5); 9(2); 10(1) |
| 26 | Touchet R., WA | C1(1); I2(3); I3(1) | 1D(1); 1K(4) |
| 27 | Abernathy Crk., WA | C1(10); C4(1); C6(1) | 1K(2); 12A(1); 12D(9) |
| 28 | Washougal R., WA | C1(4) | 1K(2); 3A(1); 12D(1) |
| 29 | Kalama R., WA | C1(12) | 1K(6); 9(1); 12D(4); N6(1) |
| 30 | Hood R., OR | C1(5);C4(1);I1(1);I2(1);I3(1) | 1D(1); 1K(6); 9(1); 12D(1) |
| 31 | Little Sheep Crk., OR | I3(16) | 1K(9); 9(1); 28(1); N6(5) |
| 32 | Clearwater R., ID | I2(9) | 1K(6); 9(3) |
| 33 | Rapid R., ID | I2(1); I3(6) | 1B(1); 1K(5); 9(1) |
| 34 | Pahsimeroi Hatchery, ID | I2(1); I3(7) | 1K(4); 9(1); 27(1); 28(2) |
| 35 | Alsea R., OR | C1(1) | 1K(1) |
| 36 | Warm Springs R., OR | C1(1);C2(2);I1(5);I2(4);I3(5) | 1D(9);1K(4);12B(1); 28(3) |
| 37 | Bake Oven Crk., | C1(1); I1(5); I2(4); I3(3) | 1K(5); 9(3); 28(3); N6(2) |
| 38 | Rogue R., OR | C1(2); C2(4) | 1K(2);12B(1);12C(2); 12D(1) |
| 39 | Buck Crk., OR | C1(1) | 1C(1) |
| 40 | Bridge Crk., OR | I1(2) | 1K(2) |
| 41 | Upper Williamson R., OR | C1(4) | 3C(1); N2(3) |
| 42 | Witham Crk., OR | C1(4); C5(1) | 1C(3); 1K(1); 3C(1) |
| 43 | Threemile Crk., OR | I1(4) | 22(4) |
| 44 | Bridge Crk., OR | C2(3) | 1J(2); 22(1) |
| 45 | Mud Crk., OR | I1(3) | 22(3) |
| 46 | Thomas Crk., OR | C1(3) | 1F(2); N6A(1) |
| 47 | Honey Crk., OR | C1(2) | 1C(2) |
| 48 | N. Fork Little Deep Crk, OR | I2(1) | 1F(1) |
| 49 | W. Little Owyhee R., OR | I1(2) | 1K(2) |
| 50 | Sheepheaven Crk., CA | C1(2) | 1K(2) |
| 51 | Hayspur Hatchery, ID | C1(16) | 1A(1);1H(1);1K(8);3A(1);B(1); 10(4) |
| 52 | South Tacoma Hatchery, WA | C1(7) | 1K(7) |
| 53 | Spokane Hatchery, WA | C1(9) | 1G(4); 1K(4); 3B(1) |
| 54 | Scott Crk., CA | C1(5) | 1I(2); 5(1); 25(2) |
| 55 | Whale Rock Res., CA | C1(6) | 3B(1); 8(5) |
| 56 | Volcano Creek, CA (Golden trout) | V(5) | 3B(5) |
| 57 | Black R., AZ (Apache trout) | A(7) | A(7) |
Genomic and mitochondrial DNA were isolated by proteinase-K digestion of tissue samples preserved in 95% ethanol and stored at room temperature. DNA was phenol/chloroform extracted from the digested sample and precipitated by addition of 0.1 volume of 8.0 M ammonium acetate and an equal volume of isopropyl alcohol. Samples were then washed with 70% ethanol suspended in TE and quantified (Sambrook et al. 1989).
