Abstract
Purpose
Acute Myeloid Leukemia (AML) is frequently associated with genetic abnormalities. Based on pre-treatment cytogenetics, patients are classified into favorable, intermediate and poor subgroups. Cytogenetics predicts treatment outcome for the favorable and poor subgroups but not for the intermediate subgroup. Polymorphisms within the nucleotide excision repair (NER) pathway may lead to inter-individual differences in DNA repair capacity (DRC) which could influence outcome.
Methods
We studied the role of 6 polymorphisms (ERCC1 Gln504Lys, XPD Lys751Gln, XPC Ala499Val, XPC Lys939Gln, XPG Asp1104His, and CCNH Val270Ala) within NER pathway on overall and disease-free survival among 170 adult de-novo AML patients with intermediate cytogenetics [diploid (n=117); non-diploid (n=53)], treated with induction chemotherapy. Kaplan-Meier methods and Cox proportional hazards models were performed.
Results
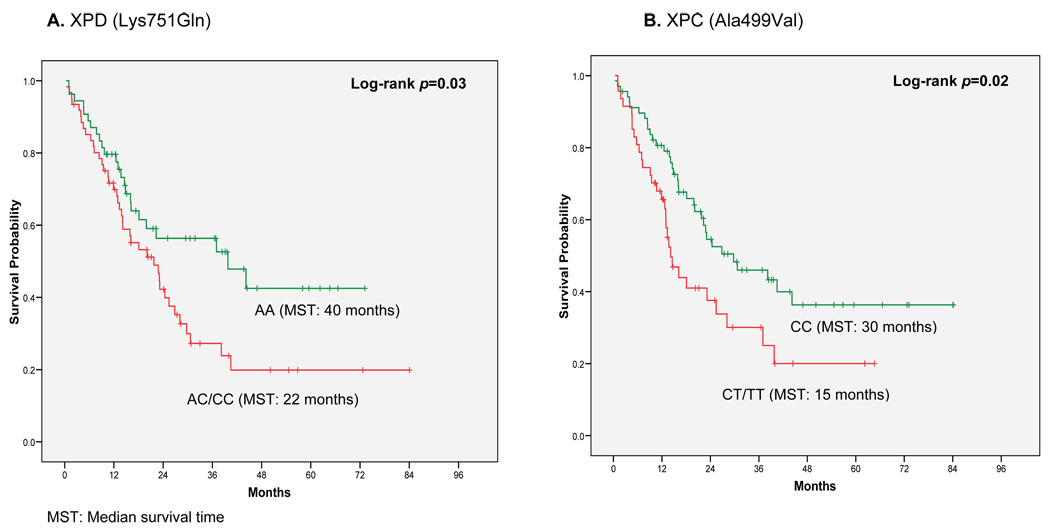
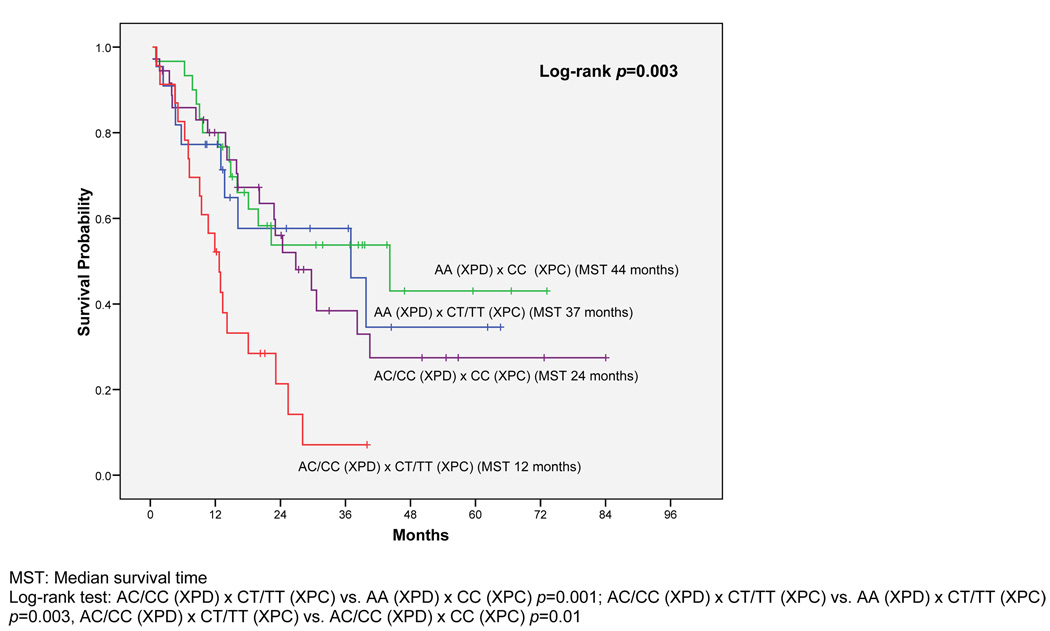
Diploid patients with the XPD AC/CC genotype survived shorter than those with the wild-type (AA) genotype (median survival 22 vs. 40 months, log-rank p = 0.03). Similarly diploid patients with XPC CT/TT genotype survived shorter than those with the wild-type (CC) genotype (median survival 15 vs. 30 months, log-rank p = 0.02). Among diploid patients, after adjusting for clinical and socio-demographic variables, patients carrying both XPD AC/CC and XPC CT/TT had a greater than two-fold increased risk of dying compared to those with the wild-type genotypes (HR=2.49; 95%CI: 1.06–5.85). No significant associations were observed for disease-free survival in AML patients.
