Abstract
Cellular senescence is the dominant phenotype over immortality. In our studies to identify senescence related genes we cloned Morf4 that induced senescence in a subset of tumor cells. Morf4 is a member of a family of 7 genes, and the Morf related genes (Mrg) on chromosomes 15 (Mrg15) and X (MrgX) are also expressed. In contrast to MORF4, MRG15 and MRGX are positive regulators of cell division. All three proteins interact with histone acetylases (HATs) and acetyltransferase (HDACs), suggesting they function in regulation of chromatin dynamics. Mrg15 knockout mice are embryonic lethal, and MEFs derived from Mrg15 null embryos proliferate poorly, enter senescence rapidly and have impaired DNA repair compared to wild type. Mrg15 null embryonic neural stem/progenitor cells also have a decreased capacity for proliferation and differentiation. Further studies are needed to determine the function of this gene family in various biological processes including neural stem/progenitor cell aging.
Keywords: cellular senescence, Mrg (Morf related genes), MRG15 (Morf related gene on chromosome 15), chromatin remodelling, neural stem/progenitor cells, proliferation, differentiation, DNA damage, aging
INTRODUCTION
Biological aging is a phenomenon resulting from the accumulation of changes in molecular, cellular and tissue organization aspects over time. The mechanisms of aging can be broadly categorized into either accumulation of damage or genetic and epigenetic control. According to the genetic basis for aging theory, aging is a continuum of growth, morphogenesis and differentiation. The question of whether cells in vivo also age like the organism that they build remained controversial until the pioneering studies of Leonard Hayflick in the early sixties. Hayflick and Moorhead showed for the first time that there is a limited division potential of human diploid fibroblasts in culture, a phenomenon termed cellular or replicative senescence.1,2 This senescence has since been observed in other cell types such as endothelial cells, T lymphocytes, epidermal keratinocytes, adrenocortical cells, smooth muscle cells, glial cells, lens epithelial cells and human pancreatic β-cells, and provides the basis for a model of cellular aging.3 However, the aging of stem cells is still under investigation.
Cellular senescence studies identify the novel MORF/MRG family of proteins
The life span of a cultured normal cell population is between 30 and 90 population doublings, although this number varies depending on the source of the tissue. After ceasing division and entering senescence cells do not die in culture. They can stay viable in this state for several years, with the regular renewal of medium. Senescent cells acquire a characteristic of enlarged and flattened morphology and increased expression of Senescence associated beta galactosidase (SA-β-gal) activity at pH 6.4 Other features include changes in gene expression, resistance to some apoptotic stimuli, inability to induce DNA synthesis in response to mitogens, loss of phosphorylated pRb, and increased expression of the cyclin dependent kinase (CDK) inhibitors p21 and p16.5,6 However, SV40 T antigen can induce DNA synthesis in these cells, but not mitosis, suggesting a pathway involving p53 and pRb contributes to the senescent state. Cells can, however, evade cellular senescence by adopting an immortalized phenotype, acquiring the ability to circumvent cell cycle control mechanisms and divide limitlessly.
Genetic analyses revealed that fusion of young and old cells yield hybrids with life spans similar to that of the old parental cells; fusion of old cell cultures with each other did not produce hybrids with a life span greater than that of either parental cell line, providing evidence for a dominant regulatory mechanism causing senescence.7 Further evidence was obtained from fusion of normal with immortal, tumor derived cells. The hybrids regained growth control and stopped dividing indicating that unlimited division potential results from recessive changes in normal growth control mechanisms and providing the first evidence for cell senescence as a mechanism for tumor suppression.7
To determine whether cellular senescence-related genes could induce senescence in an entire subset of immortalized cells, single chromosomes from normal human cells were introduced into immortal human cells by microcell-mediated chromosome transfer.8-10 From these studies, a gene encoding an intron-less transcription factor-like protein, which was named mortality factor on chromosome 4 (MORF4), was identified as a senescence inducing gene.9 Further study of the MORF4 gene led to the discovery that it was one of seven members of a family of novel transcription factor-like genes. Among these family members, only MORF4, MORF-related gene on chromosome 15 (MRG15/MORF4L1), and MORF-related gene on chromosome X (MRGX/MORF4L2), are expressed. MRG15 and X are expressed ubiquitously in all cells and tissues.9 All three proteins share an ATP/GTP binding region followed by a helix-loop-helix and a leucine zipper at the C terminus. Additionally, MRG15 and MORF4 encode a bipartite nuclear localization signal (NLS) flanked by phosphorylation sites, while MRGX contains a single NLS.9 Consistent with the NLS motif, all three proteins localize to the nucleus. MRG15 is unique from MORF4 and MRGX in that it has an N-terminal extension, which includes a chromatin organization modifier domain (chromodomain), implicating a role for it in chromatin related functions.9,11 Among the family members, only MORF4 has the ability to induce senescence in immortal human cell lines.9,unpublished data MRG15 and MRGX, in contrast, have been found to be more intimately linked to cell cycle progression and positively affect cell proliferation.12
MRG15 associated nuclear protein complexes
Proteins containing chromo domains are involved in protein-protein interactions and play important roles in chromatin remodeling that can activate or repress the transcription of a large number of genes.13,14 Studies in D. melanogaster and S. cerevisiae suggest that chromodomain-containing proteins, such as Msl-3, polycomb, HP1, and SWI/SNF, bind specific regions of chromatin rather than defined segments of DNA, and thereby interact with transcriptional repressors and activators in target gene promoters and affect gene expression.15-17 In Drosophila, Msl-3(male-specific lethal 3, a MRG15 homolog and chromodomain-containing protein), binds to multiple sites on the male X chromosome, inducing chromatin remodeling and subsequent hyper transcription of genes necessary for the regulation of dosage compensation.