Primers for the OmyY1 Y-specific marker were used to PCR amplify a 1058 bp product spanning the OmyY1 locus in males. The forward primer was GAC GTT GTG GCA ATA GAT C and the reverse primer was CGA TTA GAA AGG CCT GCT TG. To provide consistent comparisons among samples the sequences were trimmed to 969 bp for analysis. Polymerase Chain Reaction amplifications utilized 50ng of DNA in 20 μL reactions. PCR reaction ingredient concentrations were 2 mM MgCl2, 0.25 mM dNTP, 1X Mg-free PCR buffer (Invitrogen Corporation, Carlsbad, CA.), 10 pmol of forward and reverse primer, and 1 U of Taq polymerase (New England Biolabs, Inc., Ipswich, MA). OmyY1 PCR thermocycling was conducted in Amplitron-II thermocyclers with amplification parameters being initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C denaturation for 50 s, 63.5°C annealing for 50 s, and 72°C extension for 60 s, with a final elongation at 72°C for 2 min. Seven hundred bp mitochondrial D-loop products were amplified with a forward primer of CCA CTC TTT ACG CCG GTA G and a reverse primer of ACT CTT ATT GAT GGT CAG GGG CAG. Amplification parameters were initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C denaturation for 50 s, 63°C annealing for 50 s, and 72°C extension for 60 s, with a final elongation at 72°C for 2 min. PCR-amplified products were evaluated by electrophoresis through 2% agarose gels in TAE buffer.
Amplified products were purified by ExoSapIT (Amersham, Piscataway, NJ, USA) and sequenced directly. OmyY1 haplotype sequences were derived from 3 separate overlapping sequencing reactions and mitochondrial haplotype sequences were derived from single-pass sequence reactions. The OmyY1 sequencing primers were GTT CAT ATG CCA GGC TCA AC and GCT AAT GGA CGA CGC TTT TC (forward primers) and GAA AAG CGT CGT CCA TTA GC (reverse primer). The mitochondrial sequencing primer was ACC GGC CCT CTT AAC CTT A (forward primer). All DNA sequencing reactions were performed by the Washington State University Laboratory for Biotechnology and Bioanalysis. DNA sequences were analyzed using Sequencher v3.1.1 software (Gene Codes Corporation, Ann Arbor, MI). Sequences were deposited in GenBank (Table 2).
Table 2.
GenBank deposition numbers for Y-linked OmyY1 haplotypes (a) and mitochondrial D-loop haplotypes (b).
| a) Y haplotype depositions | |
|---|---|
| Y-haplotype | GenBank deposition No. |
| A | 1353602_Seq1 |
| V | 1353602_Seq2 |
| C1 | 1353602_Seq3 |
| C2 | 1353602_Seq4 |
| C3 | 1353602_Seq5 |
| C4 | 1353602_Seq6 |
| C5 | 1353602_Seq7 |
| C6 | 1353602_Seq8 |
| I1 | 1353602_Seq9 |
| I2 | 1353602_Seq10 |
| I3 | 1353602_Seq11 |
| I4 | 1353602_Seq12 |
| I5 | 1353602_Seq13 |
| b) Mitochondrial D-loop depositions | |
|---|---|
| Mitochondrial haplotype | GenBank deposition No. |
| 1A | 1353343_Seq1 |
| 1B | 1353343_Seq2 |
| 1C | 1353343_Seq3 |
| 1D | 1353343_Seq4 |
| 1E | 1353343_Seq5 |
| 1F | 1353343_Seq6 |
| 1G | 1353343_Seq7 |
| 1H | 1353343_Seq8 |
| 1I | 1353343_Seq9 |
| 1J | 1353343_Seq10 |
| 1K | 1353343_Seq11 |
| 3A | 1353343_Seq12 |
| 3B | 1353343_Seq13 |
| 3C | 1353343_Seq14 |
| 5 | 1353343_Seq15 |
| 8 | 1353343_Seq16 |
| 9 | 1353343_Seq17 |
| 10 | 1353343_Seq18 |
| 12A | 1353343_Seq19 |
| 12B | 1353343_Seq20 |
| 12C | 1353343_Seq21 |
| 12D | 1353343_Seq22 |
| 12E | 1353343_Seq23 |
| 21 | 1353343_Seq24 |
| 22 | 1353343_Seq25 |
| 25 | 1353343_Seq26 |
| 27 | 1353343_Seq27 |
| 28 | 1353343_Seq28 |
| N1 | 1353343_Seq29 |
| N2 | 1353343_Seq30 |
| N3 | 1353343_Seq31 |
| N4 | 1353343_Seq32 |
| N5 | 1353343_Seq33 |
| N6 | 1353343_Seq34 |
| N6A | 1353343_Seq35 |
| N7 | 1353343_Seq36 |
| N8 | 1353343_Seq37 |
| N9 | 1353343_Seq38 |
| A | 1353347_Seq39 |
Distribution of genetic diversity was explored using AMOVA performed in Arlequin 3.11 (Excoffier et al. 2005). Samples were partitioned into inland and coastal groups based on the traditional distribution of the coastal and inland subspecies (Figure 1; Behnke 1992). Relationships among Y and mtDNA haplotypes were reconstructed using the median-joining (MJ) method (Bandelt et al. 1999) implemented in Network 4.5.0.0 (fluxus-engineering.com).