Conclusion
By reduced DRC, this combined genotype may result in greater susceptibility to treatment effects decreasing overall survival. These findings could in the future help in selecting treatment strategies for patients with normal cytogenetics.
Introduction
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, with an estimated 13290 new cases diagnosed and 8820 deaths in 2008. (1) Five-year survival for adult AML is estimated to be only 22%,(2) even when 50–75% of patients achieve complete remission (CR) following induction.(3) Current prognostic indicators for AML include age, previous history of cancer treatment, and clinicopathologic characteristics, the most important of which is pre-treatment cytogenetics. However, significant variability in treatment response and survival remains especially among the large group of patients with normal cytogenetics.
Recent studies have suggested that inter-individual differences in DNA repair capacity may have an influence on AML pathogenesis.(4–9) Among DNA repair mechanisms, the nucleotide excision repair (NER) pathway is able to eliminate a wide variety of damage including bulky adducts such as pyrimidine dimers and other photo-products, large chemical adducts, and cross-links through recognition of distortions in DNA.(10) Substrates include products of oxidation and methylation processes caused by cancer therapy.(11) Common variants within the NER pathway may lead to inter-individual differences in DNA repair capacity which could in turn result in greater susceptibility to the genotoxic effects of treatment.
We conducted survival analysis among 170 adult de novo AML patients to explore the role of six polymorphisms in genes within the NER pathway [ERCC1 Gln504Lys (rs3212986), XPD Lys751Gln (rs28365048), XPC Ala499Val (rs2228000), XPC Lys939Gln (rs2228001), XPG Asp1104His (rs17655), and CCNH Val270Ala (rs2266690)]. All patients had intermediate cytogenetics and were treated with induction chemotherapy at the University of Texas M. D. Anderson Cancer Center (UTMDACC).
Materials and Methods
Study Population
This study included adult patients diagnosed with hematologically confirmed de novo AML from 1995–2005 at UTMDACC. Patients were classified according to the World Health Organization (WHO) classification system. A total of 251 AML patients enrolled in an ongoing case-control study were included from whom blood was collected along with a detailed demographic and lifestyle profile by personal interview. Clinical and pathological information was obtained by chart review and from the Leukemia department’s clinical database. This study was approved by the Institutional Review Board. Informed consents were obtained prior to the collection of blood, clinical and epidemiological data. All 251 patients included in this study received induction chemotherapy: 94% received cytarabine arabinoside (Ara-C) with or without idarubicin, and the remaining 6% received other anti-metabolite (nucleoside analogue) drugs. Once in CR, patients continued to receive consolidation with the same drugs at reduced doses with adjustments made based on toxicity for 3–6 months. These treatments can be categorized in three groups: 1) miscellaneous chemotherapy with or without Ara-C; 2) anthracycline treatment with or without Ara-C and 3) targeted therapies. Twenty three patients were transplanted.
CR was defined as less than or equal to 5% blasts in bone marrow following induction along with count recovery (absolute neutrophil count ≥1000 and platelet ≥100). Treatment failure was defined as death during induction therapy, while treatment resistance was defined as not achieving a CR following induction. Conventional bone marrow cytogenetics analysis was performed at the clinical cytogenetics laboratory at UTMDACC. Patients were categorized according to their pre-treatment cytogenetics as favorable [t(8;21), (Inv16)], poor [(−5, −7, −3, or complex (≥ 3 abnormalities)], or intermediate (diploid, other abnormalities not classified as favorable or poor).(12, 13) Information on the fms-like tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) and the D835 activating mutation in the tyrosine kinase domain (TKD) status was available on 84% of the patients (n = 143).
Genotyping
Genomic DNA was extracted from whole blood drawn at the time of patient registration using a QiAmp DNA blood kit (Qiagen, Valencia, CA). Genotyping was performed with the Taqman assay using protocols as described by the manufacturer (Applied Biosystems, Foster City, CA). Primer and probe sequences were obtained from the SNP500 database or used the Assays-by-Design service from Applied Biosystems. The dual-384-well GeneAmp® PCR System 9700 (Applied Biosystems) was used for PCR reactions (5 µl) that included 5 ng of sample DNA, 2.5µl of 2 × Taqman genotyping master mix (Applied Biosystems), and 900 nM each primer and 200 nM for each probe. The PCR conditions were 95°C for 10 min, followed by 50 cycles of 92°C for 30 s and 60°C for 1 min. The reacted plates were then read using the ABI Prism 7900HT Sequence Detection System and genotypes were automatically called by the built-in software. Positive and negative controls were used in each genotyping assay, and more than 5% of samples were randomly selected and run in duplicates with 100% concordance.