In the earliest studies, MRG15 was shown to associate with at least two complexes of nuclear proteins, MRG15-associated factors 1 and 2 (MAF1 and MAF2).11 MAF1 is composed of, at a minimum, MRG15, retinoblastoma protein (Rb), and PAM14, a novel interacting protein. Rb can, in fact, associate with both MRG15 and MRGX. The association with MRG15 activates the B-myb promoter in both EJ, a bladder carcinoma derived cell line, and HeLa cells whereas the association with MRGX activates the B-myb promoter in HeLa while repressing it in EJ cells.18,19 This repression is relieved by a histone deacetylase (HDAC) inhibitor, suggesting protein components of a cell determine the complexes that can be formed and how genes are regulated.19 This indicates a potential mechanism for the MRG family effect on cellular proliferation via acetylation and deacetylation of histones and indeed MAF2 was found to contain MRG15 bound to hMOF, a MYST family HAT.11 Further, it was demonstrated that MRG family proteins are components of mSin3A/Pf1/HDAC complexes.20 The MRG domain of MRG15 is bound by the plant homeodomain zinc finger protein Pf1, which acts as an adaptor between the two transcriptional repressors mSin3A and the transducin-like enhancer of split (TLE). Additionally, MRG15 alone or MORF4 or MRGX fused with the Gal4 DNA binding domain can bind the mSin3A–TLE co-repressor complex and induce transcriptional repression.20 These studies implicate MRG15/MRGX in both transcriptional activation and repression.
MRG15 and its homologue Eaf3 have now been identified as a component of the Tip60/NuA4 histone acetylase complex in human, Drosophila and yeast, respectively.21-24 It has been demonstrated that the Tip60 complex in Drosophila acetylates nucleosomal phospho-H2Av at Lys5 in a double strand break (DSB) dependent manner and enhances H2AX variant exchange following DNA damage.25 MRG15 also was found to interact with PALB2, a tumor suppressor protein, which plays a crucial role in DNA damage repair by homologous recombination. Mass spectrometry analysis and co-immunoprecipitation experiments identified MRG15, along with its homolog MRGX, as major binding partners of PALB2, via the common MRG domain. PALB2 localizes at the site of DNA breaks upon DNA damage. Immunofluorescence staining revealed that endogenous MRG15 localized to the site of DNA breaks marked by γH2AX following ionizing radiation.26 These results demonstrate a role for MRG15 in DNA repair.
MRG15 affects mouse neural stem/progenitor cell proliferation, and differentiation
A targeted gene knockout of MRG15 resulted in embryonic lethality, cell growth defects, and delayed development in many organ systems, including brain.27 The embryos were smaller than their wild-type littermates. General thinning of the neural tube was observed in embryonic day 10.5 (E10.5) null embryos and a smaller number of neural precursor cells were obtained from Mrg15 null embryos immediately after dissociation, compared to wild-type (wt) littermates, suggesting defects in Mrg15 null neural stem/precursor cell proliferation. To assess whether the thinning of neural tube was the result of decreased cell division or increased apoptosis in vivo, histology and immunostaining for the mitosis marker MMP2 and apoptosis evaluation-TUNEL assay, were performed. We found fewer MMP2 positive cells and increased TUNEL positive staining in the Mrg15 null embryonic forebrain. To exclude the effect from the in vivo environment, neural stem cells were isolated from E15.5 embryonic whole brain to evaluate the cell proliferation rate and viability in vitro using the neurosphere formation assay (NSFA). There were fewer big neurospheres formed in cultures from Mrg15 null embryos than wt control. Fewer Mrg15 null cells could incorporate BrdU in a 4 hour time frame, and this could be reversed by MRG15 adenovirus infection of the cells.28 These results indicated that cell proliferation was decreased/impaired in Mrg15 null neural stem/progenitor cells.