3. Results
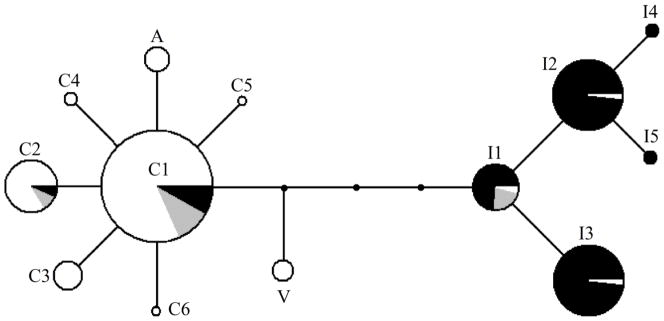
We evaluated the single copy Y-linked OmyY1 marker (Brunelli et al. 2008) for variation over 969 bases from 333 males sampled at 57 localities throughout the natural range of rainbow trout (Figure 1; Table 1). This evaluation revealed 13 Y haplotypes. Haplotype networks revealed two distinct haplogroups separated by four mutations (Figure 2). Each haplogroup generally included individuals from either the inland or coastal subspecies (Figures 1 & 2). All but 3 individuals collected from coastal localities had Y haplotypes corresponding to the coastal halpogroup and most individuals sampled from inland localities had haplotypes belonging to the inland halpogroup. Apache trout (O. apache) samples contained a distinctive Y haplotype not found at any other locality and differed from the common coastal Y haplotype (C1) by a single mutational step. Samples of California Golden trout (O. m. aguabonita) also contained a distinct Y haplotype that branched from the network at an intermediate node.
Fig. 2.
Network of Y-chromosome haplotypes in rainbow trout. Size of nodes are proportional to haplotype frequency, lines connecting nodes represent single step mutations, and undetected intermediate haplotypes are shown as small circles. Pie charts within nodes represent proportion of haplotype sampled from either coastal (white) or inland (black) localities. Haplotypes sampled from the closed basins of Oregon are shown in grey. Haplotypes from Apache trout in Arizona and Golden trout in California are indicated as “A” and “V” respectively. Haplotype labels correspond to those in Table 1.
Inland haplotypes showed some drainage-specific patterns (Table 1). The inland haplotype I1 was found only in some internally draining Oregon basins (localities 40, 43, 45), the Deschutes River drainage (localities 36–37), Hood River (locality 30), and the Touchet River (locality 26). The I2 haplotype was geographically distributed broadly, with representation in British Columbia (localities 15–16), Montana (localities 20–21) and the Oregon basins (locality 48). The I3 haplotype was widely distributed in tributaries of the Columbia (locality 25) and Snake River (localities 31, 33, 34) and predominated in Pennask Lake of British Columbia (locality 16), suggesting post-glacial invasion into the region from the Columbia River populations (McPhail 2007).
The Hood River (locality 30) and the Warm Springs and Bakeoven tributaries of the Deschutes River (localities 36 and 37) are drainages which enter the Columbia River in Oregon on either side of the traditional boundary of the Cascade Range separating the coastal and inland subspecies (Figure 1). Y sequences from individuals inhabiting the Deschutes River, an inland drainage, contained predominantly inland Y-haplotypes, while individuals inhabiting the Hood River, a coastal drainage, contained predominantly coastal haplotypes. Populations sampled from internally draining basins in Oregon included both the inland and coastal Y-haplogroups (Figure 1, inset).