We selected single nucleotide polymorphisms (SNPs) within the NER pathway, which are coding polymorphisms that result in amino acid substitutions likely to affect the resulting protein structure and function; occur at a relatively high frequency, thus affecting a relatively large segment of the general population; and have been reported to be associated with cancer development and outcome.(14)
Statistical Analysis
The main outcomes of interest were overall and disease-free survival. Overall survival (OS) was measured from the date of diagnosis to date of death or last follow-up. Disease-free survival (DFS), defined as time from achievement of CR to date of relapse, death, or last follow-up, was analyzed among those patients who achieved a CR. Survival rates and median survival times were estimated using the Kaplan-Meier method with the log-rank test used to determine statistical significance.(15, 16) For SNP analysis, we tested three different genetic models, including dominant model, recessive model and additive model. The best-fitting model was the one with the smallest P value among the three models. If the genotype counts for the variant genotype were less than five in cases or controls analysis was not performed. Univariate Cox proportional hazards analysis was used to evaluate the crude effect of each polymorphism on outcome. Multivariate Cox proportional regression analysis was used to estimate the independent effects of the polymorphisms after controlling for clinical factors and potential confounders.(17, 18) Using a backwards elimination approach, all demographic, clinical, and genotypic factors were included in the initial model, and variables that did not significantly influenced the model were dropped in a step-wise fashion, with the probability threshold for removal set at 0.10. Interaction between polymorphisms was tested by including the product term for pairs of SNPs in the model. Statistical significance for main effects and interactions was defined by a p-value of less than 0.05. The proportionality assumption was confirmed visually using the Kaplan-Meier survival plots. Among potential demographic and clinical covariates, age, white blood cells (WBC), and bone marrow blast percentage were analyzed continuously. All other variables were treated as categorical variables. Zubrov performance status was grouped into two categories (1/2 and 3/4). When the homozygous variant frequency was less than 10%, these patients were combined with the heterozygous group. Differences in clinical and demographic characteristics between genotypes were compared using chi-squared tests for categorical variables and ANOVA/Kruskall-Wallis tests for continuous variables. Hardy-Weinberg equilibrium was tested using Chi-square goodness-of-fit test for all polymorphisms. Linkage disequilibrium was determined for SNPs in close proximity using the approach described by Terwilliger and Ott.(19) The normalized disequilibrium coefficients, Lewontin’s D', was also calculated, using Chi-square tests for significance.(20) All analyses were performed using STATA version 9.0 software (STATA, College Station, TX).
Results
Among 251 adult de novo AML patients treated with chemotherapy, 31 (12%) had favorable cytogenetics, 50 (20%) had poor cytogenetics, and 170 (68%) were classified as having intermediate cytogenetics. Because prognosis varies most widely among patients with intermediate cytogenetics, we focused our analysis on this large group which included 117 cases with diploid karyotypes and 53 with non-diploid karyotypes. The non-diploid intermediate group included patients with chromosomal aberrations other than those included in the favorable and poor risk groups. For 6 patients (13%) there were insufficient metaphases for analysis. Among the rest, the most common aberrations were: +8 (38%); abnormal 11Q (15%); del (5q) 4%: and miscellaneous aberrations (43%). Table 1 shows the demographic and clinical characteristics of the patients with intermediate cytogenetics. Patients with diploid cytogenetics were older and had better clinical profiles and treatment response than those with non-diploid cytogenetics; however these results were based on a small number of patients. After a median follow-up of 16 months, 56% of diploid patients have died compared to 71% among the non-diploid group (log-rank p = 0.03).
Table 1.