The Mrg15 null neurospheres also displayed defects in the differentiation process. The neurospheres formed from the first NSFA of Mrg15 null mice could not spread well under differentiation conditions and there were fewer neurons formed from Mrg15 null neurospheres compared with wt controls.28 Thus, MRG15 affects both neural stem/progenitor cell proliferation and differentiation. This is not unexpected because MRG15 associates in complexes with the HATs Tip60 and hMOF as well as mSin3/HDAC.20-24 MRG15 is clearly involved in the regulation of gene expression by changing the acetylation status of histones surrounding target genes.
Neural stem/progenitor cell aging and the possible role of MRG15
Neurogenesis continues throughout life in the subgranular zone (SGZ) of the hippocampal dentate gyrus (DG) and in the subventricular zone (SVZ) through neural stem/progenitor cells proliferation, maintenance, and differentiation. Although neurogenesis persists in the aged brain, its rate declines with age in rats, mice, monkeys and humans.29-33 In rodents, as measured by BrdU incorporation, there was a decreased number of BrdU positive cells in both SGZ and SVZ in the old compared with young,30,34-39 indicating there is a decreased proliferation rate of neural stem/progenitor cells during aging. Other possibilities include decreased survival, increased loss of newborn neural stem/progenitor cells or its progeny lineage in aged mice or rats. However, results regarding the self-renewal and differentiation capacity of neural stem cells isolated from senescent mice remain controversial. Some studies have found that neural stem/progenitor cells had lower BrdU incorporation in senescent mice, but that the isolated cells grew well in vitro and were comparable to young mice and the cells could be induced to differentiate into various lineages.40 In contrast, other experiments demonstrate decreased BrdU incorporation rate in senescent mice as well as decreased ability for neurosphere formation in vitro, and a reduction in differentiation into neuronal phenotypes.38,41
In spite of these contradictory reports many studies have now described the critical role of epigenetic mechanisms in adult neurogenesis. Orphan nuclear receptor TLX is highly expressed in the adult mouse brain.42,43 Although TLX knockout mice are viable and appear normal at birth, the mutant mice show a loss of cell proliferation and reduced neural precursors in the neurogenic areas of adult brain. Cells isolated from adult TLX mutant mouse brains fail to proliferate in culture. This is most likely because TLX normally interacts with HDAC3 or 5 to inhibit the p21 promoter.42-46 Valproic acid (VPA, 2-propylpentanoic acid) is a well known anticonvulsant and mood stabilizer, which can block seizure-induced neurogenesis and induce neural differentiation of adult hippocampal neural progenitor cells in vitro and in vivo, by inhibiting HDAC activity and causing hyperacetylation of histones to regulate gene expression.47-51 Sirt1, a mammalian homologue of Sir2, yeast HDAC, deficient mice have been reported to exhibit severe neural defects, including exencephaly and disturbed neuroretinal morphogenesis.52-54 Sirt1 and dSir2 (the Sirt1-related protein in Drosophila melanogaster) function together with Hes1 as transcriptional co-repressors.55,56 In the developing brain, Hes1 prevents premature neuronal differentiation by repressing the activation of the pro-neuronal basic helix-loop-helix transcription factor Mash1.57 Since MRG15 associates with both HATs and HDACs,20-24 controlling gene expression or repression by modifying the histone acetylation status, it could well be important for organ specific tissue senescence and aging.12 More recently, we found MRG15 is down-regulated in old mouse livers, and size exclusion chromatography of nuclear extracts from young and old livers using immunoblotting with antibodies to MRG15 and its interacting partner, HDAC1, revealed that MRG15 co-fractionates with HDAC1 in high-molecular-weight (HMW) fractions in old livers but, in contrast, is present in low-molecular-weight (LMW) fractions away from HDAC1, in young livers (unpublished data, with N. Timchenko). HMW age specific complexes have been previously described and been shown to contain the senescence-associated proteins RB and HDAC1.58,59 In vitro, MRG15 is down regulated in senescent human fibroblasts versus young proliferating fibroblasts. Over-expression of MRG15 and/or MRGX in pre-senescent cells causes them to re-enter the cell cycle, as evidenced by an increased number of BrdU incorporating cells (unpublished data). As stated above, Mg15 null embryonic neural stem/progenitor cells have deficient proliferation and differentiation capacity compared with cells derived from wt littermates. However, this decreased proliferation capability can be compensated by MRG15 adenoviral infection as demonstrated by increased BrdU incorporation. Taken together, the data suggest MRG15 might be one potential intrinsic controller of neural stem/progenitor cell aging.