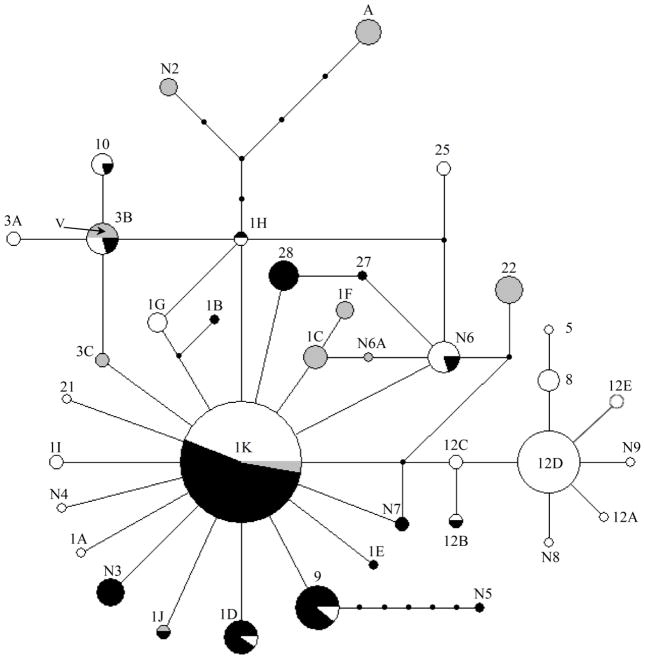
We also sequenced 610 bases of the mtDNA control region from these same males and identified 38 mitochondrial haplotypes (Figure 3; Table 1). In contrast to the pattern exhibited by Y haplotypes, little geographic structure was evident within the mtDNA network (Figure 3). The most common haplotype, 1K, accounted for approximately 45% of recovered sequences and was found roughly equally in inland and coastal localities. Mitochondrial haplotypes described in this study were designated according to previously established nomenclature developed to identify control region haplotypes (Nielsen et al. 1994, Graziano et al. 2005). Our sequence evaluation extended beyond the region used in the original nomenclature assignment and identified additional genetic variation which subdivided previously designated mtDNA haplotypes into haplotype subtypes. These subtypes are indicated with a letter following the designated name (e.g. 1K). Eight new haplotypes were found in the rainbow trout samples and samples from Apache trout also contained a new haplotype. One individual from the Yakoun River (locality 8) contained a coastal cutthroat (O. clarki clarki) mitochondrial haplotype and a coastal rainbow Y haplotype, indicating hybridization between a female cutthroat and a male rainbow trout. We did not determine if this represents an F1 hybrid or more ancient hybridization between the two species (Brown et al. 2004). Hybridization events representing both F1 and backcrosses between rainbow and cutthroat have been demonstrated from numerous localities in the Copper River, Alaska (Williams et al. 2007).
Fig. 3.
Network of mitochondrial haplotypes in rainbow trout. Size of nodes are proportional to haplotype frequency, lines connecting nodes represent single step mutations, and undetected intermediate haplotypes are shown as small circles. Pie charts within nodes represent proportion of haplotypes sampled from either coastal (white) or inland (black) localities. Haplotypes sampled from the closed basins of Oregon are shown in grey. Proportion of haplotypes in pie charts that were derived from Apache trout and Golden trout are indicated as “A” and “V” respectively. Haplotype labels correspond to those in Table 1. Haplotype names are based on the existing nomenclature (Graziano et al. 2005). Distinct haplotypes defined by variation outside the named sequences are denoted by a letter following the haplotype. A single sample with a coastal cutthroat mitochondrial haplotype was excluded from the network.
3.1. Partitioning genetic diversity
Results of AMOVA on rainbow trout samples, excluding the Golden and Apache trout species, reveal that 34.27% (P < 0.001) of the variation in the Y marker is explained by grouping populations into inland and coastal localities while just 6.06% (P < 0.001) of the variation in the mitochondrial marker is explained by this same grouping. However, samples from the internally-draining Oregon basins, which are traditionally classified as an inland form, displayed a mixture of both inland and coastal Y-markers, probably indicating a complex history of invasion by both types or introduction of coastal rainbow trout into the area. Excluding these Oregon basin samples from the AMOVA reveals that 51.3% (P < 0.001) of the variation observed in the Y marker is explained by grouping populations into coastal and inland populations. Additionally, if the Oregon basin samples are removed and haplogoups are used to represent variation within the lineages then 80.6% (P < 0.001) of the variance in the Y marker is explained by the inland/coastal split.
4. Discussion
Our results provide the first population-level evaluation of a Y-linked marker in a fish species. Our Y marker results show the clearest molecular evidence to date supporting the coastal and. inland subspecies designations in rainbow trout and suggest different sex-specific evolutionary histories in this species.