Patient Characteristics with Intermediate Cytogenetics (n = 170)
| Factor | Overall (n = 170) |
Diploid (n = 117) |
Non-Diploid (n = 53) |
|---|---|---|---|
| Median Age (Range) | 56 (18 – 79) | 57 (18–79) | 52 (21–74) |
| Age ≥ 60 | 71 (42%) | 52 (44%) | 19 (36%) |
| Gender | |||
| Male | 95 (56%) | 62 (53%) | 33 (62%) |
| Female | 75 (44%) | 55 (47%) | 20 (38%) |
| Race | |||
| White | 131 (77%) | 93 (79%) | 38 (72%) |
| Hispanic | 27 (16%) | 20 (17%) | 7 (13%) |
| African American | 11 (6%) | 3 (3%) | 8 (15%) |
| Asian/other | 1 (1%) | 1 (1%) | - |
| % BM Blast | |||
| Median (Range) | 60 (12 – 98) | 56 (15–98) | 64 (12–90) |
| WBC (leucocyte/µL) | |||
| Median (Range). | 7.0 (0.8 – 291) | 6.0 (0.8–291) | 12.1 (0.9–148) |
| > 20 | 54 (34%) | 38 (33%) | 16 (36%) |
| FLT-3 Status* | |||
| Wild type | 111 (77%) | 77 (76%) | 34 (79%) |
| + ITD | 28 (20%) | 20 (20%) | 8 (19%) |
| + TKD | 4 (3%) | 2 (4%) | 1 (2%) |
| Performance Status | |||
| 0–1 | 138 (82%) | 99 (85%) | 39 (75%) |
| 2–3 | 30 (18%) | 17 (15%) | 13 (25%) |
| Treatment Response | |||
| Complete | 137 (81%) | 99 (85%) | 38 (72%) |
| Remission | 22 (13%) | 13 (11%) | 9 (17%) |
| Resistant | 11 (6%) | 5 (4%) | 6 (11%) |
| Failure | |||
| Smoking History | |||
| Never | 74 (43%) | 53 (45%) | 21 (40%) |
| < 10 pack years | 33 (20%) | 18 (15%) | 15 (29%) |
| 10–30 pack years | 33 (19%) | 28 (21%) | 8 (15%) |
| > 30 pack years | 30 (18%) | 22 (19%) | 8 (15%) |
| Vital Status | |||
| Deceased | 109 (64%) | 66 (56%) | 38 (71%) |
| Follow-up (months) | |||
| Median (Range) | 16 (1 – 84) | 16 (1 – 84) | 16 (1 – 76) |
FLT3 status n = 143
All the polymorphisms analyzed were found to be in Hardy-Weinberg equilibrium, and no major differences were found in the genotype frequencies between patients with diploid and non-diploid cytogenetics (Table 2). XPC Ala499Val and XPC Lys939Gln were not found to be in linkage disequilibrium. We therefore chose to include both in subsequent analyses.
Table 2.
SNP Frequencies by Cytogenetics (n = 170)
| Genotype | Overall (n = 170) |
Diploid (n = 117) |
Non-Diploid (n = 53) |
|---|---|---|---|
| ERCC1 (Gln504Lys) | |||
| GG | 89 (52%) | 59 (50%) | 30 (56%) |
| GT | 69 (41%) | 48 (41%) | 21 (40%) |
| TT | 12 (7%) | 10 (9%) | 2 (4%) |
| XPD (Lys751Gln) | |||
| AA | 81 (48%) | 54 (47%) | 27 (51%) |
| AC | 71 (42%) | 51 (44%) | 20 (38%) |
| CC | 16 (10%) | 10 (9%) | 6 (11%) |
| XPC (Ala499Val) | |||
| CC | 93 (56%) | 68 (59%) | 25 (49%) |
| CT | 61 (37%) | 37 (32%) | 24 (47%) |
| TT | 12 (7%) | 10 (9%) | 2 (4%) |
| XPC (Lys939Gln) | |||
| AA | 65 (39%) | 45 (39%) | 20 (38%) |
| AC | 77 (46%) | 50 (44%) | 27 (51%) |
| CC | 26 (15) | 20 (17%) | 6 (11%) |
| XPG (Asp1104His) | |||
| GG | 98 (58%) | 67 (58%) | 31 (59%) |
| GC | 57 (34%) | 40 (34%) | 17 (32%) |
| CC | 14 (8%) | 9 (8%) | 5 (9%) |
| CCNH (Val270Ala) | |||
| TT | 105 (62%) | 75 (65%) | 30 (57%) |
| TC | 57 (34%) | 35 (31%) | 22 (41%) |
| CC | 6 (4%) | 5 (4%) | 1 (2%) |
We explored the associations between NER SNPs and CR rate, DFS, and OS among patients with intermediate cytogenetics. The overall CR rate was 81% and did not differ significantly by any of the SNPs analyzed. Table 3 shows results from univariate Cox proportional hazards regression analyses of OS and DFS by SNP among AML patients with intermediate cytogenetics. Overall, variants in XPD Lys751Gln and XPC Ala499Val were associated with statistically significant lower OS. The dominant models showed the best fit. When we stratified by diploid vs. non-diploid cytogenetics, these associations were found to be restricted to the 117 patients with diploid karyotypes. Among these patients, the risk of dying among those carrying at least one variant XPD Lys751Gln allele (AC/CC) was 73% greater than among patients with the XPD Lys751Gln wild-type (AA) genotype (HR 1.73, p = 0.03). For XPC Ala499Val, carrying one or two variant alleles (CT/TT) was associated with 78% increased mortality compared to the wild-type genotype (CC) (HR 1.78, p = 0.02). Patients with both variant alleles (TT) had a greater than two-fold increased mortality (HR 2.40, p = 0.02).
Table 3.
