Abstract
In HIV-1 infected cells, the LTR promoter, once organized into chromatin, is transcriptionally inactive in the absence of stimulation. To examine the chromosomal events involved in transcriptional activation, we analyzed histone acetylation and factor recruitment at contiguous LTR regions by a quantitative chromatin immunoprecipitation assay. In chronically infected cells treated with a phorbol ester, we found that acetylation of both histones H3 and H4 occurs at discrete nucleosomal regions before the onset of viral mRNA transcription. Concomitantly, we observed the recruitment of known cellular acetyl-transferases to the promoter, including CBP, P/CAF and GCN5, as well as that of the p65 subunit of NF-κB. The specific contribution of the viral Tat transactivator was assayed in cells harboring the sole LTR. We again observed nucleosomal acetylation and the recruitment of specific co-factors to the viral LTR upon activation by either recombinant Tat or a phorbol ester. Strikingly, P/CAF was found associated with the promoter only in response to Tat. Taken together, these results contribute to the elucidation of the molecular events underlying HIV-1 transcriptional activation.
Keywords: chromatin immunoprecipitation/histone acetyl-transferases/histones/HIV-1/long terminal repeat/nucleosomes/Tat
Introduction
In eukaryotic cells, DNA is tightly packed in a highly organized chromatin structure. The packaging of DNA essentially controls the interaction of regulatory proteins with their cis-acting elements, including those found in the promoter of genes. Chromatin structure is modulated by the covalent modifications of the N-termini of the core histones in nucleosomes and by the action of ATP-dependent chromatin remodeling complexes. In particular, histone acetylation at the promoter of genes, mediated by histone acetyltransferases (HAT), has been shown to be necessary, albeit not sufficient, for transcriptional activation (for recent reviews see Berger, 2002; Cosma, 2002; Narlikar et al., 2002). Each HAT has its own lysine specificity within the tails of histones H3 and H4, leading to the notion of a ‘histone code’ that determines the epigenetic control of transcription (Strahl and Allis, 2000).
Following infection of susceptible cells, the human immunodeficiency virus type 1 (HIV-1) proviral DNA gets integrated into the host genome. Here, chromatin conformation essentially represses transcription from the integrated long terminal repeat (LTR) promoter (reviewed in Marzio and Giacca, 1999). Experiments performed both in vivo (Verdin et al., 1993; Van Lint et al., 1996; El Kharroubi et al., 1998) and in vitro using the HIV promoter reconstituted into chromatin (Van Lint et al., 1996; Sheridan et al., 1997) have shown that, independent from the integration site, nucleosomes in the 5′ LTR are precisely positioned with respect to cis-acting regulatory elements. In the transcriptionally silent provirus, these nucleosomes define two large nucleosome-free areas. The first one is composed of the core promoter, containing three tandem Sp-1 binding sites and the TATA box sequence, and of the LTR enhancer, which is the target for the p50/p65 NF-κB heterodimer; the same region also contains the binding sites for other transcription factors including Ets-1 and USF (reviewed in Jones and Peterlin, 1994). The second open area spans the primer-binding site immediately downstream of the 5′ LTR. These two open regions are separated by a single nucleosome called nuc-1 that is specifically and rapidly destabilized during transcriptional activation. The position of nuc-1 in the close proximity of the transcription start site and its displacement during transcriptional activation suggest that chromatin plays a crucial role in the suppression of HIV-1 transcription during latency and that nuc-1 disruption is necessary for transcriptional activation (Verdin et al., 1993; Van Lint et al., 1996).
Genomic footprinting experiments performed either in activated or in silently infected cells have indicated that most of the transcription factor binding sites at the promoter, including the TATA box, the Sp1 sites, the enhancer region and the USF site, are occupied by cellular proteins independent from the activation state (Demarchi et al., 1993). This is a further indication that the transcriptional activation of the integrated LTR is not primarily restricted by DNA target site accessibility, but occurs through the modulation of chromatin conformation.
The silent proviral LTR can be switched on from post-integration latency by cell treatment with a variety of stimuli, including cytokines, antibodies or phorbol esters, which act primarily through the enhancer region of the LTR (Pomerantz et al., 1990; Jeang et al., 1993), and by the action of the viral Tat protein. Tat is a highly unusual transactivator that binds an RNA element positioned at the 5′ end of the proviral transcript (Berkhout et al., 1989; reviewed in Marcello et al., 2001b). Through this interaction, the protein activates HIV-1 transcription by promoting the assembly of transcriptionally active complexes at the LTR by multiple protein–RNA and protein–protein interactions. Three prominent types of interaction that mediate Tat transactivation have been biochemically and genetically defined: those with general transcription factors (including TBP, TAFII250 and RNA polymerase II) (reviewed in Marcello et al., 2001b); those with kinase complexes able to phosphorylate the C-terminal domain of RNA polymerase II (in particular with cyclin T1/CDK9) (see Marcello et al., 2001a, 2003, and references therein); and those with cellular proteins possessing HAT activity. In particular, we and others have shown that Tat associates with: (i) the transcriptional co-activators p300 and the highly homologous cAMP-responsive binding protein (CREB) binding protein (CBP); (ii) the p300/CBP-associated factor (P/CAF); and (iii) the general control non-derepressible-5 (GCN5) protein (Benkirane et al., 1998; Hottiger and Nabel, 1998; Marzio et al., 1998; Col et al., 2001). The Tat-mediated recruitment of HAT proteins most likely explains the changes in chromatin conformation observed at the LTR upon transactivation (Verdin et al., 1993; Van Lint et al., 1996).
In this work we exploited chromatin immunoprecipitation (ChIP) to map precisely the kinetics of histone acetylation and factor recruitment at the LTR during transcriptional activation.
Results
Histone acetylation at the LTR promoter of U1 cells upon TPA induction
The U1 monocytic cell line is a well defined model of post-integration latency (Wright et al., 1986; Folks et al., 1987). This cell line contains two copies of the integrated provirus that produce very low, albeit detectable, full-length HIV-1 mRNAs under basal conditions (Jeang et al., 1993). HIV-1 expression and viral replication is markedly induced at the transcriptional level by exposure to different mitogens, including phorbol esters (Folks et al., 1987, 1988; Demarchi et al., 1993). To analyze in detail the kinetics of provirus transcriptional activation in these cells, we measured the levels of viral RNA after induction with TPA using a quantitative RT–PCR procedure based on competitive PCR (Diviacco et al., 1992; Comar et al., 1996). Competitive RT–PCR was performed with the primer pair Nu2f/Nu2r (Table I) in the viral RNA leader sequence, and using the multicompetitor DNA fragment shown in Figure 2A, also allowing quantification of the reference cellular mRNA for β-actin. The quantitation of HIV-1 and β-actin mRNA levels at different time points (0, 1, 3 and 5 h) after TPA treatment is shown in Figure 2B. In unstimulated cells, one HIV-1 transcript was detected every ∼100 β-actin mRNA molecules; after 5 h of TPA treatment, the relative abundance of HIV-1 mRNA remarkably increased, reaching almost 50% in the β-actin mRNA levels. This is in agreement with our previously published northern blot analysis of viral transcripts in TPA-stimulated U1 cells (Demarchi et al., 1993).