4.1. Y-marker patterns
The pattern displayed by the Y marker shows remarkable consistency between sample localities and halpogroups. The only discordance detected between coastal localities and haplotypes were 3 males sampled from Hood River, Oregon (locality 30). The Hood River has been proposed to be near the boundary between the two subspecies along the Columbia River (Behnke 1992)). The Deschutes River is the next major tributary entering the Columbia River upstream of the Hood River and with a drainage east of the Cascade Mountains it is within the distribution of the inland subspecies. Individuals sampled from tributaries of the Deschutes River (localities 38 and 39) had predominantly inland haplotypes but also contained some coastal haplotypes. Therefore, the transitional zone between the inland and coastal haplogroups corresponds to the geologic boundary separating the inland and coastal rainbow trout subspecies. This transition occurs over the course of ~60 km along the Columbia River.
Several inland localities that were some distance from the subspecies boundary also had coastal haplotypes. Individuals derived from the Blackwater River of British Columbia (locality 14) all had coastal haplotypes. British Columbia was completely covered by the Laurentide ice sheet during the last glaciation, and there is geologic evidence for headwater transfer of coastal drainages into the region that eventually became the Blackwater tributary of the Fraser River during the last glacial retreat of the Pleistocene (McPhail 2007). This may explain the occurrence of coastal haplotypes in the headwaters of this inland habitat. The remaining inland areas that have coastal haplotypes are all in eastern Washington (localities 18, 24–26). Localities 18 and 25 represent areas that have documented stocking activities for the past 70 years (Small et al. 2007) and coastal haplotypes in the area may represent introgression of coastal-derived hatchery stock into the native inland populations. Locality 24, the Wells Steelhead Hatchery, also contained coastal haplotypes and may have used milt from coastal steelhead to propagate inland populations (Thorgaard 1983). A single fish in the Touchet River, Washington (locality 26) which had a coastal haplotype may reflect past straying or introductions of fish with coastal haplotypes.
It is notable that the California Golden trout contained the only intermediate Y-haplotype suggesting an ancestral form of the rainbow Y chromosome. The California Golden trout population at Volcano Creek (locality 56) is considered to be the purest population of California Golden trout and is isolated above waterfalls which formed about 10,000 years ago (Coldes et al. 2006). The retention of a distinctive Y chromosome haplotype in this population is consistent with a long-standing separation of this population. The internally-draining basins in Oregon have no contemporary connection to other river systems or the ocean. This area has been proposed to have experienced invasions by both inland and coastal forms from the upper Klamath River or the Pit River (Behnke 1992, Currens et al. 2009). Most of the internally draining Oregon basins had both coastal and inland Y haplotypes. However, some streams in the region have experienced stocking with coastal rainbow over the past 100 years which may have led to the presence of coastal haplotypes (Behnke 2007).
The lack of divergence between common coastal Y haplotypes and the Apache trout haplotype is contrasted by substantial divergence of the Apache trout mtDNA haplotype (Figure 3). The California Golden trout have a much less divergent mitochondrial haplotype, which was also found in inland and coastal localities. Localities in the Oregon basins contain complex mixtures of both inland and coastal Y-haplotypes and mtDNA haplotypes. This overlapping of inland and coastal rainbow haplotypes could represent either recent hybridization or secondary contact between the subspecies during wet phases when pluvial lakes overflowed into coastal and inland drainages (Reheis et al. 2002).
The divergence we have documented between the inland and coastal Y chromosome haplotypes is deep and appears to indicate that a long-term divergence occurred between the inland and coastal subspecies before they came back into contact.
4.2. Mitochondrial patterns
Previous examination of mitochondrial diversity in rainbow trout using RFLP showed two divergent but geographically overlapping lineages identified as clade A and B (McCusker et al. 2000). To establish comparability between previous RFLP patterns and our sequence data the authors of the previous study provided us with samples from tributaries (Kutcho Crk. and Wolverine Lake [McCusker et al. 2000]) of the Turnagain River (our locality 8) known to display an RFLP pattern for clade B. Our results support these previous observations of two mitochondrial lineages. Individuals with the common haplotype in their clade A correspond to mtDNA haplotype 1K in our study, while individuals with the common haplotype in clade B correspond to mitochondrial haplotype 12D. Although the mtDNA network generally lacks strong geographic correlations, an association does exist for the small subnetwork centered on haplotype 12D (Figure 3). These individuals contained only coastal Y-haplotypes and were collected primarily in northern coastal localities from the Columbia River to coastal Alaska including the Queen Charlotte Islands. This network also has the characteristic star-shaped pattern of recent and rapid expansion, suggesting that this haplogroup survived Pleistocene glaciations in a coastal locality and recently expanded from it.