Disease-Free and Overall Survival by NER Pathway SNPs
| Genotype | Disease-free survival |
Overall survival |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall |
Diploid |
Non-diploid |
Overall |
Diploid |
Non-diploid |
|||||||||||||
| n | HR | p | n | HR | p | n | HR | p | n | HR | p | n | HR | p | n | HR | p | |
| ERCC1 (Gln504Lys) | ||||||||||||||||||
| GG | 72 | 1.00 | 49 | 1.00 | 23 | 1.00 | 89 | 1.00 | 59 | 1.00 | 30 | 1.00 | ||||||
| GT | 55 | 0.78 | 0.24 | 41 | 0.86 | 0.54 | 14 | - | - | 69 | 0.91 | 0.64 | 48 | 0.84 | 0.51 | 21 | - | - |
| TT | 10 | 0.89 | 0.78 | 9 | 1.05 | 0.92 | 1 | - | - | 12 | 1.31 | 0.48 | 10 | 1.44 | 0.42 | 2 | - | - |
| GT/TT vs. GG Dom* | 0.79 | 0.26 | 0.88 | 0.60 | 0.64 | 0.27 | 0.96 | 0.82 | 0.91 | 0.72 | 1.11 | 0.75 | ||||||
| GG/GT vs. TT Rec* | 0.99 | 0.99 | 1.13 | 0.79 | - | - | 1.36 | 0.40 | 1.55 | 0.31 | - | - | ||||||
| XPD (Lys751Gln) | ||||||||||||||||||
| AA | 65 | 1.00 | 47 | 1.00 | 18 | 1.00 | 81 | 1.00 | 54 | 1.00 | 27 | 1.00 | ||||||
| AC | 55 | 0.83 | 0.41 | 40 | 0.87 | 0.58 | 15 | 0.76 | 0.50 | 71 | 1.57 | 0.03 | 51 | 1.81 | 0.03 | 20 | 1.35 | 0.39 |
| CC | 15 | 0.61 | 0.18 | 10 | 0.66 | 0.35 | 5 | 0.47 | 0.25 | 16 | 1.21 | 0.58 | 10 | 1.37 | 0.49 | 6 | 0.98 | 0.97 |
| AC/CC vs. AA Dom* | 0.78 | 0.24 | 0.82 | 0.43 | 0.67 | 0.30 | 1.50 | 0.04 | 1.73 | 0.03 | 1.25 | 0.49 | ||||||
| AA/AC vs. CC Rec* | 1.50 | 0.25 | 1.41 | 0.42 | 1.86 | 0.32 | 1.03 | 0.93 | 0.99 | 0.99 | 1.15 | 0.79 | ||||||
| XPC (Ala499Val) | ||||||||||||||||||
| CC | 76 | 1.00 | 59 | 1.00 | 17 | 1.00 | 93 | 1.00 | 68 | 1.00 | 25 | 1.00 | ||||||
| CT | 48 | 1.25 | 0.33 | 31 | 1.02 | 0.94 | 17 | 1.69 | 0.23 | 61 | 1.50 | 0.06 | 37 | 1.61 | 0.09 | 24 | - | - |
| TT | 9 | 1.74 | 0.15 | 7 | 1.97 | 0.12 | 2 | 1.29 | 0.75 | 12 | 1.85 | 0.08 | 10 | 2.40 | 0.02 | 2 | - | - |
| CT/TT vs. CC Dom* | 1.33 | 0.18 | 1.16 | 0.56 | 1.62 | 0.25 | 1.56 | 0.03 | 1.78 | 0.02 | 1.09 | 0.79 | ||||||
| CT/CC vs. TT Rec* | 0.62 | 0.20 | 0.51 | 0.12 | - | - | 0.63 | 0.17 | 0.49 | 0.05 | - | - | ||||||
| XPC (Lys939Gln) | ||||||||||||||||||
| AA | 51 | 1.00 | 36 | 1.00 | 15 | 1.00 | 65 | 1.00 | 45 | 1.00 | 20 | 1.00 | ||||||
| AC | 62 | 0.90 | 0.64 | 43 | 0.94 | 0.83 | 19 | 0.80 | 0.60 | 77 | 0.92 | 0.69 | 50 | 0.79 | 0.38 | 27 | 1.14 | 0.71 |
| CC | 22 | 1.14 | 0.64 | 18 | 1.19 | 0.60 | 4 | 1.41 | 0.61 | 26 | 0.63 | 0.14 | 20 | 0.69 | 0.30 | 6 | 0.29 | 0.51 |
| AC/CC vs. AA Dom* | 0.96 | 0.86 | 1.02 | 0.95 | 0.87 | 0.73 | 0.84 | 0.37 | 0.75 | 0.26 | 0.99 | 0.97 | ||||||
| AA/AC vs. CC Rec* | 0.82 | 0.45 | 0.81 | 0.47 | - | - | 1.51 | 0.13 | 1.28 | 0.45 | 2.13 | 0.21 | ||||||
| XPG (Asp1104His) | ||||||||||||||||||
| GG | 80 | 1.00 | 56 | 1.00 | 24 | 1.00 | 98 | 1.00 | 67 | 1.00 | 31 | 1.00 | ||||||
| GC | 45 | 0.61 | 0.25 | 34 | 0.57 | 0.29 | 11 | 0.68 | 0.60 | 57 | 0.59 | 0.22 | 40 | 0.28 | 0.08 | 17 | 1.34 | 0.60 |
| CC | 11 | 0.83 | 0.42 | 8 | 1.01 | 0.96 | 3 | 0.48 | 0.14 | 14 | 0.88 | 0.53 | 9 | 0.71 | 0.20 | 5 | 1.35 | 0.39 |
| GC/CC vs. GG Dom* | 0.79 | 0.26 | 0.92 | 0.72 | 0.52 | 0.14 | 0.82 | 0.32 | 0.63 | 0.07 | 1.35 | 0.36 | ||||||
| GG/GC vs. CC Rec* | 1.53 | 0.31 | 1.75 | 0.28 | - | - | 1.60 | 0.26 | 3.15 | 0.11 | 0.84 | 0.74 | ||||||
| CCNH (Val270Ala) | ||||||||||||||||||
| TT | 83 | 1.00 | 64 | 1.00 | 19 | 1.00 | 105 | 1.00 | 75 | 1.00 | 30 | 1.00 | ||||||
| TC | 48 | - | - | 29 | - | - | 19 | - | - | 57 | - | - | 35 | - | - | 22 | - | - |
| CC | 4 | - | - | 4 | - | - | - | - | 6 | - | - | 5 | - | - | 1 | - | - | |
| TC/CC vs. TT Dom* | 1.31 | 0.20 | 1.40 | 0.19 | 0.99 | 0.98 | 1.12 | 0.58 | 1.07 | 0.80 | 1.05 | 0.89 | ||||||