The accumulation of somatic DNA damage is considered another main cause of stem cell aging.60-62 Genomic stability of stem cells is essential for self-renewal and differentiation. Damaged cells undergo apoptosis or differentiation, which causes unbalanced tissue homeostasis and the aging phenotype. From studies of hematopoietic stem cells, more DNA damage was found to accumulate in cells from old versus young mice. Hematopoietic stem cell functional capacity was severely affected under conditions of stress in mice deficient in genomic maintenance pathways, leading to loss of reconstitution and proliferative potential, diminished self-renewal, increased apoptosis and ultimately, functional exhaustion.60 Similar to HSCs, when exposed to excessive genotoxic stress, melanocyte stem cells lose their stem cell immaturity and commit to differentiation.62 Deficiency in DNA damage repair, Ataxia telangiectasia (A-T) is characterized by normal brain development followed by progressive neurodegeneration.63,64 ATM is implicated in cell cycle regulation and DNA repair.64 ATM expression is abundant in proliferating neural stem/progenitor cells.65,66 Neural stem/progenitor cells in the dentate gyrus of ATM deficient adult mice show abnormal proliferation and blunted survival in vivo67. When these cells were put in culture under differentiation conditions, they could not differentiate into multi-lineages.67
MRG15 associates with the NuA4/Tip 60 histone acetyltransferase complex. It also interacts with PALB2, the DNA repair protein. From a study of MEFs, we have shown that MRG15 is very important for DNA damage repair in that Mrg15 null MEFs do not repair DNA as rapidly or efficiently as wild type control, as measured by the comet assay. Subsequent cell proliferation, determined by clonogenic assay is also impaired.68
The proliferation and differentiation impairment observed in Mrg15 null neural stem/progenitors suggests the importance of MRG15 in both neural stem/progenitor cell self-renewal and differentiation.
MORF4 as an antagonist of MRG protein activity
Studies with MORF4 have been difficult for a number of reasons. RNA expression levels are low,9 it is toxic to cells if expressed under other promoter control and generating an antibody specific to this protein has been problematic because of the high similarity to MRG15, which does not permit the use of a long stretch of amino acid sequence for antibody production. The anti-peptide antibody we have generated does not have a high affinity for MORF4 protein (unpublished results). Using an inducible system to express MORF4 we have analyzed the protein in a clone of HeLa cells and have found that MORF4 protein is highly unstable with a half-life of less than 1 h, (ms submitted). The mechanism of degradation involves the proteasome following ubiquitination.
Based on what we know about the properties of the three proteins we hypothesize that through interactions with other proteins MRG15 and MRGX act to positively regulate cell growth. The proteins most likely translocate to the nucleus through some protein-protein interaction, in addition to the NLS, as deletion of the NLS does not result in complete loss of these proteins in the nucleus. Since MORF4 is a truncated version of the MRG proteins it has the potential to act in a dominant negative manner and replace them in active complexes to inactivate or possibly disrupt them. In fact, use of a chromodomain minus mutant, which mimics MORF4, did result in loss of the MAF2 complex.11 However, we cannot rule out the possibility that MORF4 acts in a separate complex to regulate cell proliferation (Figure 1).
Figure 1. Model for MORF4 action in the cells.