4.3. Interpreting patterns
The paternally-inherited Y marker and maternally-inherited mitochondrial marker show very different evolutionary patterns in rainbow trout. The mitochondrial DNA shows a relatively homogenized pattern between the inland and coastal subspecies, while the Y marker shows a sharp division along geographic and subspecies boundaries. The divergence we have documented between the inland and coastal Y chromosome haplotypes is deep and appears to indicate that a long-term separation occurred between the inland and coastal subspecies followed by secondary contact. Several mechanisms may have contributed to the observed patterns. One general class of explanations could relate to homing and behavioral differences between the sexes.
Sex-specific reproductive differences could contribute to the strong geographic structure of Y haplotypes. After returning from the sea, males of both inland and coastal forms might show higher homing fidelity than females, resulting in a tighter geographic correlation with the Y than the mitochondrial haplotype. An alternative mechanism for achieving apparently strong male-specific homing fidelity could be the common occurrence of non-anadromous “resident” males in some populations.
Discordant patterns in the markers could also be explained by differential reproductive success between the sexes when spawning outside their home range. Straying males of each subspecies may not be readily chosen by resident females and perform poorly in the range of the other subspecies, while stray females may readily find mates and be relatively more successful in spawning. Stray anadromous females tend to be larger and more fecund than resident females, thereby assuring contribution of associated mitochondrial haplotypes (Thériault et al. 2007). Y haplotypes might also be more likely to become fixed in a population than mitochondrial haplotypes because not all males will spawn, and dominant males may spawn successfully with more than one female, effectively reducing the male effective population size and leading to fixation of Y haplotypes within regions.
A second general class of explanation relates to genomic features of the Y chromosome and mtDNA. Genes that contribute to male fitness in the coastal and inland ranges may be linked to the non-recombining portion of the Y-chromosome. Linkage of reproduction-related genes to the sex chromosomes has been reported in some species, but the male “good genes” hypothesis has not been supported in studies comparing dominance status and viability of progeny through artificial fertilization (Jacob et al. 2007). Alternatively, an inland/coastal split in the mitochondrial genome could have occurred but the high rate of mitochondrial evolution might have saturated the sequences with mutations, effectively obscuring any signal. However, this study and previous studies examining mitochondrial diversity in inland and coastal rainbow trout using other loci or RFLP have not found consistent geographic patterns similar to those displayed by the Y marker.
Our data provide the first examination of the diversity and distribution of a Y-linked marker in a fish species and demonstrate incongruence between the genetic patterns for paternally and maternally-inherited markers. Some similar observations have been made previously in mammals but the extent of the difference between Y and mitochondrial patterns in this fish species raises the question of whether such differences may prove to be widespread among fishes. The divergence in Y-chromosome sequence also corresponds to sub-species. These subspecies may not have yet speciated due to female-biased gene flow between the subspecies as indicated by mtDNA patterns. Additionally, coastal rainbow trout have been widely distributed within the range of inland rainbow trout and protection of native inland rainbow trout is a significant conservation concern (Behnke 1992). The notable difference in Y chromosome sequence between inland and coastal rainbow trout suggests that this marker is likely to be useful in identifying pure, unhybridized populations of inland rainbow trout.
Supplementary Material
Acknowledgments