| TT/TC vs. CC Rec* | - | - | - | - | - | - | 0.58 | 0.29 | 0.65 | 0.47 | - | - | ||||||
Dom* Dominant model
Rec* Recessive model
Kaplan-Meier results show that diploid patients with the XPD Lys751Gln genotype (AC/CC) survived shorter than those with the wild-type genotype (AA) (median survival 22 vs. 40 months, log-rank p = 0.03, Figure 1A). Similarly, patients with at least one XPC Ala499Val variant allele (CT/TT) had a median survival time of 15 months compared to 30 months for patients with the wild-type (CC) (log-rank p = 0.02, Figure 1B). As shown in Figure 2, when we combined these two variant genotypes, the median survival of patients carrying variants in both SNPs was only 12 months compared to 44 months for patients with both wild-type genotypes (log-rank p = 0.001). Twenty-one percent of the AML patients with normal cytogenetics carried the high-risk genotype combination.
Figure 1.
Overall Survival among 117 AML Patients with Diploid Cytogenetics: A. XPD Lys751Gln (AA vs. AC/CC) log-rank p=0.03; B. XPC Ala499Val (CC vs. CT/TT) log-rank p=0.02.0.0
Figure 2.
Joint Effect of XPD and XPC on Overall Survival among Patients with Normal/Diploid Karyotypes (n = 117)
In a Cox regression analysis we determined the combined influence of XPD Lys751Gln and XPC Ala499Val on OS. After controlling for clinical and socio-demographic characteristics (age, sex, ethnicity, WBC, FLT3-ITD, performance status, and smoking) (Table 4), carrying variant alleles for both polymorphisms was associated with a greater than two-fold increased risk of dying compared to having the wild type alleles (HR 2.49; 95% CI 1.06–5.85; p = 0.03). Other factors associated with OS in the multivariate analysis included WBC, age, and FLT3-ITD status.
Table 4.
Multivariate Analysis of Overall Survival among Patients with Normal/Diploid Karyotypes (n = 117)
| Univariate |
Multivariate |
|||
|---|---|---|---|---|
| Factor | HR (95% CI) | p | HR (95% CI) | p |
|
XPD (Lys751Gln) & XPC (Ala499Val) |
||||
| XPD AA × XPC CC (n=31) | 1.00 | - | 1.00 | - |
| XPD AA × XPC CT/TT (n=38) | 1.34 (0.68 – 2.64) | 0.39 | 1.09 (0.43 – 2.82) | 0.84 |
| XPD AC/CC × XPC CC (n=24) | 1.19 (0.53 – 2.69) | 0.67 | 1.22 (0.55 – 2.71) | 0.62 |
| XPD AC/CC × XPC CT/TT (n=24) | 3.22 (1.59 – 6.51) | 0.001 | 2.49 (1.06 – 5.85) | 0.03 |
| FLT-3 Status | ||||
| Wild | 1.00 | - | 1.00 | - |
| FLT-3/ITD | 2.87 (1.61 – 5.09) | <0.001 | 2.66 (1.26 – 5.63) | 0.01 |
| Performance Status (continuous) | 1.31 (0.95 – 1.82) | 0.09 | 0.90 (0.56 – 1.45) | 0.66 |
| WBC count, ×109 | ||||
| ≤ 20 | 1.00 | 1.00 | - | |
| > 20 | 1.69 (1.02 – 2.80) | 0.04 | 1.00 (0.99 – 1.01) | 0.87 |
| Age (continuous) | 1.04 (1.02 – 1.06) | <0.001 | 1.05 (1.03 – 1.08) | <0.001 |
Discussion
On one end of the AML spectrum, targeted therapies have been developed that are effective in treating favorable subtypes of AML such as acute promyelocytic leukemia, while at the other end of the spectrum lie the group of patients with poor cytogenetics who tend to have universally poor prognoses regardless of treatment protocol. Between these groups exists the majority of AML patients with intermediate cytogenetics for whom outcome remains uncertain. Among 170 de novo intermediate risk AML patients treated with chemotherapy, we evaluated the effects of six common SNPs involved in the NER pathway on OS and DFS. We showed that variations in two SNPs (XPD Lys751Gln and XPC Ala499Val), after adjustment for demographic and clinical characteristics, were independent predictors of decreased survival among the large group of AML patients with normal cytogenetics but not in the non-diploid group.