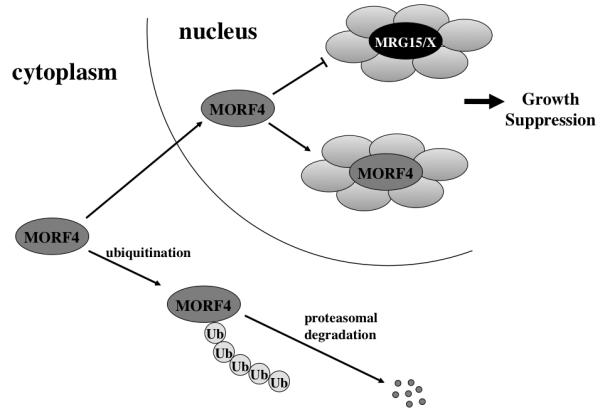
MORF4 protein is always maintained at low level in the cells by proteasomal degradation following ubiquitination in the cytoplasm. MORF4 which escapes degradation may translocate into the nucleus and inhibit MRG15/X function to achieve growth suppression. Alternatively, MORF4 may associate with an independent complex(s) in the nucleus and suppress cell growth.
CONCLUSION
The data summarized here emphasize the need for a delicate balance of proteins and their interacting partners in nuclear protein complexes in cells. A shift to excess or decrease in amount of a regulatory protein can result in cessation of cell proliferation, and if too extreme cell death. It is clear that MRG15 in particular has multiple functions in development, and in proliferation and DNA damage repair in cells including neural stem/progenitor cells and in aging liver. As we learn more about the mechanisms of action of this gene family we will obtain better insights into their function in these various biological processes.
Acknowledgements
This work was supported by Ellison Medical Foundation (O.M.P.-S), RO1 AG032134 (O.M.P.-S), The American Federation for Aging Research (K.T.), and the University of Texas Health Science Center at San Antonio (ERC) (K.T.).
REFERENCES
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 4.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noda A, et al. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 6.Hara E, et al. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira-Smith OM, Smith JR. Evidence for the recessive nature of cellular immortality. Science. 1983;221:964–966. doi: 10.1126/science.6879195. [DOI] [PubMed] [Google Scholar]
- 8.Bertram MJ, et al. Assembly of BAC contig of the complementation group B cell senescence gene candidate region at 4q33-q34.1 and identification of expressed sequences. Genomics. 1999;56:353–354. doi: 10.1006/geno.1998.5726. [DOI] [PubMed] [Google Scholar]
- 9.Bertram MJ, et al. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- 11.Pardo PS, et al. MRG15 a novel chromodomain protein is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- 12.Pena AN, Pereira-Smith OM. The Role of the MORF/MRG family of genes in cell growth, differentiation, DNA repair, and thereby aging. Ann. N. Y. Acad. Sci. 2007;1100:299–305. doi: 10.1196/annals.1395.031. [DOI] [PubMed] [Google Scholar]
- 13.Cavalli G, Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol. 1998;10:354–360. doi: 10.1016/s0955-0674(98)80011-2. [DOI] [PubMed] [Google Scholar]
- 14.Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. BioEssays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Kennison JA. The polycomb and trithorax groupproteins of drosophila: Trans-regulators of homeotic gene function. Annu. Rev. Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: new members, duplication of the chromodomain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorentz A, et al. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- 18.Leung JK, et al. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 2001;276:39171–39178. doi: 10.1074/jbc.M103435200. [DOI] [PubMed] [Google Scholar]
- 19.Tominaga K, et al. MRGX: a novel transcriptional regulator that exhibits activation or repression of B-myb promoter in a cell type dependent manner. J. Biol. Chem. 2003;278:49618–49624. doi: 10.1074/jbc.M309192200. [DOI] [PubMed] [Google Scholar]
- 20.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol. Cell. Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Y, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 22.Doyon Y, et al. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayakawa T, et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 24.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. USA. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 26.Sy SM-H, Huen MSY, Chen J. MRG15 is a novel PALB2 interacting factor involved in homologous recombination. J. Biol. Chem. 2009;284:21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tominaga K, et al. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M, et al. MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. J. Neurosci. Res. 2009;87:1522–1531. doi: 10.1002/jnr.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kempermann G, Kuhn H, Gage F. Experience induced neurogenesis in the senescent dentate gyrus. J Neurosci. 1998;18:3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cameron HA, McKay RD. Restoring production of hippocampal neurons in old age. Nat. Neurosci. 1999;2:894–897. doi: 10.1038/13197. [DOI] [PubMed] [Google Scholar]
- 32.Gould E, et al. Hippocampal neurogenesis in adult Old World primates. Proc. Natl. Acad. Sci. USA. 1999;96:5263–5267. doi: 10.1073/pnas.96.9.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kukekov VG, et al. Multipotent stem/progenitor cells with similar properties arise from two neurogenic regions of adult human brain. Exp Neurol. 1999;156:333–344. doi: 10.1006/exnr.1999.7028. [DOI] [PubMed] [Google Scholar]