Table I. Primers used for the competitive PCRs.
| Primer | Position | Primer sequence | Size of PCR product (bp) |
|
|---|---|---|---|---|
| Target | Competitor | |||
| Nu0f | HIV-1 -453 | GAAGGGCTAATTTGGTCCCA | 307 | 340 |
| Nu0r | HIV-1 -146 | GATGCAGCTCTCGGGCCTG | ||
| L1f | HIV-1 -176 | CGAGAGCTGCATCCGGAGTA | 239 | 300 |
| L1r | HIV-1 +61 | AGCTTTATTGAGGCTTAAGC | ||
| Nu1f | HIV-1 +96 | AGTAGTGTGTGCCCGTCTGT | 205 | 294 |
| Nu1r | HIV-1 +301 | TTGGCGTACTCACCAGTCGC | ||
| Nu2f | HIV-1 +304 | ATTTTGACTAGCGGAGGCTA | 202* | 278 |
| Nu2r | HIV-1 +506 | ACAGCCTTCTGATGTCTCTA | ||
| B13dx | Human chr 19 | GCCAGCTGGGTGGTGATAGA | 164 | 190 (HL3T1) or 226 (U1) |
| B13sx | CCTCAGAACCCAGCTGTGGA | |||
| B48dx | GACTGGAAACTTTTTTGTAC | 168 | 198 (HL3T) or 214 (U1) | |
| B48sx | TAGCTACACTAGCCAGTGACCTTTTTCC | |||
| CATf | CAT gene | AATCGTCGTGGTATTCACTCC | 300* | 210 |
| CATr | CCGTTGATATATCCCAATGGC | |||
Four HIV-1 genomic regions, two regions in human chromosome 19 and one region in the CAT reporter gene were selected for amplification. The position of the HIV-1 primers refers to the NY5/BRU isolate; position +1 corresponds to the first transcribed nucleotide. Each primer pair (see Figure 1) was used for the simultaneous amplification of both target DNA and competitor; the length of the respective PCR products is indicated in the corresponding columns. The asterisk also indicates the length of the amplified cDNA products, when RT–PCR was performed.
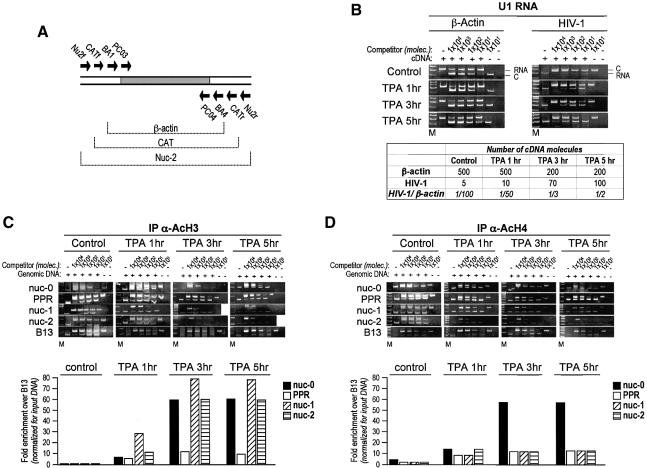
Fig. 2. Localized histone acetylation at the integrated LTR in U1 cells following TPA stimulation. (A) Schematic representation of the multicompetitor DNA used for quantification of RNA by competitive RT–PCR. (B) Induction of HIV-1 transcription in U1 cells by TPA measured using quantitative RT–PCR. Cells were either mock- or TPA- (10–7 M final concentration) treated for 1, 3 or 5 h. Decreasing amounts of competitor DNA were mixed with fixed amounts of cDNA, and PCR was performed either with β-actin or HIV-specific primers. (C) Histone H3 acetylation. Formaldehyde-crosslinked chromatin from U1 cells induced with TPA (t = 0, 1, 3 and 5 h) was immunoprecipitated with anti-acetylated histone H3 (AcH3) antibody, and the relative acetylation of histone H3 was quantitated by competitive PCR as described above. The results of fold enrichment for regions nuc-0, PPR, nuc-1 and nuc-2 over control B13, calculated on the basis of the estimated number of DNA molecules, are shown graphically in the histograms below the gels. (D) Histone H4 acetylation, as for (C), with the immunoprecipitation performed with the anti-acetylated histone H4 antibody.
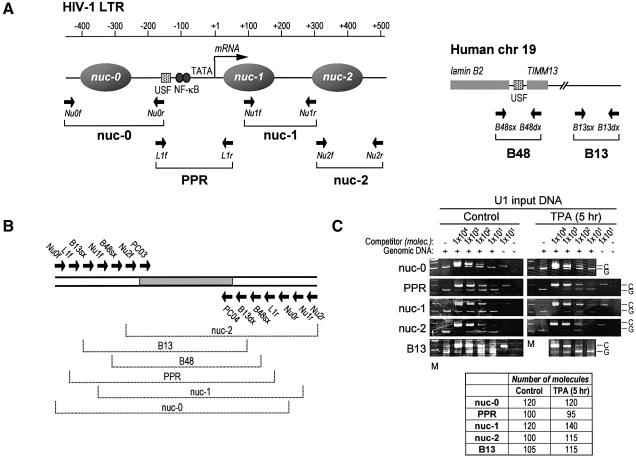
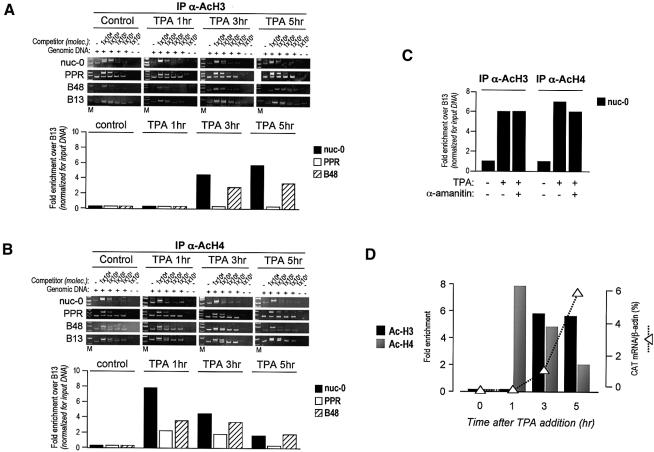
To determine whether transcriptional activation by TPA induces an increase in histone acetylation at the viral promoter in vivo, aliquots of untreated and TPA-treated cells were analyzed in parallel by ChIP at different time points after treatment. Formaldehyde crosslinked, sonicated chromatin fragments from U1 cells were immunoprecipitated at different time points (0, 1, 3 and 5 h) using antibodies directed against the N-terminal tails of acetylated histone H3 (AcH3) and histone H4 (AcH4). Six different genomic sites were investigated in U1 cells (Figure 1A). Four of these mapped to contiguous regions in the HIV-1 proviral DNA, mostly according to the positions of the nucleosomes (nuc-0, -1 and -2) and the nucleosome-free region (PPR). The other two regions mapped in the lamin B2 gene domain on chromosome 19, and were extensively studied for the identification and characterization of a human origin of DNA replication (Giacca et al., 1994; Abdurashidova et al., 2000). B13 is 5 kb away from the lamin B2 origin, in a region not containing any gene. B48 is located within the lamin B2 origin, and also encompasses the promoter region of the mitochondrial inner membrane translocase 13 (TIMM13) gene.