We thank all of the many individuals and agencies who provided the tissue and DNA samples in for this study (Supplementary Table 1). Supported by National Institute of Environmental Health Sciences grant ES012446 to James Nagler, by USDA CSREES National Research Initiative grants 2006-35205-16728 to Gary Thorgaard and Hubert Schwabl and 2008-04041 to Ruth Phillips and Gary Thorgaard.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avise JC. Mitochondrial DNA and the evolutionary genetics of higher animals. Phil Trans Roy Soc Lond B. 1986;312:325–342. doi: 10.1098/rstb.1986.0011. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Behnke RJ. Am Fish Soc Monogr No 6. American Fisheries Society; Bethesda, Maryland, USA: 1992. Native trout of western North America. [Google Scholar]
- Behnke RJ. Trout and Salmon of North America. The Free Press; New York, New York, USA: 2002. [Google Scholar]
- Behnke RJ. Redband of the Northern Great Basin. In: Schroeder RK, Hall JD, editors. Redband trout: resilience and challenge in a changing landscape. Oregon Chapter of the American Fisheries Society; Corvallis: 2007. pp. 1–9. [Google Scholar]
- Bernatchez L. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution. 2001;55:351–379. doi: 10.1111/j.0014-3820.2001.tb01300.x. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Boursot P. Discordant phylogeographic patterns between the Y chromosome and mitochondrial DNA in the house mouse: selection on the Y chromosome? Genetics. 1997;146:1019–1034. doi: 10.1093/genetics/146.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KH, Patton SJ, Martin KE, Nichols KM, Armstrong R, Thorgaard GH. Genetic analysis of interior Pacific Northwest Oncorhynchus mykiss reveals apparent ancient hybridization with westslope cutthroat trout. Trans Am Fish Soc. 2004;133:1078–1088. [Google Scholar]
- Brunelli JP, Thorgaard GH. A new Y-chromosome-specific marker for Pacific salmon. Trans Am Fish Soc. 2004;133:1247–1253. [Google Scholar]
- Brunelli JP, Wertzler KJ, Sundin K, Thorgaard GH. Y-specific sequences and polymorphisms in rainbow trout and Chinook salmon. Genome. 2008;51:739–748739. doi: 10.1139/G08-060. [DOI] [PubMed] [Google Scholar]
- Coldes JF, Stephens MR, Blumberg MA, May B. Identifying introgressive hybridization in native populations of California golden trout based on molecular markers. Trans Am Fish Soc. 2006;135:110–128. [Google Scholar]
- Consuegra S, Garcia de Leaniz C. Fluctuating sex ratios, but no sex-biased dispersal in a promiscuous fish. Evolutionary Ecology. 2007;21:229–245. [Google Scholar]
- Currens KP, Schreck CB, Li HW. Evolutionary ecology of redband trout. Trans Am Fish Soc. 2009;138:797–817. [Google Scholar]
- Danzmann RG, Morgan RPM, Jones MW, Bernatchez L, Ihssen PE. A major sextet of mitochondrial DNA phylogenetic assemblages extant in eastern North American brook trout (Salvelinus fontinalis): distribution and postglacial dispersal patterns. Can J Zool. 1998;76:1300–1318. [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and differentiation in fish: an overview of genetic, physiological and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Dittman AH, Quinn TP. Homing in pacific salmon: mechanisms and ecological basis. J Exp Biol. 1996;199:83–91. doi: 10.1242/jeb.199.1.83. [DOI] [PubMed] [Google Scholar]
- Eriksson J, Siedel H, Lukas D, Kayser M, Erler A, Hashimoto C, Hohmann G, Boesch C, Vigilant L. Y-chromosome analysis confirms highly sex-biased dispersal and suggests a low male effective population size in bonobos (Pan paniscus) Mol Ecol. 2006;15:939–949. doi: 10.1111/j.1365-294X.2006.02845.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fraser DJ, Lippe C, Bernatchez L. Consequences of unequal population size, asymmetric gene flow and sex-biased dispersal on population structure in brook charr (Salvelinus fontinalis) Mol Ecol. 2004;13:67–80. doi: 10.1046/j.1365-294x.2003.02038.x. [DOI] [PubMed] [Google Scholar]
- Graziano SL, Brown KH, Nielsen JL. Nomenclature of mitochondrial DNA haplotypes for Oncorhynchus mykiss. Trans Am Fish Soc. 2005;134:1271–1273. [Google Scholar]
- Halverson A. An Entirely Synthetic Fish. Yale University Press; New Haven: 2010. [Google Scholar]
- Hendry AP, Bohlin T, Jonsson B, Berg OK. To sea or not to sea: anadromy versus non-anadromy in salmonids. In: Hendry AP, Stearns SC, editors. Evolution Illuminated, Salmon and Their Relatives. Oxford University Press; Oxford: 2004. pp. 92–126. [Google Scholar]
- Hutchings JA, Gerber L. Sex-biased dispersal in a salmonid fish. Proc Royal Soc Lond B. 2002;269:2487–2493. doi: 10.1098/rspb.2002.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A, Nusslé S, Britschgi A, Evanno G, Müller R, Wedekind C. Male dominance linked to size and age, but not to good genes in brown trout (Salmo trutta), BMC Evol. Biol. 2007;7:207–216. doi: 10.1186/1471-2148-7-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KL, Muhlfeld CC, Sage GK, Leary RF. Genetic structure of Columbia River redband trout population in the Kootenai River drainage, Montana, revealed by microsatellite and allozyme loci. Trans Am Fish Soc. 2002;131:1093–1105. [Google Scholar]