- 34.Tropepe V, et al. Transforming growth factor-alpha Null and senescent mice show deceased neural progenitor cell proliferation in the forebrain subependyma. J Neurosci. 1997;17:7850–7859. doi: 10.1523/JNEUROSCI.17-20-07850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nacher J, et al. NMDA receptor antagonist treatment inreases the production of new neurons in the aged rat hippocampus. Neurobiol Aging. 2003;24:273–284. doi: 10.1016/s0197-4580(02)00096-9. [DOI] [PubMed] [Google Scholar]
- 36.McDonald HY, Wojtowicz JM. Dynamics of neurogenesis in the dentate gyrus of adult rats. Neurosci Lett. 2005;385:70–75. doi: 10.1016/j.neulet.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Montaron MF, et al. Lifelong corticosterone level deterines age-related decline in neurogenesis and memory. Neurobil Aging. 2005;27:645–654. doi: 10.1016/j.neurobiolaging.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao MS, Hattiangady B, Shetty AK. The window and mechanisms of major age-related decline in the production of new neurons within the dentate gyrus of the hippocampus. Aging Cell. 2006;5:545–558. doi: 10.1111/j.1474-9726.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- 40.Ahlenius H, et al. Neural stem amd progenitor cells retain their potential for proliferation and differemtiation into functional neurons despite lower number in aged brain. J. Neurosci. 2009;29:4408–4419. doi: 10.1523/JNEUROSCI.6003-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maslov AY, et al. Neural stem cell detection, characerization, and age-related changes in the subventricular zone of mice. J. Neurosci. 2004;24:1726–1733. doi: 10.1523/JNEUROSCI.4608-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 43.Qu Q, Shi Y. Neural stem cells in the developing and adult brains. J. Cell. Physiol. 2009;221:5–9. doi: 10.1002/jcp.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monaghan AP, et al. Defective limbic system in mice lacking the taillessgene. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 45.Land PW, Monaghan AP. Expression of transcription factor, tailless, is required for formation of superficial fortical layers. Cereb Cortex. 2003;13:921–931. doi: 10.1093/cercor/13.9.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun G, et al. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl. Acad. Sci. USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottlicher M, et al. Volproic acid defines a novel class of HDAC inhibitors inducing differentiation of rtansformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phiel CJ, et al. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 49.Hsieh J, et al. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jessberger S, et al. Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. J. Neurosci. 2007;27:5967–5975. doi: 10.1523/JNEUROSCI.0110-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukuchi M, et al. Valproic acid induces up- or down-regulation of gene expression responsible for the neuronal excitation and inhibition in rat cortical neurons trough its epignetic actions. Neurosci. Res. 2009;65:35–43. doi: 10.1016/j.neures.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog(SIRT1)- deficient mice. Proc. Natl. Acad. Aci. USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McBurney MW, et al. The Mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prozorovski T, et al. Sirt 1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 55.Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(sp1) bHLH repressors in segmentation and sex determination. Cell. 2002;109:447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- 56.Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 57.Ishibashi M, et al. Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-hlix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 58.Bandyopadhyay D, et al. Dynamic assembly of chromatin complexes during cellular senescence: implications for the growth arrest of human melanocytic nevi. Aging Cell. 2007;6:577–591. doi: 10.1111/j.1474-9726.2007.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunaief JL, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 60.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of harmatopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 61.Ruzankina Y, Asare A, Brown EJ. Replicative stress, stem cells and aging. Mech. Ageing Dev. 2008;129:460–466. doi: 10.1016/j.mad.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inomata K, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 63.Sedgewick R, Boder E. Ataxia-telangiectasia. Handbook of clinical Neurology. 1991:347–423. doi: 10.1016/B978-0-444-51892-7.00019-X. [DOI] [PubMed] [Google Scholar]
- 64.Barlow C, et al. Atm-deficient mice: A paradigm of Ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 65.Chen G, Lee EY-H. The product of the ATM gene is a 370-kDa nuclear phosphoprotein. J. Biol. Chem. 1996:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- 66.Herzog KH, et al. Requirement for Atm in ionizing radiattion-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 67.Allen DM, et al. Ataxia telangiectasia mutated is essential during adult neurogenesis. Genes Dev. 2001;15:554–566. doi: 10.1101/gad.869001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia SN, et al. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 2007;581:5275–5281. doi: 10.1016/j.febslet.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