Fig. 1. Competitive PCR for quantitative ChIP. (A) Schematic structure of the integrated HIV-1 LTR promoter (left side) and of the control genomic region on human chromosome 19 (right side). On the LTR sequence, the positions of the nucleosomes are indicated, alongside those of some of the relevant transcriptional control elements; the transcription start site is indicated by an arrow pointing rightward. On both regions, the position of the primer pairs used for PCR amplification is indicated by arrows. (B) Schematic representation of the multicompetitor DNA used for DNA quantification by competitive PCR. This DNA fragment contains all the primer recognition sites, as indicated, arranged in order to generate PCR amplification products of different but comparable size to those obtained by the amplification of genomic DNA. The gray area indicates the original DNA fragment used as a core for competitor construction, flanked by the β-globin primers PCO3 and PCO4. (C) Quantification by competitive PCR of crosslinked, non-immunoprecipitated (input) DNA from U1 cells without, or following, stimulation with TPA. After crosslinking, a fixed amount of total sonicated DNA was mixed with decreasing concentrations of the multicompetitor segment, as indicated at the top of each lane, and amplified with the primer pairs for nuc-0, PPR, nuc-1, nuc-2 and B13 (Table I). After amplification, the PCR products were resolved by polyacrylamide gel electrophoresis and stained with ethidium bromide; the specific bands were quantified by densitometric analysis. The number of DNA molecules corresponding to each genomic region was calculated according to the principles of competititve PCR by evaluating the slope of the curve fitting the values corresponding to the competitor:genomic ratios (Diviacco et al., 1992). The calculated number of DNA molecules in the analyzed sample dilution is reported in the table below the gels. The results of a representative experiment are shown. All experiments were independently repeated at least three times; the detected variation was <25%. M, molecular weight markers; C, PCR band corresponding to competitor DNA; G, PCR band corresponding to genomic or HIV DNA.
On the basis of these selected sequences, we constructed a specific multicompetitor DNA (shown in Figure 1B), in which all competitor segments are equimolar in the PCR reactions. We also verified that all primers amplified at the same rate in control amplifications using crosslinked, but not immunoprecipitated, chromatin, as shown in Figure 1C. This control was performed in each ChIP experiment to exclude an amplification bias for any of the analyzed region after protein–DNA crosslinking.
Following TPA induction in U1 cells, we found that selected segments of the proviral genome were specifically enriched by ChIP with anti-AcH3 and anti-AcH4 antibodies. One hour after TPA stimulation, H3 was found already acetylated in the region of nuc-1 (∼30-fold enrichment over the control B13 region; Figure 2C). nuc-0 and -2 appeared only marginally acetylated at this time point compared with nuc-1, although their acetylation level was increased almost 10-fold over control. The acetylation of H3 in nuc-1, as well as in nuc-0 and -2, continued to increase after 3 h of TPA treatment, with a remarkable enrichment of >70-fold for nuc-1; these levels of acetylation persisted unmodified after 5 h. Interestingly, only a relatively modest increase in acetylation (<10-fold) was observed in the PPR region, even after 5 h of TPA induction, a result that is in perfect agreement with the nucleosome positioning within the LTR region (Verdin et al., 1993; Van Lint et al., 1996). This observation also indicates that the developed ChIP procedure allows for the precise analysis of contiguous DNA regions positioned at distances <500 bp.
H4 displayed a different pattern of acetylation in the same proviral region. Enrichment for acetylated H4 in the nuc-0 area began 1 h after TPA addition; enrichment over control was 50-fold at 3 h and persisted unchanged afterwards. In contrast, H4 acetylation in the PPR, nuc-1 and nuc-2 regions was detectable as early as 1 h after TPA stimulation, but remained at the same low levels at later time points. Interestingly, in basal conditions (no TPA), H4 appeared modestly, but still reproducibly, acetylated in the nuc-0 region.
Together these results indicate that site-specific modification of chromatin at the promoter region anticipates full transcriptional activation of the HIV-1 LTR.
Nucleosomes at the LTR show distinct acetylation patterns at specific lysines of H3 and H4
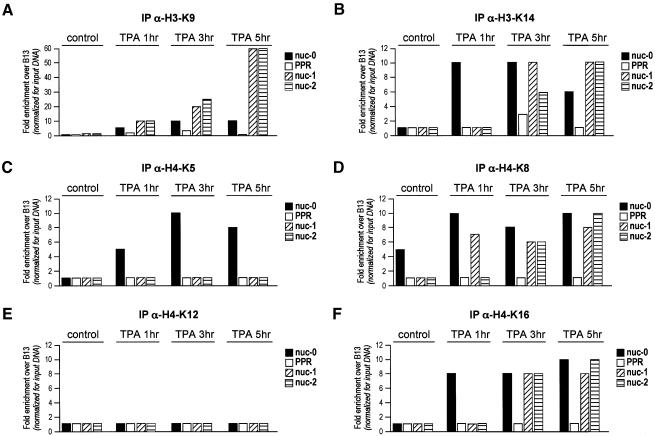
The ChIP experiments described in the previous paragraph were performed with antibodies raised against peptides of the N-terminal tail of histone 3 acetylated at K9 and K14, and histone H4 acetylated at K5, K8, K12 and K16. In order to dissect the contribution of each acetylated lysine to HIV transcription, we performed a series of ChIP experiments with antibodies against individual acetylated lysines of H3 and H4. Following TPA induction of viral transcription in U1 cells, acetylation of H3 K9 was detectable at 1 h after drug addition at all three nucleosomal regions; in contrast, K14 was acetylated prevalently at nuc-0 (Figure 3A and B). At the subsequent time points (3 and 5 h), K9 acetylation markedly increased at nuc-1 and -2 only, while K14 acetylation, although uniformy distributed in all three nucleosomal regions, remained at lower levels.
Fig. 3. Nucleosomes at the LTR show distinct acetylation patterns at specific lysines of H3 and H4. Chromatin immunoprecipitation, performed as in Figure 2, was performed using antibodies against acetylated Lys9 (A) and Lys14 (B) of histone H3, and against acetylated Lys5 (C), Lys8 (D), Lys12 (E) and Lys16 (F) of histone H4.
Global H4 acetylation had shown a marked increase at Nuc-0 (up to 60-fold) with modest, albeit detectable acetylation (10-fold) of the other two regions (Figure 2D). In an early phase after TPA stimulation, the acetylation of Nuc-0 was mainly attributable to the acetylation of K5, K8 and K16 (Figure 3C, D and F). In contrast, the lower levels of acetylation of the other two nucleosomal regions were mostly due to the acetylation of K8 and K16. No increase in K12 acetylation was observed for any of the regions tested (Figure 3E).
Taken together, these results indicate that, with respect to the transcription start site, both the upstream and the downstream nucleosomal regions display peculiar patterns of acetylation, temporally preceding the onset of transcription.
Factor recruitment at the viral LTR upon TPA induction of U1 cells
We went on to exploit the ChIP methodology to study the binding of transcription factors and coactivators to the LTR promoter in the same experimental conditions.