- Koizumi I, Yamamoto S, Maekawa K. Female-biased migration of stream-dwelling Dolly Varden in the Shiisorapuchi River, Hokkaido, Japan. J Fish Biol. 2006;68:1513–1529. [Google Scholar]
- Kondo M. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 2006;16:815–826. doi: 10.1101/gr.5016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker MR, Parkinson E, Taylor EB. Mitochondrial DNA variation in rainbow trout (Oncorhynchus mykiss) across its native range: testing biogeographical hypotheses and their relevance to conservation. Mol Ecol. 2000;9:2089–108. doi: 10.1046/j.1365-294x.2000.01121.x. [DOI] [PubMed] [Google Scholar]
- McMillan JR, Katz SL, Pess GR. Observational evidence of spatial and temporal structure in a sympatric anadromous (winter steelhead) and resident rainbow trout mating system on the Olympic Peninsula, Washington. Trans Am Fish Soc. 2007;136:736–748. [Google Scholar]
- McPhail JD. The freshwater fishes of British Columbia. University of Alberta Press; Edmonton: 2007. [Google Scholar]
- Narum SR, Contor C, Talbot A, Powell MS. Genetic divergence of sympatric resident and anadromous forms of Oncorhynchus mykiss in the Walla Walla River, USA. J Fish Biol. 2004;65:471–488. [Google Scholar]
- Nielsen JL, Gan C, Thomas WK. Differences in genetic diversity of mitochondrial DNA between hatchery and wild populations of Oncorhynchus. Can J Fish Aquat Sci. 1994;51(Suppl 1):290–297. [Google Scholar]
- Olsen JB, Wuttig K, Fleming D, Kretschmer EJ, Wenburg JK. Evidence of partial anadromy and resident-form dispersal bias on a fine scale in populations of Oncorhynchus mykiss. Conserv Genet. 2006;7:613–619. [Google Scholar]
- Pidancier N, Jordan S, Luikart G, Taberlet P. Evolutionary history of the genus Capra (Mammalia, Artiodactyla): discordance between mitochondrial DNA and Y-chromosome phylogenies. Mol Phylogenet Evol. 2006;40:739–749. doi: 10.1016/j.ympev.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Quinn TP. The Behaviour and Ecology of Pacific Salmon and Trout. University of Washington Press; Seattle: 2005. [Google Scholar]
- Reheis M, Sarna-Wojcicki AM, Reynolds RL, Repenning CA, Mifflin MD. Pliocene to middle Pleistocene lakes in the western Great Basin: Ages and connections. In: Hershler R, Madsen D, Currey D, editors. Great Basin Aquatic Systems History. Smithsonian Press; Washington D.C: 2002. pp. 53–108. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genetics Dev. 2004;14:634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Seamons TR, Bentzen P, Quinn TP. The mating system of steelhead, Oncorhynchus mykiss, inferred by molecular analysis of parents and progeny. Environ Biol Fishes. 2004;69:333–344. [Google Scholar]
- Small MP, McLellan JG, Loxterman J, Von Bargen JF, Frye A, Bowman C. Fine-scale population structure of rainbow trout in the Spokane river drainage in relation to hatchery stocking and barriers. Trans Am Fish Soc. 2007;136:301–317. [Google Scholar]
- Smith JJ, Voss SR. Amphibian sex determination: segregation and linkage analysis using members of the tiger salamander species complex (Ambystoma mexicanum and A. t. tigrinum) Heredity. 2009;102:542–548. doi: 10.1038/hdy.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Björnerfeldt S, Leonard J, Hailer F, Hedhammar Å, Ellegren H, Vilà C. Unequal contribution of sexes in the origin of dog breeds. Genetics. 2006;172:1121–1128. doi: 10.1534/genetics.105.042358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault V, Bernatchez L, Dodson JJ. Mating system and individual reproductive success of sympatric anadromous and resident brook charr Salvelinus fontinalis under natural conditions. Behav Ecol Sociobiol. 2007;62:51–65. [Google Scholar]
- Thorgaard GH. Chromosomal differences among rainbow trout populations. Copeia. 1983;1983:650–662. [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ, Huelsenbeck J. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Utter F. Patterns of subspecific anthropogenic introgression in two salmonid genera. Rev Fish Bio Fish. 2001;10:265–279. [Google Scholar]
- Williams I, Reeves GH, Graziano SL, Nielsen JL. Genetic investigation of natural hybridization between rainbow and cutthroat trout in the Copper River Delta, Alaska. Trans Am Fish Soc. 2007;136:926–942. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.