DNA repair enzymes in the NER pathway are able to recognize and eliminate a wide variety of damage including bulky adducts including those induced by chemotherapy. Therefore, it is possible to hypothesize that common variants within the NER pathway may lead to inter-individual differences in DNA repair capacity, which could in turn result in greater susceptibility to the genotoxic effects of treatment.
The XPD gene encodes a DNA helicase involved in NER. The common XPD Lys751Gln polymorphism is known to affect protein function and alter cellular responses to specific types of DNA damage. Few studies have explored the role of DNA repair SNPs in AML pathogenesis. While one study found no association between XPD genotype and outcome of childhood AML,(21) investigations among adult AML patients after chemotherapy treatment have reported that XPD 751 as well as other XPD variants were associated with AML survival. Allan et al., found that among 341 patients more than 60 years old, the XPD 751 Gln/Gln genotype was associated with worse DFS and OS.(6) In 2007, Kuptsova et al.(4) reported that in patients older than 55 years of age, the XPD Asp312Asn and Lys751Gln genotypes were associated with treatment outcome. Patients with XPD GlnC/Asp312A haplotypes had significantly decreased OS and those with XPD GlnC/Asp312G haplotype had better CR. Only one previous study has analyzed the role of this variant among patients with intermediate cytogenetics.(5) An analysis of 110 patients that included 4 SNPs in NER genes showed that the XPD Gln variant was associated with relapse and OS only in univariate analysis. A sub-analysis of the 70 patients with normal cytogenetics showed similar results than in the entire series.
As a whole, these previous data as well as our new results indicate that variations in genes of NER pathway play an important role in AML outcome. Differences shown in the role of some of these variants by type of outcome could be related to patient’s characteristics such as age, the functional DNA repair capability of the variants as well as interactions with specific therapeutic agents used in different stages of treatment.
The XPD variant has been shown to affect protein function and to modulate cellular response to genotoxins suggesting that its functionality may be exposure- and pathway-specific, affecting both cell death and DNA repair.(14) In AML, due to the effect of chemotherapy, any functional effect of this variant in myeloid cells, both leukemic and normal, may be amplified producing an altered clinical response. The already reported poorer prognosis and resistant disease associated with the XPD 751 variant is consistent with the hypothesis that the 751 glutamine variant protects against cell death relative to the wild type. Allan et al. (6) have suggested two mechanisms by which this variant may affect apoptosis in response to chemotherapy. One is through its direct involvement in signaling cell death by affecting the ability of XPD to signal p53-dependent apoptosis. The other mechanism suggests an indirect modulation of myeloid cell death by reducing the efficiency of NER in repairing DNA lesions induced by chemotherapy drugs.
None of the aforementioned studies included XPC in their analyses. XPC binds to HR23B and forms the XPC-HR23B complex, which is involved in DNA damage recognition and/or in altering chromatin structure to allow access by damage-processing enzymes. Some epidemiological studies had been conducted to explore the association of XPC polymorphisms with the risk of solid tumors like lung cancer, bladder cancer and head and neck cancer.(22, 23) However no study looked at an association with the risk of hematopoietic cancers or cancer outcome. The XPC 939 variant has been associated with reduced repair of single-strand breaks measured by the Comet assay,(24) however the functional significance of the 499 variant that leads to amino acid substitution is still uncertain.
Our results also showed a joint effect between these two SNPs, with patients who carry both variants having a 2.49 fold increased mortality compared to those with the wild genotypes that is independent of clinical characteristics including FLT3-ITD status. The high prevalence (21%) of this high risk genotype combination suggests that this could be a relevant prognostic marker with a potential clinical utility.
It is now postulated that multiple genes act independently, collectively, or interact with each other to influence carcinogenesis. Combination of certain genotypes may be more efficient in discriminating risk factors than a single locus genotype.
Conclusion
In conclusion, results from this study suggest that a combination of polymorphic variants in NER repair enzymes may modulate AML outcome in patients with diploid cytogenetics. Our study has limitations due to the relatively small sample size, lack of toxicity information, and limited number of SNPs. Future large studies should include multiple SNPs of genes involved in the different DNA repair pathways as well as in xenobiotic and apoptotic pathways necessary to validate the association between variants in DNA repair genes and outcome and to clarify the underlying mechanisms involved in AML progression (e.g., DFS, OS, resistant disease).
Acknowledgements of research support
We acknowledge Sammer Gokhale, Mandy Chan, Vivianne Velez-Bravo, Sherry Pierce, and Yessica Nunez for data collection. This study would not have been possible without the cooperation of our participants. Work was supported by NCI grants CA100632, CA115180, and ES007784.
Footnotes
Results have been presented at Frontiers in Cancer Prevention Research, November 16–19, 2008. The contents of this manuscript have not been copyrighted or published previously; and are not under consideration for publication elsewhere.