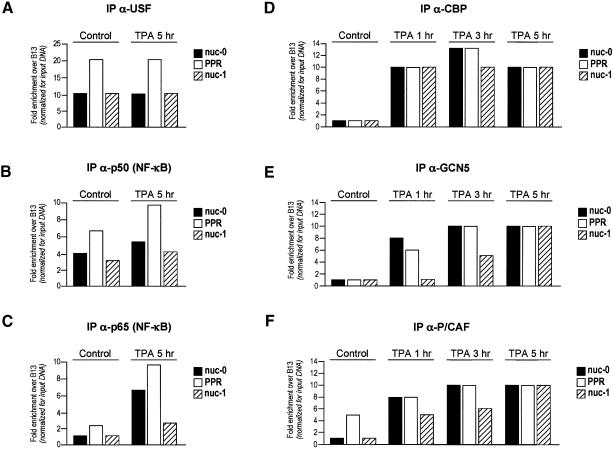
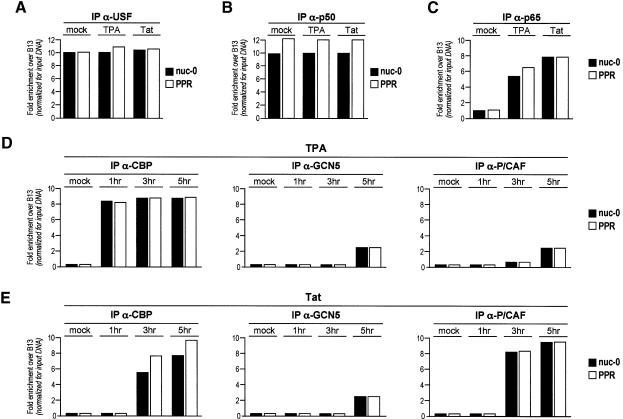
The upstream stimulatory factor (USF) and NF-κB are known to bind the HIV-1 LTR within the PPR region (nt –166 and –104 upstream of the transcription start site, respectively; Demarchi et al., 1993; d’Adda di Fagagna et al., 1995). ChIP analysis of the PPR region together with its boundaries (nuc-0 and -1) using specific anti-USF, -p50 and -p65 antibodies showed that both USF and p50 bind the PPR region in both control and TPA-induced U1 cells (Figure 4A and B). In contrast, the binding of the p65 subunit of NF-κB to the PPR region was strongly enhanced upon TPA induction (Figure 4C). These data are well in agreement with our previous observations obtained by in vivo footprinting experiments that indicated persistent occupation of the USF and NF-κB site, irrespective of the activation state of the promoter (Demarchi et al., 1993), and that transcriptional activation followed the induction of p65 (Pazin et al., 1996).
Fig. 4. Factor recruitment to the integrated viral promoter in U1 cells following TPA treatment. The binding of transcription factors USF (A), p50 (B) and p65 (C), and the time-dependent recruitment of histone acetyltransferases CBP (D) GCN5 (E) and P/CAF (F) (1, 3 and 5 h after TPA induction) to the viral regions nuc-0, PPR and nuc-1, and to the control B13 region, were analyzed by quantitative ChIP, as described in the legend of Figure 2. The results of the estimated enrichments for each transcription factor in the regions nuc-0, PPR and nuc-1 over the control B13 region are presented. Calculations were made as in Figure 2.
The observation that transcriptional induction of the LTR correlates with nucleosome acetylation prompted us to investigate the recruitment of known HATs to the promoter region. Using antibodies specific for CBP, P/CAF (p300/CBP associated factor) and hGCN5, we tested the interaction of these factors with nuc-0, nuc-1 and PPR in control and TPA-induced U1 cells. CBP was immunoprecipitated with the same efficiency at all three promoter regions after 1 h of induction, and remained bound also at later time points (Figure 4D). On the other hand, GCN5 showed specific binding preferences for different viral regions: 1 h after TPA induction its presence was detected at nuc-0 and PPR, and at 3 h after induction it was detected in all the viral regions examined (nuc-0 PPR and nuc-1). Nonetheless, the detected amount of this HAT at nuc-1 remained lower than at nuc-0 and PPR. Finally, at 5 h after induction, GCN5 demonstrated an equal distribution over all three viral regions (Figure 4E). The recruitment of P/CAF to the promoter was peculiar in that it was the only HAT that was detectable at the PPR before induction. After induction, further recruitment of P/CAF was time-dependent, showing maximum binding at 5 h after TPA treatment (Figure 4F). As in the case of GCN5, P/CAF initially showed stronger binding at nuc-0 and PPR, and only afterwards appeared to bind nuc-1 with the same efficiency. In any case, it should be considered that the spread association of acetyltransferases with all three regions investigated probably reflects the fact that these transcriptional co-activators do not contact DNA directly, contrary to both histones and transcription factors.
Localized histone acetylation at the viral LTR promoter in HL3T1 cells upon induction with TPA or recombinant Tat
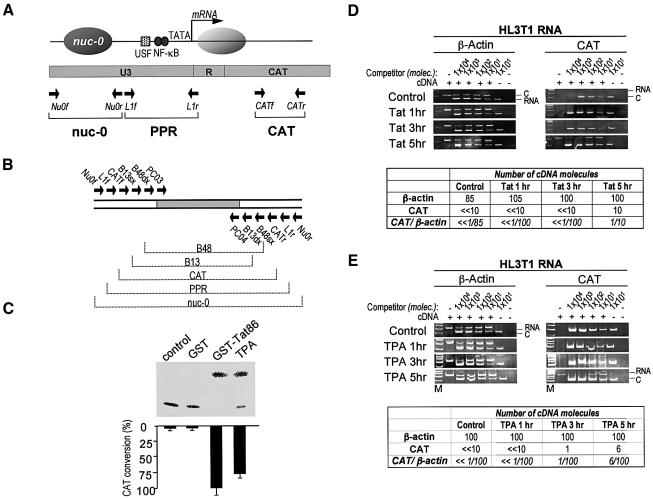
In order to analyze the specific contribution of the Tat transactivator to the activation process, we performed an array of ChIP experiments in HL3T1 cells. This is a widely exploited HeLa cell derivative that harbors several copies (see below) of an integrated LTR-CAT (chloramphenicol acetyltransferase) cassette that is silent in basal conditions, but can be readily activated by a variety of stimuli, including treatment with TPA (Wright et al., 1986; Felber and Pavlakis, 1988; Marzio et al., 1998). In addition, exogenous recombinant Tat protein is efficiently internalized by these cells in a transcriptionally active form through an active endocytosis pathway (Marzio et al., 1998; Tyagi et al., 2001; Fittipaldi et al., 2003). The induction of the LTR promoter in HL3T1 cells by extracellullar recombinant glutathione S-transferase (GST)-Tat (5 µg/ml) or by TPA (1 × 10–7 M) is shown in the CAT assay of Figure 5C.
Fig. 5. Induction of HIV-1 transcription in HL3T1 cells. (A) Schematic structure of the integrated LTR-CAT cassette. Converging arrows indicate the position of the specific primers used for amplification. (B) Multicompetitor used for the PCR quantification of DNA from HL3T1 cells. (C) Transcriptional activity of the LTR-CAT promoter upon stimulation with GST, GST–Tat or TPA as measured by CAT assays. GST or GST–Tat were delivered to cells at a final concentration of 5 µg/ml, while final TPA was 1 × 10–7 M. Five hours after treatment, cells were extensively washed with PBS and new medium was added, and the CAT assay was performed 20 h later. (D) Kinetic analysis of LTR activation in Tat-treated HL3T1 cells. One, 3 and 5 h after stimulation, total RNA was extracted and reverse-transcribed with random primers. A fixed amount of cDNA from each time point was mixed with increasing amounts of competitor molecules and used for competitive PCR amplification with CAT- or β-actin primers. The quantification was performed as in Figure 2. (E) Kinetic analysis of LTR activation in control cells and in cells treated for 1, 3 and 5 h with TPA.
The precise kinetics of transcriptional activation in HL3T1 cells was measured by the competitive PCR amplification of cellular RNA using the primer pairs shown in Figure 5A and the multicompetitor shown in Figure 4B, as described above. In the absence of stimuli, no HIV-CAT mRNA emanating from the HIV-1 promoter could be detected. Five hours of treatment with recombinant Tat caused a remarkable increase in CAT mRNA levels, which reached ∼10% of the cellular β-actin mRNA (Figure 5D). TPA caused a detectable (but modest) increase of CAT mRNA after 3 h of stimulation; the levels of mRNA increased further after 5 h, reaching 6% of β-actin mRNA levels (Figure 5E). Thus, the kinetics of TPA induction of HL3T1 cells is similar to that of HIV-1 mRNA induction in U1 cells, although the latter respond better in terms of RNA production (up to 50% of β-actin mRNA levels).
Initially, ChIP experiments were carried out to identify the patterns of histone acetylation after induction with TPA or with exogenous Tat protein. Four different genomic regions were examined by competitive PCR, two of which are specific viral regions, nuc-0 and PPR (Figure 5A), while the other two are the B48 and B13 genomic regions in the lamin B2 gene domain, as described above. In order to quantify the PCR products precisely, a novel multicompetitor DNA corresponding to these regions was constructed (Figure 5B).
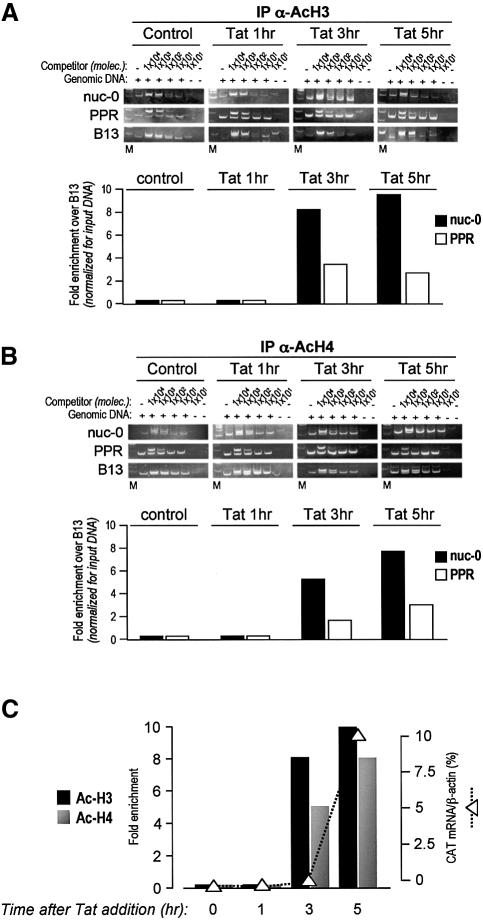
After the addition of TPA, histone acetylation in the nuc-0 region was clearly detected at earlier time points with respect to HIV-1 RNA detection. This was particularly evident for histone H4, which appeared to be already acetylated 1 h after TPA addition (8-fold increase over control cells), with acetylation progressively diminishing at 3 and 5 h (Figure 6B). The rate of H3 acetylation at nuc-0 was different, with acetylated histone H3 being undetectable at 1 h, appearing at 3 h (>4-fold increase) and increasing slightly further (>5-fold) at 5 h after TPA addition (Figure 6A). Thus, the acetylation of both histones at nuc-0, and in particular of histone H4, appears to precede the onset of LTR transcription, as shown graphically in Figure 6D. Later, ongoing transcription is paralleled by the persistent acetylation of H3, while the acetylation of H4 progressively diminishes and returns to basal levels. These results were also confirmed by ChIP analysis at 7 and 9 h after transcriptional induction, two time points at which H3, but not H4, continued to be acetylated (data not shown). TPA also caused a moderate increase in the acetylation of both H3 and H4 in the B48 region (Figure 6A and B). This is not surprising since B48 encompasses both an origin of DNA replication and the TIMM-13 gene promoter.
Fig. 6. Localized histone acetylation at the integrated LTR in HL3T1 cells following TPA treatment; acetylation precedes the onset of transcription. (A and B) Cells were treated for 1, 3 or 5 h with TPA, and the immunoprecipitated chromatin was analyzed by PCR. The competitive PCR analysis of two viral and two genomic regions immunoprecipitated with anti-acetylated H3 and H4 antibodies was performed as described above. (C) Cells were treated for 1.5 h with α-amanitin prior to TPA treatment (for 3.5 h); chromatin was then immunoprecipitated with anti-acetylated H3 and H4 antibodies as described above and analyzed for enrichment in the nuc-0 region. (D) Graphs show the kinetics of histone acetylation at nuc-0 and the levels of transcription at different time points after TPA addition.
In order to substantiate further the notion that histone acetylation occurs before mRNA production from the viral LTR promoter, we looked at histone acetylation in HL3T1 cells that were pre-treated for 1.5 h with the inhibitor of RNA polymerase II α-amanitin, before the addition of TPA for 3.5 h. As shown in Figure 6C, pre-treatment with this drug did not affect the acetylation of either histones 3 or 4 at the nuc-0 region, thus indicating that nucleosome acetylation precedes, and is independent from, the actual onset of transcription.
Changes in histone acetylation were also measured by ChIP at 0, 1, 3 and 5 h after the addition of the Tat recombinant protein to the cell culture medium. Also in this case, the acetylation of both H3 and H4 in the nuc-0 region was detected at a time point (3 h) at which no transcripts were evident in the cell (∼8-fold increase over control for H3 and ∼6-fold increase for H4; Figure 7A and B). Five hours after Tat addition, immunoprecipitation for acetylated histones H3 and H4 determined a 10- and 8-fold enrichment over control samples, respectively. Thus, unlike TPA treatment, transcriptional activation by Tat is concomitant with a parallel increase in acetylation of both H3 and H4. In further support of the specificity of the data described above, no acetylation in the B48 region was detected after treatment with Tat.
Fig. 7. Localized histone acetylation at the integrated LTR in HL3T1 cells following treatment with recombinant Tat. (A and B) Acetylation of histones H3 and H4 upon Tat stimulation, shown as in Figure 6. (C) Graphic comparison of the kinetics of histone acetylation and CAT mRNA synthesis upon Tat activation of the integrated LTR-CAT cassette.
Factor recruitment at the viral LTR upon induction with TPA and recombinant Tat in HL3T1 cells
We next set out to determine factor binding at the viral promoter, concomitant with the detected changes in chromatin acetylation and RNA transcription, by inducing HL3T1 cells with TPA or with recombinant Tat. In a first set of experiments, we performed quantitative ChIP with antibodies against USF and the p50 and p65 subunits of NF-κB. As shown in Figure 8A, we confirmed that binding of USF is constitutive under all the conditions examined. Binding of the p50 subunit of NF-κB was constitutive as well, since there were no differences between control and induced cells. In contrast, and as in the case of U1 cells, p65 was recruited to the promoter after induction by either Tat or TPA, thus confirming that the p50-p65 heterodimer bound to the viral enhancer contributes significantly to viral transcriptional activation (Figure 8B and C). As far as these transcription factors are concerned, no significant differences were detected between induction with Tat or with TPA.