Disclaimers: No potential conflicts of interest.
References
- 1.American Cancer Society: Cancer Facts & Figures 2008. Atlanta: American Cancer Society; 2008. [Google Scholar]
- 2.Ries LAG, Melbert D, Krapcho M, et al., editors. SEER Cancer Statistics Review. Bethesda, MD: National Cancer Institute; 1975–2005. [Google Scholar]
- 3.Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood. 2005;106:1154–1163. doi: 10.1182/blood-2005-01-0178. [DOI] [PubMed] [Google Scholar]
- 4.Kuptsova N, Kopecky KJ, Godwin J, et al. Polymorphisms in DNA repair genes and therapeutic outcomes of AML patients from SWOG clinical trials. Blood. 2007;109:3936–3944. doi: 10.1182/blood-2006-05-022111. [DOI] [PubMed] [Google Scholar]
- 5.Monzo M, Brunet S, Urbano-Ispizua A, et al. Genomic polymorphisms provide prognostic information in intermediate-risk acute myeloblastic leukemia. Blood. 2006;107:4871–4879. doi: 10.1182/blood-2005-08-3272. [DOI] [PubMed] [Google Scholar]
- 6.Allan JM, Smith AG, Wheatley K, et al. Genetic variation in XPD predicts treatment outcome and risk of acute myeloid leukemia following chemotherapy. Blood. 2004;104:3872–3877. doi: 10.1182/blood-2004-06-2161. [DOI] [PubMed] [Google Scholar]
- 7.Seedhouse C, Faulkner R, Ashraf N, et al. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clin Cancer Res. 2004;10:2675–2680. doi: 10.1158/1078-0432.ccr-03-0372. [DOI] [PubMed] [Google Scholar]
- 8.Seedhouse C, Bainton R, Lewis M, et al. The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood. 2002;100:3761–3766. doi: 10.1182/blood-2002-04-1152. [DOI] [PubMed] [Google Scholar]
- 9.Voso MT, Fabiani E, D'Alo' F, et al. Increased risk of acute myeloid leukemia due to polymorphisms in detoxification and DNA repair enzymes. Ann Oncol. 2007;18:1523–1528. doi: 10.1093/annonc/mdm191. [DOI] [PubMed] [Google Scholar]
- 10.Lei L. Nucleotide excision repair. In: Wei Q, Lei L, Chen DJ, editors. DNA Repair, Genetic Instability, and Cancer. Singapore: World Scientific Publishing Co. Pte. Ltd; 2006. pp. 65–85. [Google Scholar]
- 11.Das-Gupta EP, Seedhouse CH, Russell NH. DNA repair mechanisms and acute myeloblastic leukemia. Hematol Oncol. 2000;18:99–110. doi: 10.1002/1099-1069(200009)18:3<99::aid-hon662>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Estey EH. Therapeutic options for acute myelogenous leukemia. Cancer. 2001;92:1059–1073. doi: 10.1002/1097-0142(20010901)92:5<1059::aid-cncr1421>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 13.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 14.Hu Z, Wang LE, Wei Q. Molecular epidemiology of DNA repair and cancer susceptibility - A review of population-based studies. In: Wei Q, Li L, Chen DJ, editors. DNA Repair, Genetic Instability, and Cancer (ed 2006) Singapore: World Scientific Publishing Co. Pte. Ltd; 2006. pp. 315–343. [Google Scholar]
- 15.Kaplan EL, Meier P. Nonparametric estimations from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 16.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 17.Cox DR. Regression models and life tables. J Roy Stat Soc. 1972;34(2):187–220. [Google Scholar]
- 18.Cox DR, Oakes D. Analysis of Survival Data. London: Chapman and Hall; 1984. [Google Scholar]
- 19.Terwilliger J, Ott J. Handbook of Human Genetic Linkage. Boston: Johns Hopkins Press; 1994. [Google Scholar]
- 20.Lewontin RC. The interaction of selection and linkage. I. general considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehta PA, Alonzo TA, Gerbing RB, et al. XPD Lys751Gln polymorphism in the etiology and outcome of childhood acute myeloid leukemia. A children's oncology group report. Blood. 2006;107:39–45. doi: 10.1182/blood-2005-06-2305. [DOI] [PubMed] [Google Scholar]
- 22.An J, Liu Z, Hu Z, et al. Potentially functional single nucleotide polymorphisms in the core nucleotide excision repair genes and risk of squamous cell carcinoma of the head and neck. Cancer Epidemiol Biomarkers Prev. 2007;16:1633–1638. doi: 10.1158/1055-9965.EPI-07-0252. [DOI] [PubMed] [Google Scholar]
- 23.Qiu L, Wang Z, Shi X, et al. Associations between XPC polymorphisms and risk of cancers. A meta-analysis. Eur J Cancer. 2008;44:2241–2253. doi: 10.1016/j.ejca.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 24.Vodicka P, Kumar R, Stetina R, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25:757–763. doi: 10.1093/carcin/bgh064. [DOI] [PubMed] [Google Scholar]