Fig. 8. Factor binding to the LTR promoter in HL3T1 cells induced with either TPA or Tat. Immunoprecipitation of crosslinked chromatin with antibodies against transcription factors USF (A), p50 (B) and p65 (C) after TPA or Tat treatment in HL3T1 cells for 5 h. The time course of the binding of histone acetyltransferases GCN5, CBP and P/CAF to the viral regions nuc-0, PPR and nuc-1 was analyzed by quantitative ChIP after TPA (D) or Tat (E) treatment. Calculations were made as in Figure 2.
Subsequently, we reasoned that the differences in the kinetics and pattern of histone acetylation that were detected after treatment with Tat or TPA might be attributable to the differential binding of HATs to the LTR. Therefore, we investigated the kinetics of recruitment of CBP, P/CAF and hGCN5 after transcriptional activation by ChIP, in the same conditions used for the analysis of histone acetylation. As shown in Figure 8D, we found that as early as 1 h after treatment with TPA, an antibody against CBP could immunoprecipitate the crosslinked LTR region (enrichment > 8-fold over control for both nuc-0 and PPR), and that CBP recruitment continued at the same levels at 3 and 5 h. In contrast, enrichment for both GCN5 and P/CAF was less pronounced for both regions (∼2.5-fold for both GCN5 and P/CAF for nuc-0). After treatment with recombinant Tat, histone acetylation and transcriptional activation at 3 and 5 h were paralleled by a marked recruitment of both CBP and P/CAF to both nuc-0 and PPR (Figure 8E). Also in this case, enrichment for GCN5 appeared to be less pronounced (∼2-fold) and to occur at a later time point.
Discussion
In this work we examined the chromatin modifications occurring at the integrated LTR promoter in response to transcriptional activation.
First, we demonstrated that a significant increase in histone acetylation occurs at the promoter in U1 cells upon activation with phorbol esters. This increase was detected even before viral mRNA levels started to rise significantly. However, the patterns and extent of histones H3 and H4 acetylation were not identical. Histone H3 acetylation seemed to be uniformly distributed in all the nucleosomes examined (nuc-0, -1 and -2), with acetylation becoming detectable as early as 1 h after promoter induction, and being maintained at high intensity as long as mRNA levels were steady. By using antibodies against specific lysine residues, we could gain deeper insight into the complexity of the changes. Rapidly following TPA stimulation, nucleosomes showed an increase in histone H3 acetylation, with Lys14 being exclusively acetylated at nuc-0, and Lys9 being acetylated preferentially at the downstream regions nuc-1 and -2. Lysine 14 acetylation thus appeared to spread from nuc-0 to the adjacent nucleosomal regions at 3 h after TPA stimulation, hence rendering both nuc-1 and nuc-2 acetylated on both H3 lysines, an event that has recently been associated with the specific recruitment of TAFII250 to the IFN-β promoter (Agalioti et al., 2002).
The high levels of Lys9 acetylation, observed at nuc-1 and -2 at 5 h after stimulation, correlate with the spread distribution of GCN5 and P/CAF along the viral promoter. The maintenance of high levels of acetylation at a promoter was suggested to require the continuous presence of acetyltransferases at this location, otherwise the patterns of histone acetylation are rapidly restored to their basal levels (Katan-Khaykovich and Struhl, 2002). Accordingly, we could detect that CBP bound to all promoter regions from the first hour of induction of U1 cells. However, we cannot exclude the possibility that the transcription complex intermittently cycles the viral promoter, as has been shown to be the case with the estrogen receptor transcription complex in the presence of continuous stimulation by estrogen (Shang et al., 2000).
Histone H4 proved to have a very complex pattern of acetylation, especially at nuc-0. This nucleosome, which lies upstream of the transcription start site, showed exclusive acetylation at lysines 5 and 16 as early as 1 h after TPA stimulation. Lysine 8 at nuc-0 was found to be acetylated even before stimulation of U1 cells, a finding consistent with the notion that low levels of transcription are always detectable in U1 cells (see below).
These differences in the H3 and H4 acetylation of the LTR nucleosomes might possibly account for the differences detected by enzymatic digestion of the LTR chromatin, which showed selective modification of nuc-1 after transcriptional activation (Verdin et al., 1993; Van Lint et al., 1996). In this respect, it has recently been proposed that H4 acetylation is needed for the recruitment of a large protein complex that loosens DNA from histones locally, while H3 acetylation is required instead for the recruitment of a second complex that pushes the core of histones away while transcription proceeds (Agalioti et al., 2002). Histone acetylation is a prerequisite for the retention of the SWI/SNF chromatin remodeling complex at the promoter region (Hassan et al., 2002), thus further supporting the possibility that early and specific H4 changes in the nuc-0 region are needed also for chromatin remodeling. In this respect, it is worth noting that acetylation of histone H4 Lys8 leads to the firm association of SWI/SNF through the bromodomain of its BRG1 component (Agalioti et al., 2002). The difference in the distribution patterns of H3 and H4 acetylation that we observed might reflect these possibilities, with H3 acetylation occurring at downstream locations along with the transcription complex. Further studies, however, need to be performed in order to decipher completely the HIV LTR histone code.
An unexpected finding was that P/CAF was already present in the PPR region, but not at nucleosome positions, in untreated U1 cells. This observation suggests that P/CAF is most likely recruited to the promoter through a pathway other than CBP and GCN5. In this respect, it should be considered that U1 cells express a Tat protein that is mutated in its N-terminal activation domain, but maintains an intact P/CAF-interacting domain (Bres et al., 2002; Mujtaba et al., 2002; Quivy and Van Lint, 2002). Therefore, it is possible to conceive that this mutated form of Tat still recruits P/CAF at the viral promoter. According to this interpretation, the presence of P/CAF at the promoter would not be sufficient to allow activated transcription, but might still account for the basal level of transcription observed in U1 cells, concomitant with low levels of acetylation of histone H4 Lys8. Consistent with this notion, neither P/CAF nor Lys8 acetylation were present at the viral LTR in non-stimulated HL3T1 cells.
An ordered recruitment of transcription factors, as well as the transcription activation timing, are gene-specific events (Cosma, 2002). Tat is known to interact with and recruit histone acetyltransferases to the viral promoter (Benkirane et al., 1998; Hottiger and Nabel, 1998; Marzio et al., 1998; Col et al., 2001). In this study we have observed that Tat activation of HL3T1 cells is associated with histone H3 and H4 acetylation at Nuc-0, and with promoter occupancy by CBP and P/CAF. We have also observed that, in conditions under which the promoter is completely silent, its activation by Tat or TPA correlates with the recruitment of HATs and with the acetylation of histones well before the actual onset of transcription. This notion is also reinforced by the observation that α-amanitin, while blocking transcription, did not interfere nucleosomal acetylation. It is also interesting to note that, upon Tat induction, we were able to detect the presence of two HATs, CBP and P/CAF. GCN5 was detected at the promoter only in trace amounts in the late phases of induction. Our data are consistent with the finding that, in several promoters, including PH08, CATD and IFN-β, CBP and P/CAF associate with Pol II and might be involved in modulating its activity by modifying the nucleosomes that mask the TATA box or the start site (Agalioti et al., 2000; Shang et al., 2000; Reinke et al., 2001; for a recent review see Cosma, 2002).
Recombinant Tat was consistently more powerful than TPA in inducing transcriptional activation from the HIV-1 promoter in HL3T1 cells. Transcription, as measured by mRNA levels, followed the increase in H3 acetylation in both U1 and HL3T1 cells in response to Tat, reflecting the constant presence of CBP at the promoter. However, in conditions under which Tat was not present and activation occurred only via TPA, H4 acetylation at nuc-0 rapidly decreased after the initial burst. This observation is also at variance with the behavior of H4 acetylation in U1 cells, where nuc-0 was readily acetylated but remained high at later time points. We can only speculate that P/CAF is responsible for the elevated H4 acetylation levels over longer time periods, since in its absence the initial increase in H4 acetylation was not maintained in HL3T1 cells. Hence, the absence of P/CAF, under conditions of TPA stimulation, could be responsible for a suboptimal activation of the LTR. In line with this interpretation, He and Margolis showed that activation of LTR expression by Tat resulted in the displacement of HDAC1 and in the increased acetylation of H4 (He and Margolis, 2002). HDAC1 is recruited to the promoter in a complex with YY1 and LSF, and these factors together have a repressive effect on LTR-driven transcription, as shown earlier by the same group (Coull et al., 2000). Thus, conceivably, Tat appears able to transactivate the HIV-1 LTR to the highest levels, also by relieving an HDAC1 block and inducing P/CAF. Further experiments aimed at the specific study of HDAC recruitment at the LTR appear to be necessary to elucidate this mechanism in detail.
Reactivation of HIV-1 from post-integration latency has profound clinical implications, which exhort a thorough understanding of the underlying molecular mechanisms (Pierson et al., 2000). The overall picture emerging from the results obtained in this work indicate that transcriptional activation is a complex process involving remarkable changes in chromatin conformation that follow the specific recruitment of both activation-specific transcription factors and transcriptional co-activators. In this context, it is worth noting that Tat appears to play a highly specific role in this process through its multifaceted interactions with different cellular partners.
Materials and methods
Cell culture and treatments
The U1 monocytic cell line (obtained from the AIDS Research Reagent Program, National Institute of Allergy and Infectious Diseases, and the National Institutes of Health), was maintained at low-passage level from frozen stock in RPMI 1640 containing 20 mM l-glutamine and 10% fetal calf serum (FCS). Cells growing at a density of 5 × 105 were treated with 10–7 M 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma) for 1, 3 or 5 h. At the indicated time points, cells were washed twice with phosphate-buffered saline (PBS), and then either collected for RNA purification or resuspended in fresh medium and crosslinked with formaldehylde for 10 min at 37°C for ChIP.
HL3T1 cells are a HeLa derivative containing an integrated CAT gene under the control of the HIV-1 LTR promoter (Felber and Pavlakis, 1988). HL3T1 cells (2 × 107) were grown to ∼60% confluence in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS, and treated with 40 µg of recombinant GST–Tat86 (2 µg/ml final concentration). Alternatively, HL3T1 cells were treated with 1 × 10–7M TPA. In the experiments with α-amanitin, HL3T1 cells were incubated in medium containing α-amanitin (25 µg /ml) for 1.5 h before TPA treatment.
Purification of recombinant GST–Tat86
For the preparation of GST and GST–Tat86, exponentially growing cultures of Escherichia coli BL21 (DE3)pLysS*, harboring the expression plasmid, were induced with 1 mM isopropyl-b-d-thiogalactopyranoside for 4 h at 30°C. Soluble GST and GST–Tat86 were prepared as described previously (Marzio et al., 1998; Tyagi et al., 2001).
Reverse transcription and competitive RT–PCR
Cells were treated with Tat or TPA as described above. At different time points, total cellular RNA was extracted with an RNAeasy Minikit (Qiagen). RNA (1 µg) was treated with DNase (Gibco-BRL) and reverse-transcribed using 20 U MoMLV RT (Gibco-BRL), and 1 µM of random hexameric primers (Gibco-BRL). The synthesized cDNA was quantified using competitive RT–PCR as described previously (Comar et al., 1996).
Chromatin immunoprecipitation
After treatment with Tat or TPA for the indicated time periods, cells were fixed by adding a fixing solution (11% formaldehyde, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA pH 8.0, 50 mM Tris–HCl pH 8.0) directly to the cell culture medium at 1% final dilution. Cross-linking was allowed to proceed for 10 min at 37°C and was stopped by the addition of glycine at a final concentration of 0.125 M. Fixed cells were washed once in ice-cold PBS, once in buffer B1 (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris–HCl pH 8.0) and once in buffer B2 (1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl, 10 mM Tris–HCl pH 8.0). Cells were pelleted by centrifugation and resuspended in RIPA 50 buffer [50 mM NaCl, 20 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.5% Nonidet P-40 (NP-40), 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS), protease inhibitors]. Chromatin was sheared by sonication (20 pulses, 30 s each) on ice and centrifuged to pellet debris. Immunoprecipitations were carried out using the antibodies indicated (4 µl of each antibody) [anti-acetylated antibodies (total and lysine-specific) from Upstate Biotechnology; anti-CBP, anti-P/CAF and anti-GCN5 from Santa Cruz] at 4°C overnight. Immune complexes were collected with protein A–Sepharose CL-4B (Pharmacia), and beads were washed three times with 1 ml RIPA 150 buffer (same as RIPA 50 but with 150 mM NaCl). Protein–DNA complexes were resuspended in 200 µl of TE buffer and digested with 5 U of DNase-free RNase (Roche) for 30 min at 37°C. The samples were treated for 3 h at 56°C with 300 µg/ml Proteinase K (Sigma) in 0.5% SDS, 100 mM NaCl, and for 6 h at 65°C to revert crosslinks. DNA was extracted with phenol/chloroform/isoamyl alcohol, ethanol precipitated and resuspended in water for PCR quantification.
Competitive PCR
Quantification of the immunoprecipitated material was performed by competitive PCR on the eluted DNA after thermal reversion of protein–DNA crosslinks. The DNA competitors used in the quantitative–competitive PCR reactions were single DNA fragments designed to generate PCR products of different length to those obtained by amplification of genomic DNA with the same primers (Figures 1B, 2A and 4B) (Marzio et al., 1998). The competitive PCR quantifications were carried out by the addition of increasing amounts of the multi-competitor to a fixed volume of immunoprecipitated DNA, followed by PCR amplification of aliquots of the mixture with the appropriate primer pairs. Quantifications were obtained by measuring the competitor:target ratios after densitometric scanning of the respective PCR products (Diviacco et al., 1992). The oligonucleotide primers for amplification of HIV-1 DNA and RNA as well as for the controls are listed in Table I. The multicompetitor DNA templates were constructed by the amplification of a competitor DNA for human β-globin quantification (Diviacco et al., 1992) using long PCR primers containing, at their 5′-tails, sequences corresponding to the primers of interest, as described previously (Comar et al., 1996).
Acknowledgments
Acknowledgements
The authors are grateful to B.Boziglav and M.E.Lopez for technical support, and to S.Kerbavcic for excellent editorial assistance. This work was supported by grants from the National Research Programme on AIDS of the Istituto Superiore di Sanità, from the Ministero Istruzione Universita’ e Ricerca, from the ‘Progetto Genomica Funzionale’ of the ‘Consiglio Nazionale delle Ricerche’, Italy and from the Human Frontier Science Program to M.G. and A.M.
References
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T. and Thanos,D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell, 103, 667–678. [DOI] [PubMed] [Google Scholar]
- Agalioti T., Chen,G. and Thanos,D. (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111, 381–392. [DOI] [PubMed] [Google Scholar]
- Benkirane M., Chun,R.F., Xiao,H., Ogryzko,V.V., Howard,B.H., Nakatani,Y. and Jeang,K.T. (1998) Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem., 273, 24898–24905. [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2002) Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev., 12, 142–148. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Silverman,R.H. and Jeang,K.T. (1989) Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell, 59, 273–282. [DOI] [PubMed] [Google Scholar]
- Bres V., Tagami,H., Peloponese,J.M., Loret,E., Jeang,K.T., Nakatani,Y., Emiliani,S., Benkirane,M. and Kiernan,R.E. (2002) Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J., 21, 6811–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col E., Caron,C., Seigneurin-Berny,D., Gracia,J., Favier,A. and Khochbin,S. (2001) The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J. Biol. Chem., 276, 28179–28184. [DOI] [PubMed] [Google Scholar]
- Comar M., Marzio,G., D’Agaro,P. and Giacca,M. (1996) Quantitative dynamics of HIV type 1 expression. AIDS Res. Hum. Retroviruses, 12, 117–126. [DOI] [PubMed] [Google Scholar]
- Cosma M.P. (2002) Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell, 10, 227–236. [DOI] [PubMed] [Google Scholar]
- Coull J.J., Romerio,F., Sun,J.M., Volker,J.L., Galvin,K.M., Davie,J.R., Shi,Y., Hansen,U. and Margolis,D.M. (2000) The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol., 74, 6790–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Marzio,G., Gutierrez,M.I., Kang,L.Y., Falaschi,A. and Giacca,M. (1995) Molecular and functional interactions of transcription factor USF with the long terminal repeat of human immunodeficiency virus type 1. J. Virol., 69, 2765–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi F., D’Agaro,P., Falaschi,A. and Giacca,M. (1993) In vivo footprinting analysis of constitutive and inducible protein-DNA interactions at the long terminal repeat of human immunodeficiency virus type 1. J. Virol., 67, 7450–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviacco S., Norio,P., Zentilin,L., Menzo,S., Clementi,M., Biamonti,G., Riva,S., Falaschi,A. and Giacca,M. (1992) A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene, 122, 313–320. [DOI] [PubMed] [Google Scholar]
- El Kharroubi A., Piras,G., Zensen,R. and Martin,M.A. (1998) Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol. Cell. Biol., 18, 2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber B.K. and Pavlakis,G.N. (1988) A quantitative bioassay for HIV-1 based on trans-activation. Science, 239, 184–187. [DOI] [PubMed] [Google Scholar]
- Fittipaldi A., Ferrari,A., Zoppe,M., Arcangeli,C., Pellegrini,V., Beltram,F. and Giacca,M. (2003) Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem., 278, 34141–34149. [DOI] [PubMed] [Google Scholar]
- Folks T.M., Justement,J., Kinter,A., Dinarello,C.A. and Fauci,A.S. (1987) Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science, 238, 800–802. [DOI] [PubMed] [Google Scholar]
- Folks T.M., Justement,J., Kinter,A., Schnittman,S., Orenstein,J., Poli,G. and Fauci,A.S. (1988) Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J. Immunol., 140, 1117–1122. [PubMed] [Google Scholar]
- Giacca M. et al. (1994) Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA, 91, 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.H., Prochasson,P., Neely,K.E., Galasinski,S.C., Chandy,M., Carrozza,M.J. and Workman,J.L. (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell, 111, 369–379. [DOI] [PubMed] [Google Scholar]
- He G. and Margolis,D.M. (2002) Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol., 22, 2965–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger M.O. and Nabel,G.J. (1998) Interaction of human immuno deficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol., 72, 8252–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K.T., Berkhout,B. and Dropulic,B. (1993) Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J. Biol. Chem., 268, 24940–24949. [PubMed] [Google Scholar]
- Jones K.A. and Peterlin,B.M. (1994) Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem., 63, 717–743. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich Y. and Struhl,K. (2002) Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev., 16, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A., Cinelli,R.A., Ferrari,A., Signorelli,A., Tyagi,M., Pellegrini,V., Beltram,F. and Giacca,M. (2001a) Visualization of in vivo direct interaction between HIV-1 TAT and human cyclin T1 in specific subcellular compartments by fluorescence resonance energy transfer. J. Biol. Chem., 276, 39220–39225. [DOI] [PubMed] [Google Scholar]
- Marcello A., Zoppe,M. and Giacca,M. (2001b) Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life, 51, 175–181. [DOI] [PubMed] [Google Scholar]
- Marcello A., Ferrari,A., Pellegrini,V., Pegoraro,G., Lusic,M., Beltram,F. and Giacca,M. (2003) Recruitment of human Cyclin T1 to nuclear bodies trough direct interaction with the PML protein. EMBO J., 22, 2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G. and Giacca,M. (1999) Chromatin control of HIV-1 gene expression. Genetica, 106, 125–130. [DOI] [PubMed] [Google Scholar]
- Marzio G., Tyagi,M., Gutierrez,M.I. and Giacca,M. (1998) HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl Acad. Sci. USA, 95, 13519–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S., He,Y., Zeng,L., Farooq,A., Carlson,J.E., Ott,M., Verdin,E. and Zhou,M.M. (2002) Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell, 9, 575–586. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Pazin M.J., Sheridan,P.L., Cannon,K., Cao,Z., Keck,J.G., Kadonaga,J.T. and Jones,K.A. (1996) NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev., 10, 37–49. [DOI] [PubMed] [Google Scholar]
- Pierson T., McArthur,J. and Siliciano,R.F. (2000) Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol., 18, 665–708. [DOI] [PubMed] [Google Scholar]
- Pomerantz R.J., Trono,D., Feinberg,M.B. and Baltimore,D. (1990) Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell, 61, 1271–1276. [DOI] [PubMed] [Google Scholar]
- Quivy V. and Van Lint,C. (2002) Diversity of acetylation targets and roles in transcriptional regulation: the human immunodeficiency virus type 1 promoter as a model system. Biochem. Pharmacol., 64, 925–934. [DOI] [PubMed] [Google Scholar]
- Reinke H., Gregory,P.D. and Horz,W. (2001) A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell, 7, 529–538. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sheridan P.L., Mayall,T.P., Verdin,E. and Jones,K.A. (1997) Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev., 11, 3327–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Tyagi M., Rusnati,M., Presta,M. and Giacca,M. (2001) Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem., 276, 3254–3261. [DOI] [PubMed] [Google Scholar]
- Van Lint C., Emiliani,S., Ott,M. and Verdin,E. (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J., 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Paras,P.,Jr and Van Lint,C. (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J., 12, 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C.M., Felber,B.K., Paskalis,H. and Pavlakis,G.N. (1986) Expression and characterization of the trans-activator of HTLV-III/LAV virus. Science, 234, 988–992. [DOI] [PubMed] [Google Scholar]